Abstract
The zinc cluster transcription factor Upc2p mediates upregulation of ergosterol biosynthesis genes in response to ergosterol depletion in the fungal pathogen Candida albicans. One mechanism of acquired resistance to the antifungal drug fluconazole, which inhibits ergosterol biosynthesis, is constitutively increased expression of the ERG11 gene encoding the drug target enzyme. A G648D mutation in Upc2p has recently been shown to cause hyperactivity of the transcription factor, resulting in overexpression of ergosterol biosynthesis genes and increased fluconazole resistance. In order to investigate if gain-of-function mutations in Upc2p are a common mechanism of ERG11 upregulation and fluconazole resistance, we sequenced the UPC2 alleles of four ERG11-overexpressing, fluconazole-resistant C. albicans isolates and matched susceptible isolates from the same patients. In three of the isolate pairs, no differences in the UPC2 alleles were found, suggesting that mechanisms other than Upc2p mutations can cause ERG11 overexpression. One resistant isolate had become homozygous for a UPC2 allele containing a G1927A substitution that caused an alanine-to-threonine exchange at amino acid position 643 of Upc2p. Replacement of one of the endogenous UPC2 alleles in a fluconazole-susceptible strain by the UPC2A643T allele resulted in ERG11 overexpression and increased fluconazole resistance, which was further elevated when the A643T mutation was also introduced into the second UPC2 allele. These results further establish gain-of-function mutations in UPC2, which can be followed by loss of heterozygosity for the mutated allele, as a mechanism of ERG11 overexpression and increased fluconazole resistance in C. albicans, but other mechanisms of ERG11 upregulation also exist.
The antimycotic agent fluconazole, which inhibits ergosterol biosynthesis, is used widely to treat infections by the fungal pathogen Candida albicans. C. albicans can develop resistance to fluconazole by various mechanisms, including mutations in the ERG11 gene encoding the drug target enzyme, overexpression of ERG11, and upregulation of efflux pumps that transport the drug out of the cells. Often, several of these mechanisms are combined to result in high-level fluconazole resistance and therapy failure (for a review, see reference 11).
In recent years, transcription factors that regulate the expression of genes mediating fluconazole resistance in C. albicans have been identified. Tac1p (transcriptional activator of CDR genes) controls expression of the ATP-binding cassette (ABC) transporters CDR1 and CDR2, whereas Mrr1p (multidrug resistance regulator) regulates expression of the MDR1 gene encoding an efflux pump of the major facilitator superfamily (3, 12). Gain-of-function mutations in these transcription factors result in constitutive upregulation of their target genes and increased drug resistance (1, 2, 4, 12, 22). Upc2p, which like Tac1p and Mrr1p belongs to the zinc cluster transcription factor family that is specific for fungi, regulates the expression of ERG11 and other ergosterol biosynthesis genes in response to ergosterol depletion (8, 10, 13, 20, 23). Recently, a G1943A mutation, which causes an amino acid substitution at position 648 in Upc2p from glycine to aspartate (G648D), was found in one of the two UPC2 alleles of a fluconazole-resistant C. albicans isolate (S2) that exhibited increased expression of ERG11 and other ERG genes as well as of UPC2 itself compared to a matched, fluconazole-susceptible isolate (S1) from the same patient (5). Introduction of the UPC2G648D allele into the fluconazole-susceptible C. albicans strain SC5314 resulted in constitutive upregulation of ERG11 and other Upc2p target genes and increased fluconazole resistance, demonstrating that gain-of-function mutations in this transcription factor can contribute to the development of drug resistance in C. albicans (5).
While mutations in Tac1p and Mrr1p are the cause of overexpression of CDR1/2 and MDR1, respectively, in all fluconazole-resistant C. albicans isolates investigated to date, the G648D mutation is the only example of an activating mutation in UPC2 causing ERG11 upregulation in a clinical C. albicans isolate so far. ERG11 overexpression has been observed with other fluconazole-resistant C. albicans isolates, but the mechanism of this upregulation remained unknown (6, 14, 21). Therefore, we set out to determine if mutations in UPC2 might be a common mechanism of ERG11 overexpression in C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 15% glycerol at −80°C and subcultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g glucose, 15 g agar per liter) at 30°C. For routine growth of the strains, YPD liquid medium was used. For selection of nourseothricin-resistant transformants, 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) was added to YPD agar plates. To obtain nourseothricin-sensitive derivatives in which the SAT1 flipper cassette was excised by FLP-mediated recombination, transformants were grown overnight in YPM medium (10 g yeast extract, 20 g peptone, 20 g maltose per liter) without selective pressure to induce the MAL2 promoter that controls expression of the caFLP gene in the SAT1 flipper cassette. Cells (100 to 200) were then spread on YPD plates containing 20 μg/ml nourseothricin and grown for 2 days at 30°C. Nourseothricin-sensitive clones were identified by their small colony size and confirmed by restreaking on YPD plates containing 100 μg/ml nourseothricin as described previously (17).
