Abstract
An oligonucleotide-based DNA microarray was developed to evaluate expression of genes for efflux pumps in Acinetobacter baumannii and to detect acquired antibiotic resistance determinants. The microarray contained probes for 205 genes, including those for 47 efflux systems, 55 resistance determinants, and 35 housekeeping genes. The microarray was validated by comparative analysis of mutants overexpressing or deficient in the pumps relative to the parental strain. The performance of the microarray was also evaluated using in vitro single-step mutants obtained on various antibiotics. Overexpression, confirmed by quantitative reverse transcriptase PCR, of RND efflux pumps AdeABC, due to a G30D substitution in AdeS in a multidrug-resistant (MDR) strain obtained on gentamicin, and AdeIJK, in two mutants obtained on cefotaxime or tetracycline, was detected. A new efflux pump, AdeFGH, was found to be overexpressed in a mutant obtained on chloramphenicol. Study of MDR clinical isolates, including the AYE strain, whose entire sequence has been determined, indicated overexpression of AdeABC and of the chromosomally encoded cephalosporinase as well as the presence of several acquired resistance genes. The overexpressed and acquired determinants detected by the microarray could account for nearly the entire MDR phenotype of the isolates. The microarray is potentially useful for detection of resistance in A. baumannii and should allow detection of new efflux systems associated with antibiotic resistance.
Multidrug-resistant (MDR) strains of Acinetobacter baumannii have emerged in recent decades. This opportunistic pathogen is responsible for severe infections, particularly hospital-acquired pneumonia and bloodstream, urinary tract, and wound infections, and has become of worldwide concern (13). As in other bacterial species, multidrug resistance can be achieved by two mechanisms: (i) horizontal transfer of genetic information and (ii) mutation of endogenous genes. Acquired resistance determinants that are carried by plasmids (18, 28), transposons (23, 29), and integrons (33, 43) have been described for Acinetobacter spp. Determination of the genomic sequence of several A. baumannii strains has improved our knowledge of the ways in which A. baumannii can develop antibiotic resistance (1, 21, 38, 45). An 86-kb resistance island, AbaR1, found in strain AYE, contains as many as 25 antibiotic and 20 antiseptic and heavy metal resistance genes (16). Variants of this island are integrated at the same chromosomal locus in a significantly high proportion of MDR strains (37). In addition to these acquired resistance genetic elements, alterations in endogenous functions are involved in resistance, such as overexpression of chromosomally encoded β-lactamases ADC and OXA-51-like; loss of porins CarO and Omp33-36 contributing to carbapenem resistance; mutation in the GyrA and ParC fluoroquinolone targets; and overexpression of efflux systems (13).
Efflux systems are components of the bacterial membrane that are thought to play a role in homeostasis of the cell and extrusion of toxic compounds (32). They can also be involved in cell-to-cell communication via quorum-sensing systems (22) and in bacterial pathogenicity (32). Efflux pumps of the resistance nodulation cell division (RND) superfamily, widespread in gram-negative bacteria, confer multidrug resistance to their host when overexpressed (34). In most instances, the level of resistance remains moderate; however, reduced intracellular accumulation of antibiotics provides a delay for the selection of high-level resistance by drug inactivation or target alteration. Efflux thus can act in synergy with other mechanisms to achieve high-level resistance (34).
Two RND efflux systems, AdeABC and AdeIJK, have been characterized for A. baumannii (9, 24). They are composed of an efflux protein (AdeB or AdeJ) which interacts with a membrane fusion protein (AdeA or AdeI) and an outer membrane factor (AdeC or AdeK) to facilitate drug export across both the inner and the outer membranes. The adeABC operon is cryptic in approximately 80% of A. baumannii strains (24), and its overexpression leads to diminished susceptibility to aminoglycosides, cefepime, fluoroquinolones, chloramphenicol, and tetracycline-tigecycline. Consequently, the AdeABC efflux pump constitutes a major mechanism of multiple antibiotic resistance in A. baumannii, and its clinical significance has been established (31). AdeIJK is present in all A. baumannii strains and confers decreased susceptibility to β-lactams, fluoroquinolones, chloramphenicol, rifampin (rifampicin), erythromycin, clindamycin, and tetracycline-tigecycline. The toxicity observed with Escherichia coli and A. baumannii when the adeIJK operon was cloned and overexpressed suggests its effect on resistance is minimal (9). Another pump belonging to the MATE family, AbeM, has been characterized (39), but its role in the antibiotic resistance of A. baumannii remains to be determined. Analysis of the genome sequence of strain AYE revealed the presence of 41 other genes putatively associated with efflux (16).
Microarrays are powerful tools for large-scale screening of the genomic or transcriptomic content of a cell population. Several genomic microarrays have been developed to detect acquired antibiotic resistance genes (5) and transcriptomic microarrays have been used to compare spontaneous mutants with the parental strain, highlighting the role of overexpression of efflux genes in antibiotic resistance (14).
Overexpression of efflux systems is an important factor leading to antibiotic resistance but is not easily detectable at both the phenotypic level, because of the low level of resistance conferred, and the genotypic level, because the regulatory mutations most often remain unknown. To study the mechanisms of multidrug resistance in A. baumannii, particularly those involving efflux, we have developed a microarray that allows quantification of the expression of resistance genes, including intrinsic and acquired determinants.
(An initial report of this work was presented by S. Coyne et al. at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, abstr. C1-096, 2009.)
MATERIALS AND METHODS
Bacterial strains, growth conditions, and antibiotic susceptibility testing.
