Abstract
There is a need for new microbicidal agents with therapeutic potential due to antibiotic resistance in bacteria and fungi. In this study, the structure-microbicidal activity relationship of amino acid residues 14 to 31 (sequence 14-31) from the N-terminal end, corresponding to the antibacterial α-helix of human lactoferrin (LF), was investigated by downsizing, alanine scanning, and substitution of amino acids. Microbicidal analysis (99% killing) was performed by a microplate assay using Escherichia coli, Staphylococcus aureus, and Candida albicans as test organisms. Starting from the N-terminal end, downsizing of peptide sequence 14-31 showed that the peptide sequence 19-31 (KCFQWQRNMRKVR, HL9) was the optimal length for antimicrobial activity. Furthermore, HL9 bound to lipid A/lipopolysaccharide, as shown by neutralizing endotoxic activity in a Limulus assay. Alanine scanning of peptide sequence 20-31 showed that Cys20, Trp23, Arg28, Lys29, or Arg31 was important for expressing full killing activity, particularly against C. albicans. Substituting the neutral hydrophilic amino acids Gln24 and Asn26 for Lys and Ala (HLopt2), respectively, enhanced microbicidal activity significantly against all test organisms compared to the amino acids natural counterpart, also, in comparison with HL9, HLopt2 had more than 10-fold-stronger fungicidal activity. Furthermore, HLopt2 was less affected by metallic salts than HL9. The microbicidal activity of HLopt2 was slightly reduced only at pH 7.0, as tested in the pH range of 4.5 to 7.5. The results showed that the microbicidal activity of synthetic peptide sequences, based on the antimicrobial α-helix region of LF, can be significantly enhanced by optimizing the length and substitution of neutral amino acids at specific positions, thus suggesting a sequence lead with therapeutic potential.
Steadily increasing antimicrobial drug resistance has become a worldwide problem. For instance, resistance to multiple antibiotics can be found among pathogens such as staphylococci, pneumococci, Pseudomonas species, and extended-spectrum β-lactamase (ESBL)-producing strains of Enterobacteriaceae. Also, resistance to antimycotics, such as the azoles, has climbed significantly. Thus, there is a need for new treatment therapies.
Human lactoferrin (LF), a single-chain iron-binding glycoprotein, is a major protein in milk and mucosal secretions. It is also found in the secondary granulae of neutrophils. It is a multifunctional molecule participating in the innate host defense, expressing both antimicrobial and antiinflammatory activities in vivo (16-18, 34).
Iron depletion and destabilization of the bacterial cell wall are two mechanisms suggested to be involved in the antimicrobial activity of lactoferrin. In Gram-negative bacteria, destabilization could partly be due to its binding capacity to lipid A and porins (3, 4, 10, 25, 37). Binding to lipopolysaccharide (LPS) has been suggested to release LPS from the bacterial membrane. A third mechanism of its antimicrobial activity is the release of microbicidal fragments upon enzymatic hydrolysis of LF (4).
The pepsin-derived bactericidal LF fragment, lactoferricin (Lfcin), consists of residues 1-47 or 49 from the N-terminal end, comprising two antimicrobial domains (residues 1-11 and 18-40) (4, 19, 26). Another recently discovered antimicrobial region of the LF molecule comprises residues 153-183 (32). A smaller peptide sequence of LFcin (residues 21-31), corresponding to a part of the first α-helix of the LF molecule, has been studied in more detail (5). This 11-amino-acid-long peptide exhibited bactericidal activity against Escherichia coli and caused disruption of the outer membrane of E. coli O111 (6, 27). The amphipathic α-helix region also comprises a part of the LPS and glycosaminoglycan binding region of LF, as determined by site-directed mutagenesis at positions 28-34 in the LF molecule (9, 23). The amino acid residues 35-40 constitute a β-sheet region in the native LF molecule (2, 15). Although LFcin may not have a well-developed secondary structure in solution, it may adopt a helical structure when bound to membranes, analogous to other antimicrobial peptides (15).
In the present study, the aim was to investigate the structure-microbicidal activity relationship of peptide fragments based on the antibacterial α-helix (amino acid residues 14-31) of LF, using E. coli, Staphylococcus aureus, and Candida albicans as test organisms. The experimental strategy included downsizing the helical region, followed by alanine scanning and amino acid substitutions.
MATERIALS AND METHODS
Bacterial and fungal strains.
The following bacterial strains and yeast strain were used in the microplate assay: Klebsiella pneumoniae (CCUG 9997), Enterococcus faecalis (ATCC 19433), Staphylococcus epidermidis (CCUG 18000A), Pseudomonas aeruginosa (CCUG 551), Staphylococcus aureus 1800 (CCUG 1800), methicillin-resistant Staphylococcus aureus (MRS3526), E. coli O6K5 and O14, and Candida albicans (ATCC 64549). All microorganisms were cultured in brain heart infusion medium (BHI) on a shaker overnight at 37°C. A volume of the culture was transferred to a new tube with BHI and incubated for two more hours on the shaker. The microorganisms were harvested at the log phase of growth, washed once, and suspended in the broth used for the microplate assay.
LF.
Lactoferrin from human milk was purchased from Sigma (Saint Louis, MO). The level of iron saturation was approximately 7%.
Peptide synthesis: Fmoc solid-phase peptide synthesis of human lactoferrin bactericidal domain (HLBD) mimics.
Peptides were synthesized using a continuous-flow Applied Biosystems Pioneer automated peptide synthesizer and standard 9H-fluoren-9-ylmethoxy carbonyl (Fmoc) protection group protocols. The peptides were synthesized on a 0.1- to 0.2-mmol scale, using the resins 4-hydroxymethylphenoxy-acetic acid polyethylene glycol-polystyrene (PAC-PEG-PS; Applied Biosystems) (0.21 mmol/g) for the peptide acids and Fmoc-5-(4-aminophylline-3, 5-dimethoxyphenoxy) valeric acid-PEG-PS (Fmoc-PAL-PEG-PS) (0.20 mmol/g) for the peptide amides.
Side chains were protected with the piperidine-stable tert-butyl (Ser and Thr), tert-butyl ester (Glu), tert-butyloxycarbonyl (Lys and Trp), triphenylmethyl (Asn, Cys, Gln, and His), and 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Arg) groups.