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Relevant characteristics or genotypea | Reference or source |
|---|---|---|---|
| Clinical isolates | |||
| 1002 | FLUS isolate from patient 9 | 14 | |
| 3795 | FLUR isolate from patient 9 | 14 | |
| 580 | FLUS isolate from patient 14 | 14 | |
| 2440 | FLUR isolate from patient 14 | 14 | |
| 945 | FLUS isolate from patient 15 | 14 | |
| 1619 | FLUR isolate from patient 15 | 14 | |
| 5044 | FLUS isolate from patient 28 | 14 | |
| 5052 | FLUR isolate from patient 28 | 14 | |
| SC5314 | Wild-type reference strain | 7 | |
| Strains carrying introduced UPC2 alleles | |||
| SCUPC2R11A and -B | SC5314 | UPC2G648D-SAT1-FLIP/UPC2-2 | This study |
| SCUPC2R12A | SCUPC2R11A | UPC2G648D-FRT/UPC2-2 | This study |
| SCUPC2R12B | SCUPC2R11B | UPC2G648D-FRT/UPC2-2 | This study |
| SCUPC2R13A | SCUPC2R12A | UPC2G648D-FRT/UPC2G648D-SAT1-FLIP | This study |
| SCUPC2R13B | SCUPC2R12B | UPC2G648D-FRT/UPC2G648D-SAT1-FLIP | This study |
| SCUPC2R14A | SCUPC2R13A | UPC2G648D-FRT/UPC2G648D-FRT | This study |
| SCUPC2R14B | SCUPC2R13B | UPC2G648D-FRT/UPC2G648D-FRT | This study |
| SCUPC2R21A and -B | SC5314 | UPC2-SAT1-FLIP/UPC2-2 | This study |
| SCUPC2R22A | SCUPC2R21A | UPC2-FRT/UPC2-2 | This study |
| SCUPC2R22B | SCUPC2R21B | UPC2-FRT/UPC2-2 | This study |
| SCUPC2R23A | SCUPC2R22A | UPC2-FRT/UPC2-SAT1-FLIP | This study |
| SCUPC2R23B | SCUPC2R22B | UPC2-FRT/UPC2-SAT1-FLIP | This study |
| SCUPC2R24A | SCUPC2R23A | UPC2-FRT/UPC2-FRT | This study |
| SCUPC2R24B | SCUPC2R23B | UPC2-FRT/UPC2-FRT | This study |
| SCUPC2R31A and -B | SC5314 | UPC2A643T-SAT1-FLIP/UPC2-2 | This study |
| SCUPC2R32A | SCUPC2R31A | UPC2A643T-FRT/UPC2-2 | This study |
| SCUPC2R32B | SCUPC2R31B | UPC2A643T-FRT/UPC2-2 | This study |
| SCUPC2R33A | SCUPC2R32A | UPC2A643T-FRT/UPC2A643T-SAT1-FLIP | This study |
| SCUPC2R33B | SCUPC2R32B | UPC2A643T-FRT/UPC2A643T-SAT1-FLIP | This study |
| SCUPC2R34A | SCUPC2R33A | UPC2A643T-FRT/UPC2A643T-FRT | This study |
| SCUPC2R34B | SCUPC2R33B | UPC2A643T-FRT/UPC2A643T-FRT | This study |
| Reporter strains expressing GFP from the ERG11 promoter | |||
| SCEG2A and -B | SC5314 | UPC2-1/UPC2-2 | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R12EG2A | SCUPC2R12A | UPC2G648D-FRT/UPC2-2 | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R12EG2B | SCUPC2R12B | UPC2G648D-FRT/UPC2-2 | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 | |||
| SCUPC2R14EG2A | SCUPC2R14A | UPC2G648D-FRT/UPC2G648D-FRT | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R14EG2B | SCUPC2R14B | UPC2G648D-FRT/ UPC2G648D-FRT | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 | |||
| SCUPC2R22EG2A | SCUPC2R22A | UPC2-FRT/UPC2-2 | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 | |||
| SCUPC2R22EG2B | SCUPC2R22B | UPC2-FRT/UPC2-2 | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R24EG2A | SCUPC2R24A | UPC2-FRT/UPC2-FRT | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R24EG2B | SCUPC2R24B | UPC2-FRT/ UPC2-FRT | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 | |||
| SCUPC2R32EG2A | SCUPC2R32A | UPC2A643T-FRT/UPC2-2 | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 | |||
| SCUPC2R32EG2B | SCUPC2R32B | UPC2A643T-FRT/UPC2-2 | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R34EG2A | SCUPC2R34A | UPC2A643T-FRT/UPC2A643T-FRT | This study |
| ERG11-1/erg11-2::PERG11-GFP-caSAT1 | |||
| SCUPC2R34EG2B | SCUPC2R34B | UPC2A643T-FRT/ UPC2A643T-FRT | This study |
| erg11-1::PERG11-GFP-caSAT1/ERG11-2 |
SAT1-FLIP denotes the SAT1 flipper cassette. The UPC2 and ERG11 alleles in strain SC5314 can be distinguished by EcoRI and HindIII restriction site polymorphisms, respectively. The UPC2 allele containing the polymorphic EcoRI site at position +1593 was arbitrarily designated UPC2-2 (see Table 4), and the ERG11 allele containing the polymorphic HindIII site at position +347 was designated ERG2-2.