The bacterial strains used in this study are listed in Table 1. Cells were grown at 37°C in brain heart infusion broth and agar (Difco Laboratories, Detroit, MI). Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France), and MICs were determined by the Etest procedure (AB Biodisk, Solna, Sweden).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| CIP 70-10 | Susceptible reference strain | 3 |
| BM4546 | CIP 70-10 AdeST153M spontaneous mutant, MDR | 25 |
| BM4547 | CIP 70-10 AdeRP116L spontaneous mutant, MDR | 25 |
| BM4454 | Clinical strain overexpressing AdeABC | 24 |
| BM4579 | BM4454 ΔadeIJK | 9 |
| BM4651 | BM4454 ΔadeABC | 9 |
| BM4652 | BM4454 ΔadeABC ΔadeIJK | 9 |
| BM4587 | Susceptible clinical strain | This study |
| BM4665 | BM4587 spontaneous mutant obtained on gentamicin, MDR | This study |
| BM4666 | BM4587 spontaneous mutant obtained on cefotaxime, MDR | This study |
| BM4667 | Susceptible clinical strain | This study |
| BM4668 | BM4667 spontaneous mutant obtained on tetracycline, MDR | This study |
| AYE | MDR clinical strain | 16 |
| BM4675 | MDR clinical strain | This study |
| BM4676 | MDR clinical strain | This study |
Production of MDR mutants.
Spontaneous MDR mutants BM4665 and BM4666 were obtained from wild-type strain BM4587, and mutant BM4668 was obtained from strain BM4667, on gradient plates (40) containing either gentamicin, cefotaxime, or tetracycline (Table 1). Colonies growing at concentrations higher than the normal MIC were tested for antibiotic susceptibility by diffusion, and those exhibiting resistance to several drug classes were selected for further studies.
DNA manipulations.
A. baumannii genomic DNA was extracted as described previously (36). Amplification of DNA was performed in a GeneAmp PCR system 9700 (Perkin-Elmer Cetus, Norwalk, CO) with Taq (MPbio, Illkirch, France) or Phusion (Finnzymes, Espoo, Finland) DNA polymerase. Amplification of large DNA fragments was achieved using the Expand Long Template PCR system (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer's recommendations. PCR elongation times and temperatures were adjusted according to the expected sizes of the PCR products and the nucleotide sequences of the primers (Table 2), respectively. Sequence determination was carried out with a CEQ 2000 DNA analysis system automatic sequencer (Beckman Instruments, Inc., Palo Alto, CA).
TABLE 2.
Oligonucleotides used in PCR and qRT-PCR
| Gene | Primer | Sequence (5′→3′) | Positiona | Size (bp) |
|---|---|---|---|---|
| rpoB | rpoB Rev | ATGCCGCCTGAAAAAGTAAC | 1960-1979 | 154 |
| rpoB For | TCCGCACGTAAAGTAGGAAC | 2114-2095 | ||
| adeA | adeA For | ATCGCTAACAAAGGCTTGAA | 1024-1043 | 159 |
| adeA Rev | CGCCCCCTCAGCTATAGAA | 1183-1163 | ||
| adeB | adeB For | CTTGCATTTACGTGTGGTGT | 2907-2926 | 168 |
| adeB Rev | GCTTTTCTACTGCACCCAAA | 3075-3056 | ||
| adeC | adeC For | TACACATGCGCATATTGGTG | 1157-1176 | 117 |
| adeC Rev | CGTAAAATAACTATCCACTCC | 1274-1244 | ||
| adeI | adeI For | CAAATGCAAATGTAGATCTTGG | 511-532 | 210 |
| adeI rev | AAACTGCCTTTACTTAGTTG | 702-721 | ||
| adeJ | adeJ For | GGTCATTAATATCTTTGGC | 1142-1160 | 221 |
| adeJ Rev | GGTACGAATACCGCTGTCA | 1363-1345 | ||
| adeK | adeK For | TTGATAGTTACTTGACTGTTC | 1264-1285 | 162 |
| adeK Rev | GGTTGGTGAACCACTGTATC | 1407-1426 | ||
| ampC | ampC For | CAGTAATTCAGAACAGATTGTG | 927-947 | 183 |
| ampC Rev | GCGCTCTTCATTTGGAATAC | 1110-1091 | ||
| adeS | adeS For | TATGAAAAGTAAGTTAGGAAT | 1-19 | 1,072 |
| adeS Rev | TTAGTTATTCATAGAAATTTT | 1073-1053 | ||
| adeR | adeR For | AATGCTTAATACACTGACTTTAGCCGGTGG | −265-−236 | 1,429 |
| adeR Rev | TCTAGCAGAGAGGTCGC | 1101-1084 | ||
| ISAba1 | ISAba1ab | ATGCAGCGCTTCTTTGCAGG | 462-471 | 379 |
| ISAba1bb | AATGATTGGTGACAATGAAG | 822-841 | ||
| ISAba4 | ISAba4ac | ATTTGAACCCATCTATTGGC | 140-159 | 611 |
| ISAba4bc | ACTCTCATATTTTTTCTTGG | 751-732 | ||
| blaOXA-23 | oxa23+ | ATGAATAAATATTTTACTTG | 1-20 | 821 |
| oxa23− | TTAAATAATATTCAGCTGTT | 822-803 | ||
| tniA | tniA For | CAATGTGGTTGTCGGTTATGA | 54-74 | 162 |
| tniA Rev | ATTCAAACTCTCAGTTAGCGT | 216-196 | ||
| ATPase | ATPase3′ | AATGGCGGTACATCAATATGT | 1215-1195 | 492 |
| ATPase5′ | GAAACTCTTGAAGTTGCAAGT | 723-743 |
RNA isolation.