The Fmoc α-amino protecting group was removed by treatment with 20% piperidine in dimethylformamide (DMF) for 7 min. All couplings proceeded in DMF, with a four-times excess of activated amino acid over the growing peptide chain, using 4 eq of O-benzotriazole-1-yl-1,1,3,3-tetramethyluroniumtetrafluoroborate/diisopropylethylamine (1:2, mol/mol) and 4 eq of amino acid. A sixfold excess of hydroxybenzotriazole was added in the coupling of the cysteine residue.
Peptides were capped by acylation of the amino terminal by treatment with a 0.3 M solution of acetic anhydride in DMF for 10 min.
Final deprotection and cleavage of the peptide from the resin was achieved by treatment with a mixture of 9.25 ml trifluoroacetic acid (TFA), 250 μl water, 250 μl ethanedithiol, and 250 μl triisopropylsilane per gram of peptide resin for 2 h at room temperature. The resin was removed by filtration, and the peptide was precipitated twice by cold diethyl ether, centrifuged, and resuspended in fresh diethyl ether to extract the scavengers and TFA. The peptides were dissolved in water and lyophilized.
A cyclic disulfide-bridged peptide was prepared from crude, unpurified material. Approximately 200 mg of peptide was dissolved in 1 liter of deaerated water. Ammonium hydrogen carbonate was added to adjust the pH to 6 to 7, and the mixture was left with stirring and air contact for approximately 1 day and then lyophilized.
All peptides were purified by reverse-phase high-performance liquid chromatography (HPLC) and eluted with isochratic mixtures of isopropanol and 0.1% TFA. After lyophilization, the peptides were dissolved in distilled water and the purities of the peptides were determined, by HPLC, to be ≥95%. The masses of the peptides were analyzed by electrospray mass spectrometry, using a VG Zab spectrometer sector instrument, and the experimentally determined molecular masses were within 1 mass unit of the theoretical ones for the peptides.
MMC99.
Washed cells were suspended in diluted BHI medium (1/100 of 3.4%, pH 6.7 to 6.9) (BHIdil) or suspended in 1% Bactopeptone (BP) (pH 7.0) (Difco). The absorbance of the suspension was measured by spectrophotometry at 620 nm and diluted to a concentration corresponding to 4 × 106 cells/ml. Peptides serially diluted in BHIdil or BP by twofold steps (unless otherwise stated) were added to the wells of a microtiter plate in duplicate or triplicate (200 μl per well). The bacterial or yeast cell suspensions were added in 10-μl volumes to give a final concentration of approximately 2 × 105 cells/ml. The concentration of the inoculum suspension was always checked by viable counts. The microplate was incubated at 37°C in a humid chamber for 2 h unless stated otherwise. Five microliters was taken from each well and added as a drop onto a blood agar plate and incubated, overnight, at 37°C. The viable count in each area as formed by the drop was recorded, and the concentration of CFU/ml was calculated. A killing activity of between 95 and 99.9% (corresponding to 1 to 50 CFU per drop area) could be calculated. Thus, no growth corresponded to less than 200 CFU/ml, indicating a killing of more than 99.9%. The minimum concentration of a peptide causing ≥99% reduction of the inoculum was defined as MMC99.
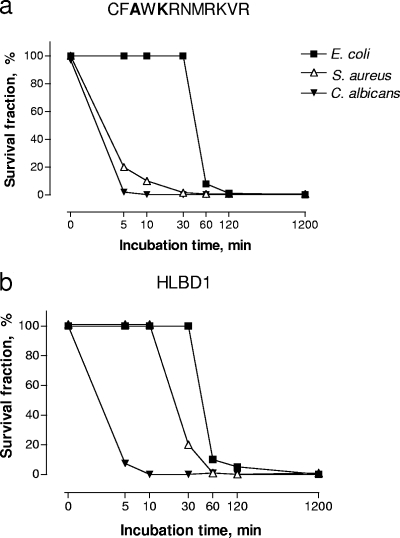
In one experiment analyzing the kinetics of killing (see Fig. 2), serial dilutions of the samples of the microplate assay were performed and cultured.
FIG. 2.
Kinetics of the killing activity of HLBD1 and peptide CFAWKRMRKVR. The peptides were incubated with E. coli strain O6K5, S. aureus strain 1800, and C. albicans in BP for 20 h using 200 μg/ml (65.4 μM) of HLBD1 and 100 μg/ml (62.7 μM) of peptide CFAWKRMRKVR. Samples for viable count were collected directly after mixing and at 5, 10, 30, 60, and 120 min. All samples were run in duplicate.
Limulus assay.
Peptides were analyzed for their capacity to neutralize LPS (O111:B4, O55:B5; Sigma) and lipid A (monophosphoryl and diphosphoryl, L6895, L0774; Sigma) by the Limulus assay (LAL activity) (coamatic endotoxin, chromogenic endpoint; Chromogenix, Sweden). Pyrogen-free Tris buffer (0.05 M, pH 7.3) was used as diluent. Also, two LPS-neutralizing agents, polymyxin B (Sigma) and a peptide sequence of the bactericidal/permeabilization-increasing protein (BPI peptide) (KISGKWAQKRFLKMSGNF; MedProbe), were included in the analysis. The peptides (200 μl) were serially diluted in two- and fivefold steps, and each dilution was added to glass tubes containing LPS/lipid A in Tris buffer (200 μl). The concentration of LPS/lipid A monophosphoryl was 0.3 ng/ml, and that of lipid A diphosphoryl was 0.15 ng/ml (approximately 0.300 to 1.0 EU/ml). The tubes were incubated for 1 h at 37°C. The remaining Limulus activity after the incubation was analyzed according to instructions in the manufacturer's manual.
Statistics.
Chi-square analyses for trends were used for comparing a series of peptides with decreasing/increasing lengths of the peptide sequence (Table 1). The Friedman test with Dunn's multiple comparison test was used for comparison of matched samples of several peptides against one specific peptide (Tables 2 and 3).
TABLE 1.