Quantitative real-time RT-PCR.
Total RNA was isolated from the clinical C. albicans isolates as previously described (5). An aliquot of the RNA preparations was used for quantitative real-time reverse transcription (RT)-PCR studies. First-strand cDNAs were synthesized from 2 μg of total RNA in a 21-μl reaction volume by using a SuperScript first-strand synthesis system for RT-PCR (Invitrogen) in accordance with the manufacturer's instructions. Quantitative real-time PCRs were performed in triplicate using a 7000 sequence detection system (Applied Biosystems, Inc., Foster City, CA). Independent PCRs were performed using the same cDNA for both the genes of interest and the 18S rRNA gene with SYBR green PCR master mix (Applied Biosystems, Inc.). Gene-specific primers were designed for the gene of interest and the 18S rRNA gene by using Primer Express software (Applied Biosystems, Inc.) and an Oligo analysis and plotting tool (Qiagen, Valencia, CA) and are shown in Table 2. The PCR conditions consisted of AmpliTaq Gold activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. A dissociation curve was generated at the end of each PCR cycle, using software provided with the 7000 sequence detection system, to verify that a single product was amplified. The change in fluorescence of SYBR green I dye in every cycle was monitored by the system software, and the cycle threshold (CT) above the background for each reaction was calculated. The CT value of the 18S rRNA gene was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of a calibrator (e.g., an azole-susceptible isolate sample of each matched set) was subtracted from the ΔCT value of each sample to obtain a ΔΔCT value. The gene expression level relative to the calibrator (change in expression) was expressed as 2−ΔΔCT. The ΔCT values were also used to calculate standard error values.
TABLE 2.
Primers used in this studya
| Primer | Sequence |
|---|---|
| ACT19 | 5′-ATATACCGCGGACATTTTATGATGGAATGA-3′ |
| ERG13 | 5′-CGACATTATTAGGGCCCTTTGAGAACAGCC-3′ |
| ERG14 | 5′-CAATAGCCATATTGTCGACTGATCTTCTTG-3′ |
| ERG15 | 5′-AATACTGCAGCAACTTTCTTTCGATTCAGTG-3′ |
| ERG16 | 5′-CTAAGAGCTCGAATCCTGGTCCTATATTAGC-3′ |
| UPC2-3A | 5′-AACAGAGCTCTACGTTATTCAGCTTTCC-3′ |
| UPC2-3B | 5′-GCTTCATTAGCACAGTTGCCCATC-3′ |
| UPC2-4A | 5′-TTATGGGCCCACAGTAACGAATCACATTTGTG-3′ |
| UPC2-4B | 5′-GCATTCAATACTTGCCTTTAGTGC-3′ |
| ERG11-f | 5′-TTTAGTTTCTCCAGGTTATGCTCAT-3′ |
| ERG11-r | 5′-ATTAGCTTTGGCAGCAGCAGTA-3′ |
| UPC2-f | 5′-TCCATCCTTGACCCCTAGTCCT-3′ |
| UPC2-r | 5′-CGGCTGAGTTTTGATGTCTTGA-3′ |
| 18S-f | 5′-CACGACGGAGTTTCACAAGA-3′ |
| 18S-r | 5′-CGATGGAAGTTTGAGGCAAT-3′ |
Restriction sites introduced into the primers are underlined. Primers UPC2-3B and UPC2-4B were used for sequencing UPC2 internal regions. The primer pairs ERG11-f and ERG11-r, UPC2-f and UPC2-r, and 18S-f and 18S-r were used for quantitative real-time PCR and yielded 135-bp, 100-bp, and 52-bp products, respectively.
Plasmid constructions.