RNA was isolated as described previously (27). Briefly, exponentially growing cells (optical density at 600 nm of 0.8 to 1.2) were harvested, and the pellet was resuspended in 12.5 mM Tris-5 mM EDTA-10% glucose. Cells were sheared mechanically in the presence of acid phenol (pH 4.6) and glass beads (0.2- to 0.3-mm diameter; Sigma-Aldrich, St. Louis, MO) using a Fastprep apparatus (Bio 101). After centrifugation, Trizol reagent (Invitrogen, Leek, The Netherlands) was added to the supernatant. Total RNA was extracted twice with chloroform-isoamyl alcohol (24:1 [vol/vol]), precipitated in 0.7 volume of isopropanol and washed twice with 70% ethanol. Treatment with RNase-free DNase (Ambion, Austin, TX) was performed according to the manufacturer's recommendations. RNA was quantified by determining absorbance at 260 and 280 nm; its purity and integrity were analyzed on agarose gels.
Microarray construction.
Oligonucleotide probes (70-mer) were designed with ArrayOligoSelector (4) to represent each open reading frame annotated as an A. baumannii coding sequence in the GenBank database (http://www.ncbi.nlm.nih.gov/) using the complete genome sequence of AYE as a reference. BLAST analysis of probes against themselves or the AYE genome enabled nonspecific binding effects to be eliminated. The 205 probes selected were synthesized and spotted in triplicate on UltraGAPS glass slides (Corning Life Sciences, Corning, NY) using a Chipwriter Pro Virtek arrayer (Bio-Rad, Hercules, CA). After printing, arrays were treated as recommended by the slide manufacturer.
Synthesis of cDNA and microarray hybridization.
cDNAs were synthesized from 10 μg of total RNA and labeled with Cy3 or Cy5 cyanin (GE Healthcare, Uppsala, Sweden) using the SuperScript indirect cDNA labeling system (Invitrogen). Labeled cDNAs of the studied and reference strains were mixed at equal concentrations (250 pmol of cyanin) and hybridized on the microarray. Slides were scanned (GenePix 4000 A scanner; Axon), and images were analyzed (GenePix5 software; Axon).
Data analysis.
Script analyses were developed with the software R (R Development Core Team; http://www.R-project.org). For each spot, the fluorescence value corresponded to the median pixel intensity within the spot and no background was subtracted. Three independent biological replicates and a dye swap for each replicate to overcome cyanin bias were performed. Loess normalization (46) was applied to all spots, on a slide-by-slide basis. To determine differentially expressed genes from replicates pooling log2 ratios, a paired t test was performed. Low numbers of observations per spot and of spots per slide (615) did not permit calculation of a specific variance and clusters of spots with equal variance, respectively. For these reasons, it was assumed that the variance of the log2 ratios was the same for all spots, and spots displaying extreme specific variance (too small or too large) were excluded from the statistical analysis (17). A spot was considered differentially expressed when the Bonferroni method-adjusted P value was lower than 0.05. The mean and standard deviation were calculated for genes corresponding to significantly differentially expressed spots and with a change of <−1.5 or >1.5.
Quantitative reverse transcriptase (qRT) PCR.
cDNA synthesis was performed for 60 min at 42°C using 0.1 to 0.2 μg of RNA, 0.8 μl of avian myaloblastosis virus reverse transcriptase (Roche Diagnostic GmbH, Mannheim, Germany), and 2 μl of random primer according to the manufacturer's instructions. Real-time PCR was performed with a 20-μl reaction volume containing 2 μl of cDNA, 1× SYBR PCR master mix (Roche), and gene-specific primers, 0.5 mM each (Table 2). Amplification and detection of specific products were performed using the LightCyler sequence detection system (Roche) with the following cycle profile: 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, 58°C for 10 s, and 72°C for 30 s.
Microarray data accession number.
The microarray data obtained in this study were deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) according to MIAME standards under accession number E-MEXP-2254.
RESULTS AND DISCUSSION
Design of the microarray.
An oligonucleotide-based DNA microarray was developed to compare the expression levels of efflux genes of an isogenic MDR mutant and its parental strain or those of clinical isolates and a reference strain. The GenBank database was screened for A. baumannii gene sequences since no annotated genome was available at the beginning of the study. A total of 205 genes was selected for the microarray. The list of genes and corresponding probes is available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MEXP-2254. The probes were designed to detect the 47 efflux-related genes of strain AYE (16), which comprise six RND (including adeABC and adeIJK), seven MF, and two MATE systems; 10 genes for outer membrane proteins, e.g., those encoding the pore-forming protein HMP-AB (19), the 33- to 36-kDa protein (10), and CarO (30); and 8 genes involved in biofilm formation, notably the csu operon (41) and its two-component regulatory system (42). Fifty-five antibiotic resistance genes from gram-negative bacteria were added, several of them not yet described for A. baumannii. They represent the most clinically relevant resistance mechanisms, i.e., the chromosomal cephalosporinase, class A β-lactamases, metallo-β-lactamases and oxacillinases, including carbapenemases; aminoglycoside modifying enzymes (11 acetyl-, 4 nucleotidyl-, and 3 phosphotransferases), 16S RNA methylases, tetracycline efflux pumps and ribosomal protection proteins, rifampin ADP ribosyltransferase (Arr-2), chloramphenicol acetyltransferases and efflux pumps, and plasmid-borne quinolone resistance gene qnr. Genes conferring resistance to arsenic (ars) and mercury (mer) heavy metals were also included. Finally, genes involved in mobility or transfer of genetic elements, such as integrases, insertion sequences, or transposases, and several genes present in the 86-kb resistance island of strain AYE, such as trxB, lspA, and uspA, were added. These sequences should enable detection of the backbone structures of mobile genetic elements carrying antibiotic resistance genes.