Microbicidal activities of peptide sequencesa
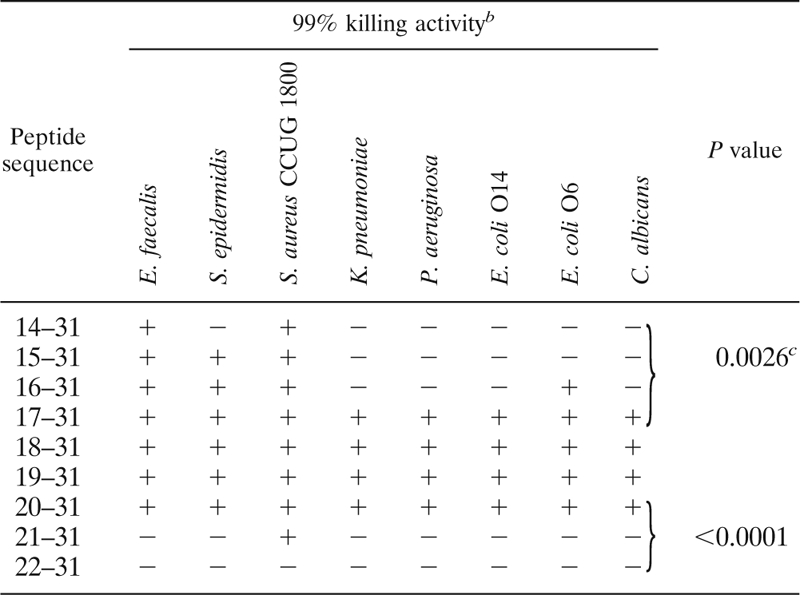 |
Shown are microbicidal activities of peptide sequence 14-31 (QPEATKCFQWQRNMRKVR) and of the series of peptides resulting from the removal of one amino acid at a time from the N-terminal end of peptide sequence 14-31 to the sequence 22-31. Microbial strains were incubated in BHIdil for 2 h at 37°C in the presence of 25 μg/ml of each peptide, except for E. coli O6 and C. albicans, which were incubated with 10 μg/ml. All samples were run in duplicate.
+, at least 99% killed by 25 μg/ml or 10 μg/ml for E. coli O6 and C. albicans; −, <99 % killed.
Values were determined using the chi-square test for trends.
TABLE 2.
Antimicrobial activities of peptide sequencesa
| Peptide sequence | MMC99 μg/ml |
P valueb | |||||
|---|---|---|---|---|---|---|---|
|
E. coli |
S. aureus |
C. albicans |
|||||
| 2 hc | 20 h | 2 h | 20 h | 2 h | 20 h | ||
| 18-31 | 50 | 25 | 50 | 100 | >200 | 50 | |
| 19-31 (HL9) | 25 | 12 | 25 | 50 | 200 | 50 | <0.05b |
| 20-31 | 100 | 50 | >100 | >100 | >200 | 100 | |
Shown are antimicrobial activities of peptide sequences containing amino acid residues 18-31 (TKCFQWQRNMRKVR), 19-31 (KCFQWQRNMRKVR), and 20-31 (CFQWQRNMRKVR) using twofold serial dilutions, starting with 100 or 200 μg/ml in BP, and incubated for 2 and 20 h with E. coli O6K5, S. aureus 1800, or C. albicans. All samples were run in duplicate.
Values were determined using the Friedman test with Dunn's multiple comparison test.
h, hours of incubation.
TABLE 3.
Microbicidal activities of peptidesa
| Amino acid position | Residue replaced by alanine | MMC99 μg/ml |
P valueb | |||||
|---|---|---|---|---|---|---|---|---|
| BHIdil |
BP |
|||||||
| E. coli | S. aureus | C. albicans | E. coli | S. aureus | C. albicans | |||
| 20 | C | >25 | 14 | 12 | >100 | >100 | >100 | |
| 21 | F | 12 | 7 | 12 | ND | ND | ND | |
| 22 | Q | 6 | 3.5 | 6 | 50 | 100 | 25 | |
| 23 | W | 25 | 14 | 12 | >100 | >100 | >100 | |
| 24 | Q | 12 | 3.5 | 12 | 50 | 100 | 50 | |
| 25 | R | 12 | 7 | 12 | ND | ND | ND | |
| 26 | N | 6 | 3.5 | 6 | 25 | 50 | 25 | <0.05 |
| 27 | M | 12 | 3.5 | 25 | 50 | >100 | 50 | |
| 28 | R | 25 | 7 | 25 | >100 | >100 | >100 | |
| 29 | K | 12 | 3.5 | 25 | 100 | >100 | >100 | |
| 30 | V | 12 | 3.5 | 25 | 50 | >100 | 50 | |
| 31 | R | 12 | 7 | 25 | >100 | >100 | >100 | |
| None | None | 12 | 7 | 12 | 100 | >100 | 50 | |
Shown are microbicidal activities of peptides derived from amino acid residues 20-31 (CFQWQRNMRKVR) by alanine scan. Peptides were incubated with C. albicans, E. coli O6K5, and methicillin-resistant S. aureus MRS3525 in BHIdil or BP. The peptide concentrations were serially diluted in twofold steps, starting with 25 or 28 μg/ml (S. aureus) using BHIdil and 100 μg/ml using BP as growth medium. All samples were run in duplicate. ND, not determined.
Values were determined using the Friedman test with Dunn's multiple comparison test. Each Ala-modified peptide was compared with the natural peptide sequence.
In the microbicidal microplate assay, peptide solutions serially diluted twofold and differing by twofold titer steps (a fourfold difference in MMC99) were significantly different.
RESULTS
Optimal amino acid length for microbicidal activity.
In order to find an optimal peptide length within the peptide sequence 14-31 with regard to antimicrobial activity, constituting the complete first α-helix in the LF molecule, amino acid residues were removed one by one, starting from the N-terminal end. The reason for keeping the C terminus intact was that the latter third of the peptide sequence comprised the main body of positively charged amino acid residues with one Arg at the C-terminal end (position 31). The antimicrobial activity of the series of peptides were screened against Enterococcus faecalis, Staphylococcus epidermidis, S. aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, E. coli O14, E. coli O6K5, and C. albicans (Table 1).
The most active peptides with regard to the number of microbial species or strains that were killed to 99% were the ones corresponding to residues 17-31, 18-31, 19-31, and 20-31 (Table 1). The trends of increasing antimicrobial activity from sequence 14-31 to 17-31 and decreasing from sequence 20-31 to 22-31 were statistically significant (chi-square test for trends) (Table 1).
Next, three of the four optimal peptide lengths were analyzed by twofold serial dilutions in the more concentrated medium (BP) (Table 2). The most active peptide in BP was sequence 19-31 (HL9), since this peptide, in contrast to sequence 18-31, differed significantly from sequence 20-31. The killing activity of the peptides increased with time for C. albicans (Table 2).
It should be noted that, in general, replacing BHIdil with BP reduced the antimicrobial activity of the peptides to various degrees against E. coli, S. aureus, and C. albicans (Tables 1 and 2).