The UPC2 alleles of the clinical C. albicans isolates were amplified with the primers UPC2-3A and UPC2-4A, which bind in the upstream and downstream region, respectively, of the UPC2 gene (all primers used in this study are listed in Table 2). The PCR products were digested at the introduced SacI and ApaI sites and cloned into the vector pBluescript KSII, generating plasmids pUPC2-580-1, pUPC2-580-2, pUPC2-2440-1, pUPC2-2440-2, pUPC2-945-1, pUPC2-945-2, pUPC2-1619-1, pUPC2-1619-2, pUPC2-1002-1, pUPC2-1002-2, pUPC2-3795-1, pUPC2-3795-2, pUPC2-5044-1, pUPC2-5044-2, and pUPC2-5052 (a plasmid's name reflects the cloned UPC2 allele and the C. albicans isolate from which it was obtained).
Plasmids pUPC2R1, pUPC2R2, and pUPC2R3, which were used to sequentially replace the resident UPC2 alleles in strain SC5314 by mutated UPC2 alleles or a nonmutated control construct with the help of the SAT1 flipper cassette (see Fig. 1), were generated as follows. pUPC2R1 was generated by amplifying the UPC2G648D-TACT1 fragment from plasmid pUPC2K3 (5) with the primers UPC2-3A and ACT19 and substituting the SacI/SacII-digested PCR product for the UPC2 upstream region in the previously described plasmid pUPC2M2 (5). To obtain pUPC2R2, a SacI-BglII fragment with the wild-type UPC2 allele from plasmid pUPC2K2 (5) was substituted for the corresponding fragment in pUPC2R1. pUPC2R3 was constructed by substituting an Nco-NdeI fragment containing the G1927A mutation from pUPC2-5052 for the corresponding fragment in pUPC2R1.
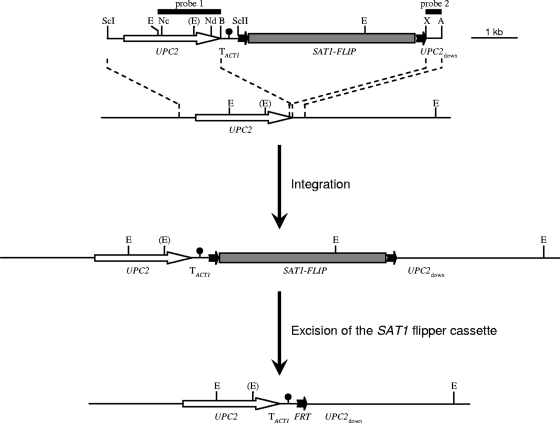
FIG. 1.
Integration of different UPC2 alleles into one or both endogenous UPC2 loci of the C. albicans wild-type strain SC5314, followed by recycling of the SAT1 flipper cassette (gray rectangle flanked by black arrows). The structure of the inserts from plasmids pUPC2R1, pUPC2R2, and pUPC2R3, which contain the UPC2G648D allele, the nonmutated wild-type UPC2 allele, and the UPC2A643T allele, respectively, is shown on top. Relevant restriction sites are indicated as follows: A, ApaI; B, BglII; E, EcoRI; Nc, NcoI; Nd, NdeI; ScI, SacI; ScII, SacII; X, XhoI. The EcoRI site shown in parentheses is present only in the UPC2-2 allele of strain SC5314 and in the UPC2 allele contained in pUPC2R3. The black circle represents the transcription termination sequence of the ACT1 gene (TACT1); the FLP recombination target sites (FRT) are symbolized by the black arrows. The EcoRI-BglII fragment (probe 1) and the XhoI-ApaI fragment (probe 2) from pUPC2R2 were used as probes in Southern hybridization experiments to confirm correct genomic insertion of the constructs and subsequent excision of the SAT1 flipper cassette.
A PERG11-GFP reporter fusion was generated by amplifying an ERG11 upstream fragment from genomic DNA of strain SC5314 with the primers ERG13 and ERG14 and substituting the ApaI/SalI-digested PCR product for the OPT1 upstream fragment in plasmid pOPT1G22 (16) to obtain pERG11G1. The ERG11 downstream region was then amplified with the primers ERG15 and ERG16 and the PstI/SacI-digested PCR product used to replace the OPT1 downstream region in pERG11G1, resulting in pERG11G2 (see Fig. 3).
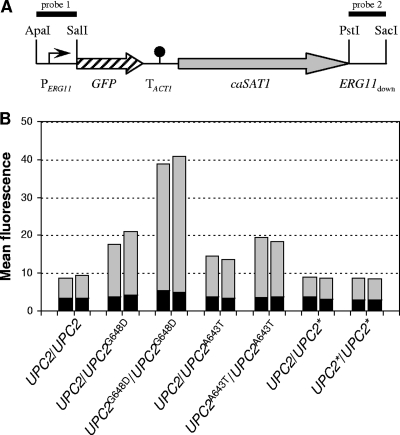
FIG. 3.