Thirty-five housekeeping genes, such as gyrA, parC and rpoB, as well as genes used for an MLST scheme (2) and a multilocus PCR typing method (15), and 20 genes linked to metabolism functions were added to avoid normalization bias due to the low density of the chip.
Validation of the microarray.
Microarray validation was first performed by comparing strain BM4454, which overexpresses the AdeABC efflux system (24), with derivative mutants BM4579, BM4651, and BM4652, in which adeIJK, adeABC, and both systems, respectively, had been inactivated by insertion-deletion (9). Comparative analysis of the data obtained with BM4454 and BM4579 (BM4454 ΔadeIJK) revealed that, as expected, the adeI and adeJ genes were not expressed in the mutant (Table 3). Since integration of the cassette partially deletes the adeI and adeJ genes but not adeK, no differences were observed for adeK expression. These results were confirmed by qRT-PCR using specific primers (Table 2). Comparison of the BM4454 and BM4651 (BM4454 ΔadeABC) transcriptomes confirmed inactivation of adeA and adeB in the latter. The aac(3)-IVa cassette that was inserted to inactivate this system was confirmed to be expressed in BM4651. Similarly, adeC was overexpressed due to a transcriptional fusion with aac(3′)-IVa in the mutant (Table 3) (9). The expression of other efflux genes was not affected by inactivation of adeABC. Comparative analysis of the transcriptomes of BM4454 and BM4652 (BM4454 ΔadeABC ΔadeIJK) confirmed the inactivation of adeA, adeB, adeI, and adeJ and overexpression of aac(3)-IVa and adeC. Differential expression of adeA and adeC was not found to be statistically significant by the microarray but was confirmed by qRT-PCR (Table 3).
TABLE 3.
Differential quantification of gene expression in isogenic strains of A. baumannii
| Strain |
Change (fold)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Parent | Mutant | adeA | adeB | adeC | adeI | adeJ | adeK | aac(3)-IVa |
| BM4454 | BM4579 | NS | NS | NS | −3.64 | −3.60 | NS | NA |
| BM4651 | −4.21 | −3.25 | 1.84 | NS | NS | NS | 3.56 | |
| BM4652 | −1.17b | −1.50 | 1.38b | −1.80 | −2.70 | NS | 12.88 | |
| CIP70-10 | BM4546 | 2.17 | 2.74 | NS | NS | NS | NS | NS |
| BM4547 | NS | NS | NS | NS | NS | NS | NS | |
| BM4587 | BM4665 | 1.79 | 3.04 | 2.09 | NS | NS | NS | NA |
| BM4666 | NS | NS | NS | 1.65 | 2.35 | 1.16b | NS | |
| BM4667 | BM4668 | NS | NS | NS | 1.53 | 2.42 | 1.04b | NS |
Positive and negative values correspond to genes with, respectively, increased or decreased expression in the mutant strain in comparison with the parental strain. NS, not statistically significant. NA, not applicable.
Not statistically significant by the microarray but confirmed by qRT-PCR.
Validation was also performed by studying AdeABC-overexpressing mutants BM4546 and BM4547 obtained from strain CIP 70-10 following point mutations in adeS and adeR, respectively (Table 1) (25). The two-component system AdeRS controls the expression of adeABC, AdeR being a transcriptional regulator and AdeS a histidine kinase sensor (25). The regulatory operon is located upstream from adeABC and transcribed in the opposite direction. Overexpression of adeA and adeB in BM4546, but not in BM4547, was observed (Table 3). qRT-PCR showed 10- and 4-fold increases in adeB expression in BM4546 and BM4547, respectively. These results are in agreement with BM4546 having an amount of adeB mRNA larger than that in BM4547 as detected by Northern hybridization (25), despite the fact that the two mutants exhibit similar resistance phenotypes. No changes in the expression of the adeR and adeS genes were detected, indicating that the two-component system was not self-regulated. Expression of several housekeeping genes, such as cpn60 and gltA in BM4546 and recA in BM4547, was modified, suggesting collateral effects due to AdeRS mutations or AdeABC overexpression. However, no differences in growth levels were observed between the strains (data not shown).
Detection of adeABC and adeIJK overexpression.
BM4665, a spontaneous resistant mutant of susceptible clinical isolate BM4587 obtained on gentamicin (Table 1), was found to be less susceptible to aminoglycosides (12-fold increase in MICs), fluoroquinolones, chloramphenicol, and tetracycline-tigecycline (Table 4). No changes in β-lactam susceptibility were observed, except for cefepime. Strain BM4665 overexpressed the adeABC operon (1.8- to 3-fold increase) (Table 3) but no other efflux genes, and the MDR phenotype of BM4665 was in agreement with overexpression of the pump (24). A 10- to 30-fold increase in expression of adeABC was found by qRT-PCR. Overexpression of the AdeABC system can be due either to mutations in the adeRS two-component system (25) or to the insertion of ISAba1 upstream from the adeABC operon (35). In order to elucidate the overexpression of adeABC in BM4665, the sequence of adeRS was determined, revealing a Gly-to-Asp substitution at position 30 of AdeS. This residue is located in the periplasmic input domain of the sensor. Mutations in this domain of several histidine protein kinases, such as NarX (7), BvgS (26), and VanSD (11), have been described and lead to constitutive expression of the regulated genes. Thus, the G30D substitution is most likely responsible for overexpression of AdeABC in BM4665.