The importance of single amino acids for microbicidal activity. (i) Alanine scanning.
One amino acid was replaced, at each position, in the peptide sequence 20-31 by Ala to determine the effect of each individual amino acid for antimicrobial activity (Table 3). The peptides were analyzed against E. coli, S. aureus, and C. albicans.
A significant reducing effect on the killing activity against E. coli, using BHIdil, was obtained with an Ala substitution at position 20, where a Cys residue was replaced (Table 3).
In BP, a significantly reduced antimicrobial activity against C. albicans was seen, not only for the Cys replacement, but also for the Trp replacement at position 23, the replacement of Arg residues at positions 28-31 and the Lys replacement at position 29. A similar pattern was observed for E. coli, although not significantly different compared to the original peptide.
Changing the hydrophilic Gln (position 22 or 24) or Asn (position 26) to a slightly hydrophobic Ala resulted in significantly increased overall antimicrobial activity of the peptide with Ala at position 26 (P < 0.05; the Friedman test with Dunn's multiple posttest). MMC99 values for the peptide with Ala at position 22 were reduced compared to those of the control peptide for all microorganisms and media, although not significantly.
A dramatic difference in the concentration of peptide needed for 99% killing between the two growth media was seen for S. aureus (up to 30 times or more), while the difference was less for E. coli and C. albicans (four- to eightfold).
(ii) Substitution by hydrophobic or charged amino acids.
In all the following amino acid replacement experiments, the microbicidal assays were run in both BHIdil and BP. There was, however, no significant difference in antimicrobial activity between the natural peptide sequence 20-31 and the peptides with the substitutions listed in Table 4 for experiments performed in BHIdil, except for one peptide. Thus, all results described below refer to assays performed in BP.
TABLE 4.
Microbicidal activity of modified amino acid sequence 20-31 (CFQWQRNMRKVR)a
| Codeb | MMC99 μg/ml |
||
|---|---|---|---|
| E. coli | S. aureus | C. albicans | |
| Positively charged amino acids | |||
| Orn or K substitution | |||
| CFQWOrnRNMRKVR | 25 | 50 | 25 |
| CFQWKRNMRKVR | 25 | 25 | 25 |
| CFQWQROrnMRKVR | 25 | 50 | 25 |
| Hydrophobic amino acids | |||
| Nle substitution | |||
| CFQWNleRNMRKVR | 50 | 50 | 25 |
| CFQWQRNleMRKVR | 50 | 50 | 50 |
| Two or more substitutions | |||
| CFQWKRAMRKVR (HLopt2) | 12 | 12 | 12 |
| CFAWKRNMRKVR | 50 | 25 | 25 |
| CFAWQRAMRKVR | 25 | 50 | 25 |
| CFALKKAMKKVR | 25 | 50 | 25 |
| Native sequence | |||
| 20CFQWQRNMRKVR31 | 100 | >100 | 50 |
The peptides were incubated with E. coli O6K5, S. aureus 1800 and C. albicans in BP. The peptides were diluted in 2-fold steps, starting at 100 μg/ml. All samples were run in duplicate. Orn, ornithine, an amino acid not coded for by DNA and a metabolic intermediate; Nle, norleucine, a nonnatural synthetic amino acid.
Substitutions are shown in bold type.
As shown in the Ala scan (Table 3), the replacement of Asn by Ala at position 26 in the peptide sequence 20-31 was shown to increase the antimicrobial activity. Thus, whether any hydrophobic or charged amino acid would increase the killing activity in general when replacing Asn26 or another neutral hydrophilic amino acid residue in the peptide, such as Gln24, was investigated. Asn or Gln was replaced by the nonnatural hydrophobic Nle (norleucine) or positively charged Lys or Orn (ornithine, a natural amino acid not present in proteins/peptides) (Table 4).
Substitution by Orn or K increased the antimicrobial activity against E. coli and S. aureus by a four- to eightfold decrease in the MMC99 value. The Nle substitutions enhanced the killing activity significantly against S. aureus only (Table 4).
(iii) Substitutions by two or more amino acids (Lys, Ala, or Leu).
Increasing the number of positively charged amino acids by substitution at position 24 (Gln) increased the microbicidal activity. Combining this replacement with the substitution of Asn at position 26 by Ala (Table 3) gave rise to a peptide (CFQWKRAMRKVR, HLopt2) with significantly increased microbicidal activities for all three microorganisms (Table 4). Thus, the changes in the peptide increased the activity in BP at least fourfold against all microbial species. Furthermore, no significant difference in activity was recorded when comparing HLopt2 in BP with that in BHIdil (data not shown). Moving the Ala substitution from N26 to Q22 affected the antimicrobial activity negatively compared with that of the former peptide sequence (Table 4).
The two separate Ala substitutions that resulted in significantly enhanced microbicidal activity (Table 3) were introduced into the same peptide. However, the activity was not different from that observed by the Ala substitution at N26 (Tables 3 and 4).
In addition to the two Ala substitutions, introducing Lys at Gln24 and changing the Arg residues at positions 25 and 28 to Lys also gave rise to a peptide that was significantly better than the natural one against E. coli and S. aureus.
The only significant enhancing effect that any substitution (one or several) had on the antimicrobial activity compared with the natural sequence (20-31), as analyzed in BHIdil, was obtained with Orn in position 24 against C. albicans (not shown).
Amino acid substitution in HLBD1.
Based on the Ala substitutions in the peptide sequence 20-31 at Q22 or N26, or to Lys at Q24 (Tables 3 and 4), the same changes were introduced in the larger lactoferricin-like HLBD1 (Fig. 1) in order to find out whether enhanced antimicrobial activities also would be expressed in the double-sized molecule compared to the native sequence (Table 5).
FIG. 1.
Amino acid sequence of HLBD1. A synthesized lactoferricin-like cyclic peptide, HLBD1, was based on the first α/β region of the LF molecule and comprised amino acid residues 16-40. The full α/β region of the LF molecule comprises the α-helix with amino acid residues 14-31 and a following β-sheet with amino acid residues 35-40. The bracket denotes the cyclization by a disulfide bond.
TABLE 5.
Microbicidal activity of modified HLBD1 peptides
| Amino acid residue substitution | MMC99 μg/ml |
||
|---|---|---|---|
| E. coli | S. aureus | C. albicans | |
| Modified HLBD1 | |||
| Q22→A | >100 | 100 | >200 |
| N26→A | 100 | 50 | 37 |
| Q24→K | 12 | 25 | 50 |
| HLBD1 | 50 | 50 | 200 |
Peptides were incubated with E. coli O6K5, S. aureus 1800, or C. albicans in BP. The peptides were diluted in twofold steps, starting at 100 or 200 μg/ml (C. albicans). Each sample was run in duplicate, and the experiment was repeated once.