The G648D and A643T mutations in Upc2p cause constitutive upregulation of the ERG11 promoter. (A) Structure of the cassette from pERG11G2 containing the PERG11-GFP reporter fusion, which was integrated into one of the ERG11 alleles of the various C. albicans strains by homologous recombination. The ERG11 promoter (PERG11) is symbolized by a bent arrow, the GFP gene by a hatched arrow, the transcription termination sequence of the ACT1 gene (TACT1) by the filled circle, and the caSAT1 selection marker by the gray arrow. The ApaI-SalI fragment (probe 1) and the PstI-SacI fragment (probe 2) from pERG11G2 were used as probes in Southern hybridization analyses to confirm correct genomic integration. (B) Fluorescence of C. albicans strains carrying the PERG11-GFP reporter fusion in a wild-type background or mutant derivatives in which one or both endogenous UPC2 alleles were replaced by the UPC2G648D allele, the UPC2A643T allele, or a nonmutated wild-type allele (UPC2*). The strains were grown to log phase in YPD medium, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independently generated reporter strains are shown in each case. The following strains were used (see Table 1): SCEG2A and -B (UPC2/UPC2), SCUPC2R12EG2A and -B (UPC2/UPC2G648D), SCUPC2R14EG2A and -B (UPC2G648D/UPC2G648D), SCUPC2R32EG2A and -B (UPC2/UPC2A643T), SCUPC2R34EG2A and -B (UPC2A643T/UPC2A643T), SCUPC2R22EG2A and -B (UPC2/UPC2*), and SCUPC2R24EG2A and -B (UPC2*/UPC2*). The background fluorescence of the parental strains, which do not contain the GFP gene, is indicated by the black part of each column.
C. albicans transformation.
C. albicans strains were transformed by electroporation (9) with the gel-purified inserts from plasmids pUPC2R1, pUPC2R2, pUPC2R3, and pERG11G2. Nourseothricin-resistant transformants were selected as described previously (17), and single-copy integration of all constructs was confirmed by Southern hybridization with the probes shown in Fig. 1 and 3.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described previously (17). The DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, and after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence-labeled probes was performed with the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Braunschweig, Germany) according to the instructions of the manufacturer.
Drug susceptibility tests.
Fluconazole susceptibility tests were carried out in high-resolution medium (14.67 g HR medium [Oxoid GmbH, Wesel, Germany], 1 g NaHCO3, 0.2 M phosphate buffer [pH 7.2]) using a previously described microdilution method (18). Readings were done after 24 h.
FACS analysis.
Fluorescence-activated cell sorter (FACS) analysis was performed with a FACSCalibur cytometry system equipped with an argon laser emitting at 488 nm (Becton Dickinson, Heidelberg, Germany). Fluorescence was measured on the FL1 fluorescence channel equipped with a 530-nm band-pass filter. Twenty thousand cells were analyzed per sample. Fluorescence data were collected by using logarithmic amplifiers. The mean fluorescence values were determined with CellQuest Pro (Becton Dickinson) software.
RESULTS
Sequence analysis of the UPC2 alleles of ERG11-overexpressing C. albicans isolates.
To investigate if activating mutations in the transcription factor UPC2 are a common cause of ERG11 overexpression in fluconazole-resistant, clinical C. albicans isolates, we selected four pairs of fluconazole-susceptible and fluconazole-resistant isolates from HIV-infected patients in which the resistant isolate exhibited increased ERG11 mRNA levels in Northern hybridization experiments (14). Quantitative RT-PCR confirmed that the resistant isolates 3795, 2440, 1619, and 5052 had increased ERG11 transcript levels and also showed increased UPC2 expression compared to the matched susceptible isolates 1002, 580, 945, and 5044, respectively (Table 3). We then cloned and sequenced the UPC2 alleles of the eight isolates (see Materials and Methods; Table 4). All four susceptible isolates contained two polymorphic UPC2 alleles. The resistant isolates 3795, 2440, and 1619 contained the same two alleles as their matched susceptible isolates, indicating that the increased ERG11 expression in these isolates was not caused by mutations in the UPC2 coding region. However, in isolate 5052 we found only one of the two UPC2 alleles that were present in the matched susceptible isolate 5044, and direct sequencing of the amplified PCR products across a region that was polymorphic in isolate 5044 confirmed the loss of heterozygosity in isolate 5052. Interestingly, the UPC2 allele that was retained in isolate 5052 contained a G1927A substitution which was not found in any of the other isolate pairs and resulted in an alanine-to-threonine exchange at position 643 (A643T) in the encoded protein. Although the susceptible isolate 5044 contained the same allele, the facts that the A643T mutation is located in the vicinity of the previously described G648D gain-of-function mutation in the C-terminal part of Upc2p and that the resistant isolate 5052 had become homozygous for the UPC2A643T allele suggested that this genomic alteration may have caused the increased ERG11 and UPC2 expression.
TABLE 3.