TABLE 4.
Antibiotic susceptibility of A. baumannii
| Strain | MIC (μg/ml)a |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | TIM | CAZ | CTX | FEP | ATM | IPM | CHL | TET | MIN | TIG | RIF | NOR | CIP | SXT | GEN | TOB | AMK | NET | |
| BM4454 | 8 | 12 | 6 | 4 | 3 | 16 | 0.25 | >256 | 32 | 0.75 | 3 | 2 | >256 | >32 | 0.064 | 6 | 4 | 8 | 16 |
| BM4587 | 6 | 8 | 4 | 3 | 0.75 | 12 | 0.19 | 96 | 1.5 | 0.064 | 0.094 | 1.5 | 1.5 | 0.094 | 0.094 | 1 | 0.75 | 4 | 1.5 |
| BM4665 | 2 | 4 | 3 | 4 | 8 | 6 | 0.5 | >256 | 3 | 0.125 | 1.5 | 1.5 | 12 | 0.5 | 0.125 | 12 | 16 | 64 | 64 |
| BM4666 | 16 | 12 | 12 | 6 | 2 | 32 | 0.19 | >256 | 8 | 0.19 | 0.38 | 3 | 4 | 0.38 | 0.25 | 1.5 | 1 | 6 | 1.5 |
| BM4667 | 12 | 12 | 8 | 4 | 1.5 | 32 | 0.25 | 96 | 3 | 0.064 | 0.125 | 2 | 1.5 | 0.125 | 0.25 | 1.5 | 1.5 | 4 | 2 |
| BM4668 | 24 | 16 | 16 | 8 | 3 | 48 | 0.25 | >256 | 8 | 0.125 | 0.5 | 3 | 4 | 0.38 | 0.5 | 1.5 | 2 | 8 | 2 |
| CIP 70-10 | 24 | 16 | 6 | 12 | 1.5 | 48 | 0.25 | 48 | 1 | 0.047 | 0.125 | 4 | 2 | 0.125 | 0.094 | 1.5 | 2 | 6 | 2 |
| AYE | >256 | >256 | >256 | >256 | >256 | >256 | 1 | >256 | 64 | 0.5 | 1.5 | 16 | >256 | >32 | >32 | >256 | 64 | 96 | 48 |
| BM4675 | >256 | >256 | >256 | >256 | 8 | 32 | 2 | >256 | 32 | 8 | 1.5 | >32 | >256 | >32 | >32 | 24 | 12 | 96 | 32 |
| BM4676 | >256 | >256 | >256 | 32 | 32 | 32 | >32 | 128 | 12 | 1 | 1.5 | 2 | 192 | 24 | 0.125 | >256 | 96 | >256 | 12 |
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; CTX, cefotaxime; ERY, erythromycin; FEP, cefepime; GEN, gentamicin; IPM, imipenem; MIN, minocycline; NET, netilmicin; NOR, norfloxacin; RIF, rifampin; SXT, co-trimoxazole; TET, tetracycline; TIC, ticarcillin; TIG, tigecycline; TIM, ticarcillin-clavulanic acid; TOB, tobramycin.
Spontaneous MDR mutants BM4666 and BM4668 obtained from BM4587 and BM4667 on cefotaxime and tetracycline, respectively (Table 1), had similar resistance profiles characterized by decreased susceptibility to tetracycline-tigecycline, fluoroquinolones, chloramphenicol, co-trimoxazole, rifampin, and to a lesser extent, β-lactams, whereas aminoglycosides were not affected (Table 4). This phenotype was distinct from that associated with overexpression of AdeABC, suggesting another mechanism. Comparative microarray experiments revealed overexpression of adeI and adeJ (1.5- and 2.4-fold increases, respectively) in the MDR mutants (Table 3), which was confirmed by qRT-PCR, indicating a 2.5- to 5-fold increase in the expression of the adeIJK gene set in the two mutants. Determination of the sequence of the adeIJK operon and of the upstream flanking region containing its putative promoter did not reveal any mutation, and regulation of expression of adeIJK remains unknown. The level of overexpression of adeIJK was systematically lower than that observed for adeABC in microarray as well as in qRT-PCR experiments. trans-complementation of BM4579 (BM4454 ΔadeIJK) with plasmid-borne adeIJK indicated that overexpression of AdeIJK was toxic for the host (9). Taken together, these observations suggest that AdeIJK is tightly regulated and can be overexpressed only within narrow limits. The fact that the BM4666 and BM4668 spontaneous mutants overexpress adeIJK at a low level suggests that these types of mutations can occur in clinical strains. The role of this efflux system in antibiotic resistance of clinical isolates remains to be determined.
Another mutant from BM4652 (BM4454 ΔadeABC ΔadeIJK) obtained on chloramphenicol exhibited an MDR phenotype. Comparison of the transcriptome of the mutant with that of the parental strain using the microarray revealed overexpression (2.9- to 3.8-fold) of a new RND efflux system, AdeFGH, that could be responsible for this phenotype and is currently under study.
Detection of antibiotic resistance genes in clinical isolates.