The change of N26 to Ala improved the activity only against C. albicans, wherein a significant increase of fungicidal activity was obtained. The increased charge of HLBD1, by changing Q24 to Lys, improved the microbicidal activity significantly against both E. coli and C. albicans compared to that of the natural sequence.
Kinetics of the microbicidal activity.
HLBD1 and the short peptide CFAWKRNMRKVR (Table 4) with corresponding MMC99 values regarding E. coli and S. aureus were investigated with respect to killing kinetics.
The amount of time needed for killing 90% of E. coli bacteria was 1 h, irrespective of the peptide used, when analyzed at similar concentrations (63 to 65 μM) (Fig. 2). While 80% of S. aureus bacteria were killed within 30 min by HLBD1 (Fig. 2b), the smaller peptide killed 90% within 10 min (Fig. 2a). More than 90% of C. albicans bacteria were killed within 5 min by HLBD1 and 99% by the smaller peptide within the same time span.
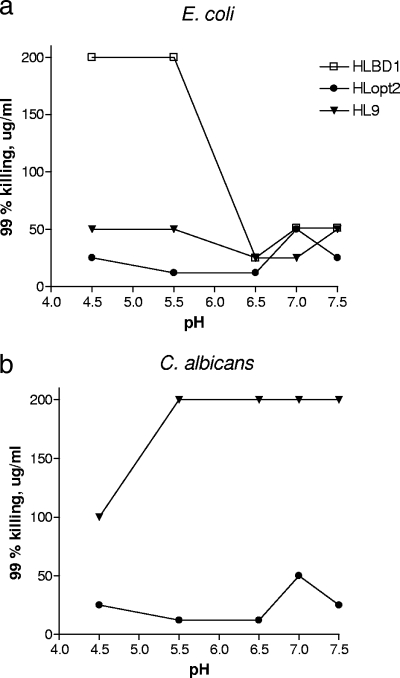
Effects of pH and metallic salts on microbicidal activity. (i) pH.
The effects of pH on the antimicrobial activities of HL9, HLopt2, and HLBD1 were analyzed within the pH range of 4.5 to 7.5 using E. coli and C. albicans (Fig. 3).
FIG. 3.
Effects of various pH levels on the microbicidal activities of HLopt2, HL9, and HLBD1. All three peptides were analyzed for 99% killing (MMC99) of E. coli (a), and HLopt2 and HL9 were analyzed for 99% killing of C. albicans (b). The microbicidal assay was run at various pH levels by adjusting the pH of BP. Twofold serial dilutions were made of each peptide starting at 100 μg/ml of HLopt2 and HL9 and at 200 μg/ml for HLBD1. All samples were run in triplicate.
HLopt2 was significantly more effective at pHs of 5.5 to 6.5 than at pH 7.0 against both test organisms (fourfold difference in MMC99) (Fig. 3a and b). For HL9, the microbicidal activity appeared to increase at pHs of 6.5 to 7.0, regarding E. coli, while an opposite effect was observed for C. albicans at a pH of ≥5.5. HLBD1 was significantly less active at pHs lower than 6.5. The MMC99 value increased eightfold at pH 5.5 compared to that at pH 6.5.
(ii) NaCl and other metallic salts.
At 100 μg per ml, both HL9 and HLopt2 expressed 99% killing activity against E. coli and C. albicans, up to 50 mM NaCl (Table 6). Doubling the salt concentration resulted in reduced killing activity. In the presence of other salts (K+, Mg2+, Ca2+), HLopt2 was consistently more active at higher concentrations of the metal ions than HL9, except against C. albicans in the presence of K+, where both peptides were equally active. Overall, CaCl2 affected the activity of the peptides the most. HLBD1, which was analyzed against E. coli at the same concentration as the other two peptides, was reduced in its activity by all salts to below that of 99% killing (not shown). No salt concentration changed the viability of the microbial inocula at 2 h of incubation in the absence of peptides.
TABLE 6.
Effects of metallic salts on the antimicrobial activities of HL9 and HLopt2 against E. coli O6K5 and C. albicans 1800a
| Salinity or metallic salt | mM | 99% killing activityb |
|||
|---|---|---|---|---|---|
|
E. coli |
C. albicans |
||||
| HL9 | HLopt2 | HL9 | HLopt2 | ||
| Salinity | |||||
| NaCl | 25 | + | + | + | + |
| NaCl | 50 | + | + | + | + |
| NaCl | 100 | − | − | − | − |
| Metallic salts | |||||
| KCl | 25 | + | + | + | + |
| KCl | 50 | + | + | + | + |
| KCl | 100 | − | + | − | − |
| MgCl2 | 1 | + | + | + | + |
| MgCl2 | 2.5 | − | + | + | + |
| MgCl2 | 5.0 | − | − | − | + |
| CaCl2 | 1 | − | + | − | + |
| CaCl2 | 2.5 | − | − | − | − |
| CaCl2 | 5.0 | − | − | − | − |
Both peptides were analyzed at a concentration of 100 μg/ml. The salts were added to BP. All samples were run in triplicate.
+, ≥99% killing of the inoculum; −, <99% killing.
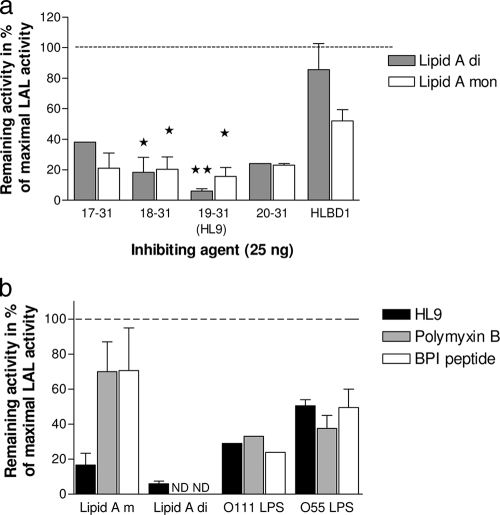
Neutralization of the Limulus activity of LPS and lipid A by peptide sequences 17-31, 18-31, 19-31 (HL9), and 20-31.