Relative ERG11 and UPC2 transcript levels in FLUS and FLUR clinical isolate pairsc
| Straina | MICFLU (μg/ml)b | ERG11 | UPC2 |
|---|---|---|---|
| 1002 (S) | 0.25 | 1 | 1 |
| 3795 (R) | >64 | 6.4 (±1.2) | 2.0 (±0.6) |
| 580 (S) | 1 | 1 | 1 |
| 2440 (R) | 32 | 4.0 (±0.1) | 5.5 (±0.2) |
| 945 (S) | 4 | 1 | 1 |
| 1619 (R) | 32 | 3.7 (±0.3) | 9.6 (±0.3) |
| 5044 (S) | 4 | 1 | 1 |
| 5052 (R) | 32 | 1.9 (±0.3) | 2.7 (±0.1) |
Matched fluconazole-susceptible (S) and -resistant (R) isolates are grouped together.
MIC data are from reference 14 (24-h readings).
ERG11 and UPC2 mRNA levels of the susceptible isolates were set as 1.
TABLE 4.
Polymorphisms in UPC2 alleles of different C. albicans strains
| Strain | Allele | Nucleotide substitutionsa |
|---|---|---|
| SC5314 (S) | 1 | T1338C, C1392T, C1410A, C1539T |
| 2 | A1203G, A1593G | |
| 1002 (S) | 1 | T1338C, C1392T, C1410A, C1539T |
| 2 | A1203G, A1593G | |
| 3795 (R) | 1 | T1338C, C1392T, C1410A, C1539T |
| 2 | A1203G, A1593G | |
| 580 (S) | 1 | T777A, T879C, C1326T, T1338C, C1392T, C1410A, C1539T |
| 2 | A234G, A276G, T387C, T425G, T747A, T777A, T879C, A1203G, A1593G | |
| 2440 (R) | 1 | T777A, T879C, C1326T, T1338C, C1392T, C1410A, C1539T |
| 2 | A234G, A276G, T387C, T425G, T747A, T777A, T879C, A1203G, A1593G | |
| 945 (S) | 1 | A1203G, A1593G |
| 2 | A234G, A276G, T387C, T425G, T747G, T777A, T879C, A1203G, A1593G | |
| 1619 (R) | 1 | A1203G, A1593G |
| 2 | A234G, A276G, T387C, T425G, T747G, T777A, T879C, A1203G, A1593G | |
| 5044 (S) | 1 | T777A, T879C, C1326T, T1338C, C1392T, C1410A, C1539T |
| 2 | A234G, A276G, T387C, T425G, T747A, T777A, T879C, A1203G, A1593G,G1927A | |
| 5052 (R) | A234G, A276G, T387C, T425G, T747A, T777A, T879C, A1203G, A1593G,G1927A |
orf19.391 in the Candida genome database (http://www.candidagenome.org/) was used as the reference sequence. The A1593G substitution (underlined) creates an EcoRI restriction site that was used to distinguish the two UPC2 alleles in strain SC5314. The sequences of the two UPC2 alleles of strain SC5314 were obtained from cloned copies (reference 15 and unpublished data). Nucleotide substitutions that result in amino acid exchanges in Upc2p are highlighted in bold. The T425G substitution, which was found in several UPC2 alleles from FLUS and FLUR isolates, causes the amino acid exchange I142S. The G1927A substitution results in the A643T gain-of-function mutation. Matched fluconazole-susceptible (S) and -resistant (R) isolates are grouped together.
The A643T mutation in Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance.
To investigate whether the A643T mutation in Upc2p causes hyperactivity of the transcription factor and enhanced fluconazole resistance, we replaced one of the endogenous UPC2 alleles of the fluconazole-susceptible C. albicans model strain SC5314 by the UPC2A643T allele with the help of the SAT1 flipper cassette (17), as outlined in Fig. 1. This strategy allowed recycling of the caSAT1 selection marker and subsequent introduction of the same mutation into the second UPC2 allele to study the effect of loss of heterozygosity. For comparison, we also introduced the previously described G648D mutation (5) into one or both UPC2 alleles of this strain in the same way. In addition, the resident UPC2 alleles were also replaced in an identical fashion by a nonmutated wild-type UPC2 allele to control for possible effects of the integration strategy. In each case, two independent series of strains were constructed to exclude unspecific phenotypic effects of the genetic manipulations. Introduction of the G1927A (A643T) and G1943A (G648D) mutations into the resident UPC2 alleles was confirmed by reamplification of the UPC2 alleles from heterozygous and homozygous mutants and sequencing of the PCR products.