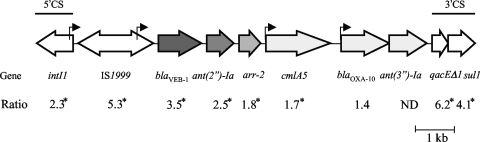
Comparison by the microarray of AYE with susceptible reference strain CIP 70-10 indicated that genes adc for the chromosomal cephalosporinase and adeABC for the efflux system were overexpressed and that, among the 70 genes of the AbaR1 resistance island, 19 out of the 33 printed on the microarray were expressed, e.g., blaVEB, ant(2′′)-Ia, cat, arr-2, aac(3)-Ia, floR, merA, IS1999, intI1, and sul1 (Table 5). Sequencing of the PCR product obtained with the ISAba1b and amp Rev primers (Table 2) confirmed the presence of a copy of ISAba1 upstream from adc that likely provides a strong promoter (20) for enhanced production of cephalosporinase in AYE. Expression of the chromosomally located blaOXA-51-type was not detected, and analysis of the AYE genome sequence did not show the presence of ISAba1 upstream from the structural gene for the β-lactamase that is required for overexpression leading to carbapenem resistance (44). Fourteen genes of the resistance island were not detected by the microarray probably due to lack of, or weak, expression. All the resistance genes present in AYE except tet(A) and blaOXA-10 were detected; tet(A) transcription is repressed by TetR in the absence of tetracycline (6), while blaOXA-10, part of a class 1 integron that also contains blaVEB, ant(2′′)-Ia, arr-2, cmlA5, and ant(3′′)Ia, was weakly expressed, although it possesses its own promoter (12). Within an integron, the expression of promoterless, cassette-associated resistance genes is influenced markedly by their position in the cassette array (12). A constant decrease in the expression of genes blaVEB, ant(2′′)-Ia, and arr-2, depending on the distance from the promoter borne by IS1999, was found by the microarray (Fig. 1). The microarray detected the expression of other genes in AbaR1, such as the mercury resistance operon and intI1 and qacEΔ1-sul1, that comprise the 5′ and 3′ conserved sequences of the class 1 integron, respectively (Table 5). Combination of (i) overexpression of the cephalosporinase and AdeABC efflux system and (ii) the presence of acquired resistance genes, most of them carried by the 86-kb island, resulted in multiple antibiotic resistance of the AYE host.
TABLE 5.
Differential quantification of gene expression in clinical isolates compared to CIP 70-10
| Function or gene | Description | Change (fold)a |
||
|---|---|---|---|---|
| AYE | BM4675 | BM4676 | ||
| Efflux | ||||
| adeA | MFP | 2.84 | 5.42 | 3.94 |
| adeB | RND | 2.80 | 3.99 | 3.58 |
| adeC | OMF | 2.28 | 2.28 | 2.72 |
| adeI | MFP | NS | NS | 1.73 |
| adeJ | RND | NS | NS | 1.83 |
| adeK | OMF | NS | NS | NS |
| APC 41_138 | d-serine/d-alanine/glycine transport protein | 2.08 | NS | NS |
| MFS 12_508 | MFS | NS | NS | 1.53 |
| RND 29_167 | RND | 2.10b | NS | NS |
| Outer membrane protein | ||||
| omp25 | Putative outer membrane protein | 1.72 | NS | NS |
| omp33-36 | 33- to 36-kDa outer membrane protein | −1.62 | NS | 1.84 |
| omp34 | 34-kDa outer membrane protein | 2.22 | NS | 14.36 |
| ompA | Outer membrane protein A | −3.01 | −2.32 | NS |
| Housekeeping and metabolism | ||||
| bfmS | Sensor kinase component of a two-component regulatory system (biofilm) | NS | NS | 1.59 |
| cpn60 | 60-kDa chaperonin | −2.00 | NS | NS |
| csuA/B | Putative pilus subunit, biofilm operon | NS | −1.72 | 1.84 |
| fusA | Translation elongation factor EF-G | NS | NS | 1.68 |
| gltA | Citrate synthase | NS | NS | 1.54 |
| ppa | Inorganic pyrophosphatase | −2.73 | NS | NS |
| rpoC | DNA-directed RNA polymerase beta subunit | −1.97 | NS | NS |
| Antibiotic resistance | ||||
| adc | Chromosomal cephalosporinase | 22.99 | 5.73 | 5.72 |
| blaOXA-23 | Class D carbapenemase | NS | NS | 29.61 |
| blaTEM-1 | β-Lactamase | NS | 1.61 | NS |
| blaVEB-1 | Extended-spectrum β-lactamase | 3.51 | NS | NS |
| aac(3)-Ia | Aminoglycoside adenylyltransferase | 2.62 | NS | NS |
| ant(2′′)-Ia | Aminoglycoside nucleotidyltransferase | 2.51 | NA | 29.39 |
| aph(3′)-VIa | Aminoglycoside phosphotransferase | NA | NS | 4.04 |
| arr-2 | Rifampin ADP-ribosylating transferase | 1.76 | NS | NS |
| catI | Chloramphenicol acetyltransferase | 3.79 | NS | NS |
| cmlA | Chloramphenicol efflux pump | 1.73 | NS | NS |
| floR | Chloramphenicol efflux pump | 1.66 | NS | NS |
| sul1 | Sulfonamide resistance | 4.10 | NS | NS |
| tet(B) | Tetracycline efflux pump | NS | 1.87 | NS |
| Heavy metal resistance | ||||
| HMD | Heavy metal detoxification | 2.50 | NS | NS |
| merA | Mercuric ion reductase | 1.79 | NS | NS |
| merR | Mercury resistance operon regulatory protein | 1.96 | NS | NS |
| Gene mobility and island | ||||
| groEL/int1 | Fusion GroEL/integrase | 2.71 | NS | NS |
| intI1 | Integrase | 2.32 | NS | NS |
| IS1999 | Insertion sequence | 5.26 | NS | NS |
| IS6100 | Insertion sequence | 1.87 | NS | NS |
| ISAba1 | Insertion sequence | NS | 1.83 | NS |
| lspA | Lipoprotein signal peptidase | 1.81 | NS | 1.56 |
| qacEdelta1 | Quaternary ammonium compound resistance protein | 6.19 | NS | NS |
| resolvase 1_318 | Resolvase | 2.56 | NS | NS |
| tniA | Transposase | NS | 2.99 | 1.59 |
| tnpM | Transposase | 1.74 | 2.38 | NS |
| uspA | Universal stress protein | NS | NS | 1.50 |
Positive and negative values correspond to genes with increased and decreased expression, respectively, in the clinical isolate in comparison with CIP 70-10. NS, not statistically significant. NA, not applicable.