Since one mechanism for the killing of E. coli has been suggested to be mediated via the binding of Lfcin to LPS in the outer membrane, this binding was studied by a neutralization test using the Limulus assay. The small peptides, spanning from 20-31 to 17-31, all neutralized the Limulus activity of lipid A by 60 to 90% at a concentration of 25 ng/ml (Fig. 4a). Using the longer peptide, HLBD1, no significant neutralizing effect was obtained for the diphosphoryl lipid A, while approximately 50% of the monophosphoryl lipid A was neutralized. A comparison of the neutralizing activity of sequence 18-31 or 19-31 with that of HLBD1 showed that the smaller peptides had significantly stronger inhibitory activities.
FIG. 4.
Neutralization of the Limulus activities of lipid A (a) and LPS (b) by the peptide sequences containing amino acid residues 20-31 to 17-31. (a) The peptides (25 ng/ml) of amino acid residues 20-31 to 17-31 were incubated with diphosphoryl and monophosphoryl lipid A, and the remaining Limulus activity was analyzed (expressed as percentage of maximal LAL activity). The experiment was repeated three times, except for peptide sequences 17-31 and 20-31 (single run for diphosphoryl lipid A; repeated once for monophosphoryl lipid A). *, P > 0.05; **, P > 0.01 (Student's t test). In panel b, the remaining activity of LPS O111 and O55 after incubation with 5 μg/ml of peptide sequence 19-31 (HL9), polymyxin B, and BPI peptide is shown. The experiment was repeated once except for O111 LPS (single run).
The most active peptide, HL9, was further compared with the known endotoxin-neutralizing agents polymyxin B and BPI peptide using LPS (Fig. 4b). The two LPSs were blocked in their Limulus activity to 71% (O111 LPS) and 50% (O55 LPS) by HL9, a neutralizing effect comparable with those of polymyxin B (76 and 51%) and BPI (58 and 61%).
DISCUSSION
The complete antimicrobial mechanism of action of LF and peptides derived from it is unknown, although considerable efforts toward understanding the relationship between structure and antimicrobial activity from studies of LF and LF peptide fragments have shed light on parts of their functions. We have previously shown that, in addition to LF, HLBD1 and a similar peptide (amino acid residues 18-40), based on the first α-helix-β-sheet region of the LF molecule, are efficient in reducing infection and inflammation in vivo in experimental E. coli-induced urinary tract infections and colitis (16, 17).
Peptides other than pepsin-derived lactoferricin may occur locally at mucosal surfaces or in tissues where various proteases may digest LF differently to as-yet-unknown peptide fragments. The antimicrobially active region in lactoferricin also included the first cationic domain of human LF comprising amino acid residues 1-11. This short 11-amino-acid-long peptide and the recently reported peptide comprising residues 153-183 of the LF molecule also have been shown to express antimicrobial activities (11, 26, 30, 32).
It is not clear whether the intact LF protein or peptides derived from it by proteolytic cleavage have the most active LF-induced antimicrobial activity in vivo.
In the present study, the peptide fragment 14-31, corresponding to the first α-helix of the LF molecule, was downsized by removing one amino acid at a time from the N-terminal end. A 13-amino-acid-long peptide (19-31, HL9) was found to be the most active with respect to the overall antimicrobial effects. Downsizing beyond residues 20-31 resulted in a considerable loss of activity. These results suggested that Lys19 and Cys20 at the N terminus were important for optimal killing activity of the natural sequence.
It is interesting to note that an amino acid sequence (Met at position 27 changed to Ile) almost identical to our antimicrobial peptide sequence 21-31, which was significantly weaker than HL9 or peptide sequence 20-31, was reported to adopt a T-shaped arrangement of a hydrophobic core composed of the N-terminal end and two clusters of basic residues perpendicular to the core (20) when interacting with LPS. The additional Cys-Lys at the N-terminal end in HL9 may facilitate or stabilize a conformation of the amphipathic peptide, which relies on three clusters of charged amino acids. Another possible conformation of HL9 includes a central β-strand with charged amino acids at the ends, as was proposed for a corresponding sequence of the 11-mer peptide (residues 20-30) of bovine LF (12). Irrespective of conformational shape, the chain size of HL9 appears to be optimal for conformation plasticity while still retaining the ability to form a defined tertiary structure in the presence of bacterial membranes. It should be noted that random conformations of the peptides are present in solution. Additionally, the presence of Lys in HL9 fits the hypothesis that positively charged amino acids add to antimicrobial activity.
The alanine scan of the shortest peptide sequence (20-31) with retained killing activity (Table 1) showed that single-amino-acid substitutions did not result in total abolishment of microbicidal activity. However, considerable reduction in antifungal activity was observed when Ala was substituted for Cys, Trp, or the charged amino acid clustered at the C-terminal end of the peptide (positions 28, 29, and 31) with regard to C. albicans, a pattern that fits the T-shaped conformational model, since Trp was found to be the central residue of the hydrophobic core and the additional Cys20 could possibly enhance the hydrophobic core (20). In general, Trp is important for its ability to cluster hydrophobic contacts around its side chain (8, 21). The importance of Trp and Arg residues for antimicrobial activity has also been shown for bovine Lfcin, residues 20-29 of bovine LF, as well as other synthetic antimicrobial peptides (7, 13, 29, 33, 36). The replacement of any of the charged amino acids also corresponded with weakened charged clusters of the upper part of the T shape. Unlike in bacteria, the cytoplasmic membranes of yeast cells contain the unique lipid ergosterol, and the cell wall is composed of chitin and β-glucan with an outer layer of mainly phosphate-containing mannoproteins. Thus, it may not be unexpected to find various patterns of antimicrobial activity.
For E. coli killing activity, the most important residue was Cys, since replacement by Ala reduced the antibacterial activity significantly.
Modifications of peptide sequence 20-31 by increasing the number of positively charged or hydrophobic amino acids enhanced the microbicidal activity in general. Even nonnatural building blocks of peptides, such as the hydrophobic Nle or basic Orn, enhanced the antimicrobial activity. Possibly, such amino acids could increase the resistance to degradation of the peptide. Thus, substituting neutral hydrophilic amino acids, such as Asp and Gln, by either single or double substitution (HLopt2) improved the microbicidal activity four- to eightfold in BP, resulting in activities close to those obtained in diluted BHI. Furthermore, comparing the modified HLopt2 with its corresponding unmodified form or the longer HL9 showed that the fungicidal activity was enhanced more than four- and 10-fold, respectively. Additionally, single-amino-acid substitutions in the double-sized HLBD1 revealed that exchange of Gln24 to Lys enhanced the killing activity significantly against E. coli and C. albicans compared to those of the unmodified peptide (Table 5) and similar levels of HLopt2 for E. coli and S. aureus (Table 4).