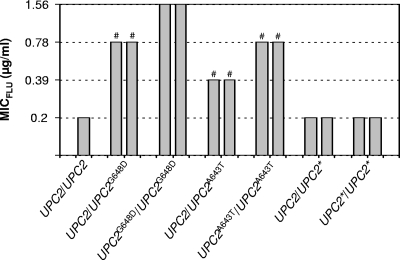
As previously reported (5), replacement of one of the wild-type UPC2 alleles by the UPC2G648D allele increased the MIC of fluconazole for the strains about fourfold (Fig. 2). Subsequent introduction of the G648D mutation into the second UPC2 allele led to a further twofold increase in resistance, demonstrating that a change from heterozygosity to homozygosity for the mutated allele confers a selective advantage in the presence of the drug. Similarly, the introduction of the A643T mutation also conferred increased fluconazole resistance, albeit not as efficiently as the G648D mutation, with a stronger effect when the mutation was present in both alleles. Replacement of the resident UPC2 alleles by a nonmutated, wild-type copy had no effect, confirming that the increased drug resistance was due to the G648D and A643T mutations.
FIG. 2.
Fluconazole susceptibilities of the wild-type parental strain SC5314 and derivatives in which one or both resident UPC2 alleles were replaced by the UPC2G648D allele, the UPC2A643T allele, or a nonmutated wild-type UPC2 allele (indicated by the asterisk). The results obtained with two independently generated strains (A and B) are shown in each case. The following strains were used (see Table 1): SC5314 (UPC2/UPC2), SCUPC2R12A and -B (UPC2/UPC2G648D), SCUPC2R14A and -B (UPC2G648D/UPC2G648D), SCUPC2R32A and -B (UPC2/UPC2A643T), SCUPC2R34A and -B (UPC2A643T/UPC2A643T), SCUPC2R22A and -B (UPC2/UPC2*), and SCUPC2R24A and -B (UPC2*/UPC2*). #, reduced growth was observed at one dilution step below the MIC.
As Upc2p is a transcription factor that regulates the expression of ERG11 and other ERG genes, we investigated whether the resistance mutations in UPC2 constitutively activated the ERG11 promoter. For this purpose, we introduced a PERG11-GFP reporter fusion into the strains expressing the various UPC2 alleles, such that GFP was expressed from one of the endogenous ERG11 promoters (Fig. 3A). The basal activity of the ERG11 promoter could be detected with GFP as a reporter gene, as transformants of the wild-type strain SC5314 carrying the reporter fusion exhibited fluorescence above the background of the nontransformed parental strain (Fig. 3B). In line with the fluconazole susceptibility tests, strains containing one copy of the mutated UPC2G648D or UPC2A643T alleles exhibited increased ERG11 promoter activity, which was further elevated when the mutation was also present in the second UPC2 allele. Again, the effect of the A643T mutation was less pronounced than that of the G648D mutation. In contrast, the control strains in which the resident UPC2 alleles had been replaced by a nonmutated allele did not show increased ERG11 promoter activity. These results demonstrate that both the G648D and A643T mutations in Upc2p result in hyperactivity of the transcription factor and constitutively elevated ERG11 expression.
DISCUSSION
In this work, we describe a second mutation in the transcription factor Upc2p in a fluconazole-resistant clinical C. albicans isolate. Although the A643T substitution had a weaker effect than the G648D mutation that was previously found in another fluconazole-resistant isolate (5), it also resulted in constitutive upregulation of the ERG11 promoter and increased fluconazole resistance when introduced into a drug-susceptible strain. These results confirm that gain-of-function mutations in Upc2p can be a cause of ERG11 overexpression and contribute to fluconazole resistance in clinical C. albicans isolates. Interestingly, the G1927A mutation that results in the A643T substitution was already present in one of the two UPC2 alleles of the susceptible isolate 5044. It can be hypothesized that the mutation was first acquired in a more susceptible progenitor of this isolate during fluconazole therapy, but isolates other than 5044 and 5052 were not available from this patient.
Our results also demonstrate that a change from heterozygosity to homozygosity for the UPC2G648D and UPC2A643T alleles further increases ERG11 expression and fluconazole resistance. The previously described fluconazole-resistant isolate S2, which had acquired the G648D mutation, was heterozygous for the mutated UPC2 allele, but continued exposure to the drug might have selected for loss of heterozygosity also in this strain. In contrast, isolate 5052 had become homozygous for the UPC2A643T allele. As the effect of the loss of heterozygosity was moderate when the mutated UPC2 alleles were expressed in strain SC5314 (Fig. 2), it cannot account for the eightfold increase in fluconazole resistance of isolate 5052 compared to the matched isolate 5044. Isolate 5052 has been reported to overexpress the ABC transporters Cdr1 and Cdr2, which may contribute to its increased drug resistance (14). In addition, it was recently shown that isolate 5052 also had acquired a gain-of-function mutation in Mrr1p and become homozygous for the mutated MRR1 allele (4). The Mrr1p mutation, which caused overexpression of the MDR1 efflux pump, is likely to be responsible for part of the increase in fluconazole resistance in isolate 5052. As UPC2 and MRR1 are located on different chromosomes (chromosome 1 and chromosome 3, respectively), the two loss-of-heterozygosity events observed in isolate 5052 may have occurred independently.