Statistically significant but not confirmed by qRT-PCR.
FIG. 1.
Differential gene expression in the class 1 integron of the AbaR1 island from strain AYE. The ratio was determined by comparing the expression levels of every gene between strains AYE and CIP 70-10. 5′CS and 3′CS, 5′ and 3′ conserved sequences of class 1 integron; *, statistically significant; ND, not determined. Arrows indicate coding sequences and sense of transcription. Bent arrows represent the promoters for the gene cassettes, with IS1999 bringing a strong promoter. Decreased intensity in gray indicates decreased overexpression from blaVEB-1 to blaOXA-10.
BM4675 and BM4676, two other unrelated MDR clinical isolates, were analyzed. BM4675 was highly resistant to fluoroquinolones, tetracyclines, rifampin, chloramphenicol, co-trimoxazole, and β-lactams and moderately resistant to aminoglycosides and exhibited decreased susceptibility to tigecycline (Table 4). The only antimicrobial agent tested that conserved in vitro activity was imipenem (Table 4). Comparison of the transcriptome of BM4675 with that of reference strain CIP 70-10 indicated that genes adc for the chromosomal cephalosporinase and adeABC for the efflux system were overexpressed (Table 5). As in AYE, a copy of ISAba1 was found upstream from adc, which probably provides the promoter for its expression. The experiment enabled detection of the β-lactam blaTEM-like and tetracycline tet(B) resistance genes. As with tet(A), regulation by the tetR repressor does not allow expression of tet(B) in the absence of tetracycline. Thus, constitutive expression of tet(B) in BM4675 could result from a lack of or a nonfunctional tetR. Overexpression of the ADC cephalosporinase and of AdeABC, combined with production of TEM and Tet(B), accounts for part of the resistance phenotype of BM4675 (Table 4). Other alterations, such as mutations in type II topoisomerases, diminished permeability, or acquired genes not spotted on the microarray, probably combined to produce the multidrug resistance phenotype.
BM4676 had high-level resistance to imipenem that was not restored by EDTA, suggesting the presence of a class D carbapenemase. The strain was also resistant to β-lactams, chloramphenicol, fluoroquinolones, and tetracycline and had decreased susceptibility to minocycline and tigecycline. Moderate-level resistance was observed for netilmicin, whereas the strain was highly resistant to the other aminoglycosides tested (Table 4). Comparative transcriptome analysis with reference strain CIP 70-10 indicated overexpression of chromosomal genes adc for the cephalosporinase and adeABC for the efflux system (Table 5). Weak overexpression of adeI and adeJ, confirmed by qRT-PCR, was observed (Table 5). As in BM4675, a copy of ISAba1 was found upstream from adc. The microarray also enabled detection of production of OXA-23 carbapenemase and of ANT(2′′)-Ia and APH(3′)-VIa aminoglycoside nucleotidyl- and phosphotransferase (Table 5). The presence of a copy of ISAba4 upstream from blaOXA-23, located by PCR using ISAba4b and oxa23- primers (Table 2), was presumably responsible for enhanced expression of this gene, as previously demonstrated (8). As observed for BM4675, the overexpressed and acquired determinants detected by the microarray could account for nearly the entire MDR phenotype of BM4676 (Table 4).
The microarray also detected the expressed transposase tnpM and tniA genes in BM4675 and the tniA, lspA, and uspA genes, coding for a transposase, a lipoprotein signal peptide, and a universal stress protein in BM4676, respectively (Table 5). These genes are part of the AbaR1 resistance island of AYE (16). PCR experiments using the ATPase3′ and tniA Rev primers (Table 2) indicated that, in both strains, tniA was inserted at the 3′ end of an ATPase gene which has been proposed to be a potential “hot spot” for integration of genomic islands in A. baumannii (37). No amplification products were obtained when long-range PCR was performed between tniA and the 5′ end of the ATPase gene using the tniA For and ATPase5′ primers (Table 2), possibly because either the 5′ end of the ATPase gene had been deleted or the primers were too distant. None of the antibiotic resistance genes described for the original AbaR1 island were detected with BM4675 and BM4676.
Differential expression of several genes coding for outer membrane proteins was observed with the three clinical isolates relative to CIP 70-10 (Table 5). These changes could impact membrane permeability and account for susceptibility decrease, in particular with imipenem, either alone or in combination with efflux systems and β-lactamase production.
Other genes, in particular housekeeping genes cpn60, ppa, and rpoC in AYE and fusA and gltA in BM4676, were found to be differentially expressed (Table 5). Modification of expression of these genes could reflect the cost of antimicrobial resistance for host fitness, as already proposed (14).