Regarding the effects of salts, it is known that LF peptides, in accordance with α and β defensins, are sensitive to high-salt environments (14, 28). Thus, an ionic strength of 100 mM NaCl weakened the lethal activity of our peptides. The strongest influence on the microbicidal activity was observed with Ca2+. Clearly, the ionic environment affects the microbicidal activity in a complex way, also including effects on the target microorganism. Regarding the pH dependence, the lethal activity of HLopt2 was unaffected at pHs of 4.5 to 6.5, while HL9 showed no significant changes within the tested pH range (pHs of 4.5 to 7.5). In contrast, the lethal effect of the longer cyclic peptide, HLBD1, was impaired at pHs below 6.5 (analyzed only for E. coli). Thus, the pattern of pH dependence appeared to be specific for each peptide.
By neutralization of the endotoxic activity measured by the Limulus assay, we showed that the short peptides (17-20-31) bound to both lipid A and LPS. The effect may be exerted by binding partly to different binding sites on lipid A and LPS, respectively. Both phosphate and carboxyl groups are available on the LPS molecule, but only phosphates on the lipid A molecule. The neutralization of endotoxin activity in vitro by the Limulus assay and by suppression of endotoxin-induced tumor necrosis factor alpha secretion in macrophages and in vivo by lethal endotoxin challenge in mice has been shown with a peptide corresponding to residues 1-33 of LF (38). The first six residues at the N terminus of the 33-mer peptide appeared critical for the antiendotoxin activity. Our results, however, indicated that the shorter peptides, comprising 12 to 15 amino acids and lacking the six N-terminal residues, retained endotoxin-neutralizing activity and were comparable with the well-known antiendotoxic agents polymyxin B and BPI peptide (24). Our results are in agreement with those of others, wherein the peptides containing residues 21-31 and another peptide very similar to residues 21-31 were found to bind to LPS (5, 12, 13, 20). The strongest interaction was obtained with HL9.
The differences between E. coli, S. aureus, and C. albicans, with respect to the kinetics of killing by short peptide, most probably reflect the difference in the construction of their cell envelopes. In two reports using synthetic peptides based on the residues 21-31 or 16-40, it was suggested that binding to LPS may be an initial event leading to penetration of the outer membranes, followed by interaction with the cytoplasmic membranes, of E. coli (1, 6). The peptide corresponding to residues 18-40 in LF was shown to interact with the cytoplasmic membranes of E. coli and cause losses of the bacterial transmembrane electrical potential and the pH gradient (1, 35). Using E. coli O111, Chapple et al. observed a time-dependent collapse of the membrane potential and membrane integrity upon exposure to peptide sequence 21-31. In agreement with our study, a lag phase of 1 to 2 h, depending on the peptide concentration, was needed for the killing of E. coli (5, 6). Our observation of binding of the peptides to LPS and lipid A supports a model in which the first step includes the interaction with LPS. This, together with the suggestion that the peptide folds and self-assembles at the outer membrane surface, indicates a two-stage process of the antibacterial activity against E. coli and could explain the longer time needed for the peptide to exert its activity against this bacterium compared to that against S. aureus and C. albicans (5).
With regard to the kinetics of the microbicidal activity towards S. aureus, the smaller size of the peptide appeared to increase the rate of killing.
The rapid killing of C. albicans by both peptides may indicate that there is a somewhat different mode of action and that the cell wall is easier to pass through. A synthetic fragment of the first cationic domain in the LF molecule from the N-terminal end comprising amino acid residues 1-11 was suggested to interact with structural elements of the plasma membrane of C. albicans and be taken up in an energy-dependent way. The following induced release of ATP, combined with the activity of the peptide, was essential for the candidacidal activity (22). A peptide corresponding to amino acid residues 17-26 from bovine LF has also been shown to enhance the candidacidal activity of antifungal drugs by inducing the ATP efflux from C. albicans (31). In a following paper, it will be shown that mitochondrial membranes are affected in the killing process.
In conclusion, the residue fragment 19-31 comprising more than half of the helix in the LF molecule was the most active microbicidal fragment against E. coli, S. aureus, and C. albicans. The Cys at the N-terminal end (position 20), a hydrophobic residue at position 23, and the charged amino acids at positions 28 and 31 were important for antimicrobial activity. However, replacement of the polar amino acid in the shorter peptide sequence 20-31 at positions Q24 by Lys and N26 by Ala not only significantly reduced the MMC99 values in comparison to the natural sequence but led to the most active peptide of all the peptides analyzed.
Acknowledgments
The excellent technical assistance of Lotta Arnäs is gratefully acknowledged.