Upc2p upregulates expression of ERG11 and other ergosterol biosynthesis genes in response to ergosterol depletion, which occurs in the presence of azole drugs and other ergosterol biosynthesis inhibitors (10, 20). It is presently unknown how Upc2p is activated under these conditions. One model suggests that a transmembrane domain in the C-terminal region of the protein acts as a cytoplasmic anchor and that proteolytic cleavage allows translocation of the transcription factor domain to the nucleus under inducing conditions, in analogy to the activation of mammalian sterol response element binding proteins (SREBPs) (20). On the other hand, Upc2p that was tagged at the C terminus with a 3× hemagglutinin (HA) epitope could be immunoprecipitated with an anti-HA antibody when bound to its target DNA (23). Therefore, at least the HA-tagged Upc2p, which was constitutively active, can bind to and activate its target genes without proteolytic processing. An alternative model suggests that inducing conditions may prevent Upc2p from interacting with a repressor or an intrinsic negative regulatory domain (23). Whatever the mechanism of Upc2p activation is, it is likely that the gain-of-function mutations in the C-terminal part of Upc2p mimic its activated state, resulting in upregulation of Upc2p target genes. Evidently, the hyperactivity of Upc2p caused by the gain-of-function mutations confers increased fluconazole resistance upon cells carrying these mutations.
In summary, this study demonstrates that gain-of-function mutations in the transcription factor Upc2p are a cause of ERG11 overexpression and increased fluconazole resistance in C. albicans. Loss of heterozygosity for a hyperactive UPC2 allele results in a further increase in drug resistance, similar to what has been found for strains carrying mutated TAC1 and MRR1 alleles (2, 4, 12). However, several ERG11-overexpressing isolates investigated in the present study did not exhibit mutations in the UPC2 coding region, suggesting that other mechanisms can cause ERG11 upregulation in fluconazole-resistant strains. As these isolates also showed increased UPC2 expression, such mechanisms could include mutations in the UPC2 promoter region or in upstream regulatory factors that control UPC2 expression (8). Alternatively, gene amplification by whole-chromosome or segmental aneuploidy could also account for increased ERG11 and UPC2 expression (19).
Acknowledgments
We thank Thomas Patterson for the gift of C. albicans isolates.
This study was supported by the Deutsche Forschungsgemeinschaft (SFB 630) and the National Institutes of Health (NIH grant AI058145).
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Coste, A., A. Selmecki, A. Forche, D. Diogo, M. E. Bougnoux, C. d'Enfert, J. Berman, and D. Sanglard. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coste, A., V. Turner, F. Ischer, J. Morschhäuser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunkel, N., J. Blass, P. D. Rogers, and J. Morschhäuser. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkel, N., T. T. Liu, K. S. Barker, R. Homayouni, J. Morschhäuser, and P. D. Rogers. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 8.Hoot, S. J., B. G. Oliver, and T. C. White. 2008. Candida albicans UPC2 is transcriptionally induced in response to antifungal drugs and anaerobicity through Upc2p-dependent and -independent mechanisms. Microbiology 154:2748-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond, and B. Turcotte. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morschhäuser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 12.Morschhäuser, J., K. S. Barker, T. T. Liu, J. Blaß-Warmuth, R. Homayouni, and P. D. Rogers. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver, B. G., J. L. Song, J. H. Choiniere, and T. C. White. 2007. cis-acting elements within the Candida albicans ERG11 promoter mediate the azole response through transcription factor Upc2p. Eukaryot. Cell 6:2231-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramírez-Zavala, B., O. Reuß, Y. N. Park, K. Ohlsen, and J. Morschhäuser. 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4:e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuß, O., and J. Morschhäuser. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795-812. [DOI] [PubMed] [Google Scholar]
- 17.Reuß, O., Å. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 18.Ruhnke, M., A. Eigler, I. Tennagen, B. Geiseler, E. Engelmann, and M. Trautmann. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selmecki, A., M. Gerami-Nejad, C. Paulson, A. Forche, and J. Berman. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68:624-641. [DOI] [PubMed] [Google Scholar]
- 20.Silver, P. M., B. G. Oliver, and T. C. White. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Znaidi, S., X. De Deken, S. Weber, T. Rigby, A. Nantel, and M. Raymond. 2007. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol. Microbiol. 66:440-452. [DOI] [PubMed] [Google Scholar]
- 23.Znaidi, S., S. Weber, O. Z. Al-Abdin, P. Bomme, S. Saidane, S. Drouin, S. Lemieux, X. De Deken, F. Robert, and M. Raymond. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]