Microarray technology is a powerful tool that can be designed for comparative genomics (presence or absence of a gene) or for transcriptomics (level of gene expression). The microarray described in this study is a combination of both approaches since it enabled us to quantify expression of chromosomal genes and to detect acquired determinants that are expressed. It represents a useful tool to screen for resistance, in particular by overexpression of efflux systems, which is difficult to detect phenotypically and genotypically. In addition, the microarray allowed detection of a new pump, AdeFGH, involved in the antibiotic resistance of A. baumannii. The large panel of antibiotic resistance genes spotted on the microarray, including some that have not yet been detected in A. baumannii, enabled detection of most of the mechanisms that are responsible for the multidrug resistance phenotype of numerous clinical isolates of this species.
Acknowledgments
We thank T. Lambert for providing clinical isolates and helpful discussions; C. Gouyette for synthesis of oligonucleotides; R. Lavenir and I. Iteman, Plateforme Génotypage des Pathogènes et Santé Publique, Institut Pasteur, for technical assistance; the Plateforme Puces à ADN for spotting the biochips; and P. E. Reynolds for reading the manuscript.
This work was supported by Institut Pasteur and Institut de Veille Sanitaire (Saint-Maurice, France).
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartual, S. G., H. Seifert, C. Hippler, M. A. Luzon, H. Wisplinghoff, and F. Rodriguez-Valera. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet, P. J. M., and P. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 4.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, L. A., S. M. Egan, and V. Stewart. 1992. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J. Bacteriol. 174:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corvec, S., L. Poirel, T. Naas, H. Drugeon, and P. Nordmann. 2007. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damier-Piolle, L., S. Magnet, S. Bremont, T. Lambert, and P. Courvalin. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Mar Tomás, M., A. Beceiro, A. Perez, D. Velasco, R. Moure, R. Villanueva, J. Martinez-Beltran, and G. Bou. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depardieu, F., M. L. Foucault, J. Bell, A. Dubouix, M. Guibert, J. P. Lavigne, M. Levast, and P. Courvalin. 2009. New combinations of mutations in VanD-type vancomycin resistant Enterococcus faecium, Enterococcus faecalis, and Enterococcus avium. Antimicrob. Agents Chemother. 53:1952-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depardieu, F., I. Podglajen, R. Leclercq, E. Collatz, and P. Courvalin. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 14.Domenech-Sanchez, A., V. J. Benedi, L. Martinez-Martinez, and S. Alberti. 2006. Evaluation of differential gene expression in susceptible and resistant clinical isolates of Klebsiella pneumoniae by DNA microarray analysis. Clin. Microbiol. Infect. 12:936-940. [DOI] [PubMed] [Google Scholar]
- 15.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. T. Pennella, C. Agasino Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddell, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagnot, S., J. P. Tamby, M. L. Martin-Magniette, F. Bitton, L. Taconnat, S. Balzergue, S. Aubourg, J. P. Renou, A. Lecharny, and V. Brunaud. 2008. CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36:986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein, F. W., A. Labigne-Roussel, G. Gerbaud, C. Carlier, E. Collatz, and P. Courvalin. 1983. Transferable plasmid-mediated antibiotic resistance in Acinetobacter. Plasmid 10:138-147. [DOI] [PubMed] [Google Scholar]
- 19.Gribun, A., Y. Nitzan, I. Pechatnikov, G. Hershkovits, and D. J. Katcoff. 2003. Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47:434-443. [DOI] [PubMed] [Google Scholar]
- 20.Heritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 21.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert, T., G. Gerbaud, and P. Courvalin. 1994. Characterization of transposon Tn1528, which confers amikacin resistance by synthesis of aminoglycoside 3′-O-phosphotransferase type VI. Antimicrob. Agents Chemother. 38:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. F., S. A. Johnson, W. J. Black, D. T. Beattie, J. J. Mekalanos, and S. Falkow. 1992. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J. Bacteriol. 174:970-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 28.Moubareck, C., S. Bremont, M. C. Conroy, P. Courvalin, and T. Lambert. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mugnier, P. D., L. Poirel, and P. Nordmann. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peleg, A. Y., B. A. Potoski, R. Rea, J. Adams, J. Sethi, B. Capitano, S. Husain, E. J. Kwak, S. V. Bhat, and D. L. Paterson. 2007. Acinetobacter baumannii bloodstream infection while receiving tigecycline: a cautionary report. J. Antimicrob. Chemother. 59:128-131. [DOI] [PubMed] [Google Scholar]
- 32.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 33.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 35.Ruzin, A., D. Keeney, and P. A. Bradford. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother. 59:1001-1004. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Shaikh, F., R. P. Spence, K. Levi, H. Y. Ou, Z. Deng, K. J. Towner, and K. Rajakumar. 2009. ATPase genes of diverse multidrug-resistant Acinetobacter baumannii isolates frequently harbour integrated DNA. J. Antimicrob. Chemother. 63:260-264. [DOI] [PubMed] [Google Scholar]
- 38.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su, X. Z., J. Chen, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49:4362-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szybalski, W., and V. Bryson. 1952. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 64:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 42.Tomaras, A. P., M. J. Flagler, C. W. Dorsey, J. A. Gaddy, and L. A. Actis. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398-3409. [DOI] [PubMed] [Google Scholar]
- 43.Turton, J. F., M. E. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 45.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J. M. Claverie, D. Raoult, C. Medigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]