This study was financially supported by A+ Science Invest AB, Gothenburg, Sweden.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Aguilera, O., H. Ostolaza, L. M. Quiros, and J. F. Fierro. 1999. Permeabilizing action of an antimicrobial lactoferricin-derived peptide on bacterial and artificial membranes. FEBS Lett. 462:273-277. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. F., H. M. Baker, G. E. Norris, D. W. Rice, and E. N. Baker. 1989. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 Å resolution. J. Mol. Biol. 209:711-734. [DOI] [PubMed] [Google Scholar]
- 3.Appelmelk, B. J., Y. Q. An, M. Geerts, B. G. Thijs, H. A. de Boer, D. M. MacLaren, J. de Graaff, and J. H. Nuijens. 1994. Lactoferrin is a lipid A-binding protein. Infect. Immun. 62:2628-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamy, W., M. Takase, K. Yamauchi, H. Wakabayashi, K. Kawase, and M. Tomita. 1992. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1121:130-136. [DOI] [PubMed] [Google Scholar]
- 5.Chapple, D. S., R. Hussain, C. L. Joannou, R. E. Hancock, E. Odell, R. W. Evans, and G. Siligardi. 2004. Structure and association of human lactoferrin peptides with Escherichia coli lipopolysaccharide. Antimicrob. Agents Chemother. 48:2190-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapple, D. S., C. L. Joannou, D. J. Mason, J. K. Shergill, E. W. Odell, V. Gant, and R. W. Evans. 1998. A helical region on human lactoferrin—its role in antibacterial pathogenesis. Adv. Exp. Med. Biol. 443:215-220. [PubMed] [Google Scholar]
- 7.Chen, P. W., C. L. Shyu, and F. C. Mao. 2003. Antibacterial activity of short hydrophobic and basic-rich peptides. Am. J. Vet. Res. 64:1088-1092. [DOI] [PubMed] [Google Scholar]
- 8.Crowhurst, K. A., and J. D. Forman-Kay. 2003. Aromatic and methyl NOEs highlight hydrophobic clustering in the unfolded state of an SH3 domain. Biochemistry 42:8687-8695. [DOI] [PubMed] [Google Scholar]
- 9.Elass-Rochard, E., A. Roseanu, D. Legrand, M. Trif, V. Salmon, C. Motas, J. Montreuil, and G. Spik. 1995. Lactoferrin-lipopolysaccharide interaction: involvement of the 28-34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli 055B5 lipopolysaccharide. Biochem. J. 312:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison, R. T., III, T. J. Giehl, and F. M. LaForce. 1988. Damage of the outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect. Immun. 56:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faber, C., H. P. Stallmann, D. M. Lyaruu, U. Joosten, C. von Eiff, A. van Nieuw Amerongen, and P. I. Wuisman. 2005. Comparable efficacies of the antimicrobial peptide human lactoferrin 1-11 and gentamicin in a chronic methicillin-resistant Staphylococcus aureus osteomyelitis model. Antimicrob. Agents Chemother. 49:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnaud, S., A. Patel, E. W. Odell, and R. W. Evans. 2004. Variation in antimicrobial activity of lactoferricin-derived peptides explained by structure modelling. FEMS Microbiol. Lett. 238:221-226. [DOI] [PubMed] [Google Scholar]
- 13.Farnaud, S., C. Spiller, L. C. Moriarty, A. Patel, V. Gant, E. W. Odell, and R. W. Evans. 2004. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol. Lett. 233:193-199. [DOI] [PubMed] [Google Scholar]
- 14.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Håversen, L., B. G. Ohlsson, M. Hahn-Zoric, L. A. Hanson, and I. Mattsby-Baltzer. 2002. Lactoferrin down-regulates the LPS-induced cytokine production in monocytic cells via NF-kappa B. Cell Immunol. 220:83-95. [DOI] [PubMed] [Google Scholar]
- 17.Håversen, L. A., L. Baltzer, G. Dolphin, L. A. Hanson, and I. Mattsby-Baltzer. 2003. Anti-inflammatory activities of human lactoferrin in acute dextran sulphate-induced colitis in mice. Scand. J. Immunol. 57:2-10. [DOI] [PubMed] [Google Scholar]
- 18.Håversen, L. A., I. Engberg, L. Baltzer, G. Dolphin, L. A. Hanson, and I. Mattsby-Baltzer. 2000. Human lactoferrin and peptides derived from a surface-exposed helical region reduce experimental Escherichia coli urinary tract infection in mice. Infect. Immun. 68:5816-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter, H. N., A. R. Demcoe, H. Jenssen, T. J. Gutteberg, and H. J. Vogel. 2005. Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob. Agents Chemother. 49:3387-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Japelj, B., P. Pristovsek, A. Majerle, and R. Jerala. 2005. Structural origin of endotoxin neutralization and antimicrobial activity of a lactoferrin-based peptide. J. Biol. Chem. 280:16955-16961. [DOI] [PubMed] [Google Scholar]
- 21.Klein-Seetharaman, J., M. Oikawa, S. B. Grimshaw, J. Wirmer, E. Duchardt, T. Ueda, T. Imoto, L. J. Smith, C. M. Dobson, and H. Schwalbe. 2002. Long-range interactions within a nonnative protein. Science 295:1719-1722. [DOI] [PubMed] [Google Scholar]
- 22.Lupetti, A., A. Paulusma-Annema, M. M. Welling, S. Senesi, J. T. van Dissel, and P. H. Nibbering. 2000. Candidacidal activities of human lactoferrin peptides derived from the N terminus. Antimicrob. Agents Chemother. 44:3257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann, D. M., E. Romm, and M. Migliorini. 1994. Delineation of the glycosaminoglycan-binding site in the human inflammatory response protein lactoferrin. J. Biol. Chem. 269:23661-23667. [PubMed] [Google Scholar]
- 24.Morrison, D. C., and D. M. Jacobs. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813-818. [DOI] [PubMed] [Google Scholar]
- 25.Naidu, S. S., U. Svensson, A. R. Kishore, and A. S. Naidu. 1993. Relationship between antibacterial activity and porin binding of lactoferrin in Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:240-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nibbering, P. H., E. Ravensbergen, M. M. Welling, L. A. van Berkel, P. H. van Berkel, E. K. Pauwels, and J. H. Nuijens. 2001. Human lactoferrin and peptides derived from its N terminus are highly effective against infections with antibiotic-resistant bacteria. Infect. Immun. 69:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odell, E. W., R. Sarra, M. Foxworthy, D. S. Chapple, and R. W. Evans. 1996. Antibacterial activity of peptides homologous to a loop region in human lactoferrin. FEBS Lett. 382:175-178. [DOI] [PubMed] [Google Scholar]
- 28.Porter, E. M., E. van Dam, E. V. Valore, and T. Ganz. 1997. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect. Immun. 65:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schibli, D. J., R. F. Epand, H. J. Vogel, and R. M. Epand. 2002. Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem. Cell Biol. 80:667-677. [DOI] [PubMed] [Google Scholar]
- 30.Stallmann, H. P., C. Faber, A. L. Bronckers, J. M. de Blieck-Hogervorst, C. P. Brouwer, A. V. Amerongen, and P. I. Wuisman. 2005. Histatin and lactoferrin derived peptides: Antimicrobial properties and effects on mammalian cells. Peptides 26:2355-2359. [DOI] [PubMed] [Google Scholar]
- 31.Tanida, T., T. Okamoto, E. Ueta, T. Yamamoto, and T. Osaki. 2006. Antimicrobial peptides enhance the candidacidal activity of antifungal drugs by promoting the efflux of ATP from Candida cells. J. Antimicrob. Chemother. 57:94-103. [DOI] [PubMed] [Google Scholar]
- 32.Viejo-Diaz, M., M. T. Andres, J. Perez-Gil, M. Sanchez, and J. F. Fierro. 2003. Potassium efflux induced by a new lactoferrin-derived peptide mimicking the effect of native human lactoferrin on the bacterial cytoplasmic membrane. Biochemistry (Moscow) 68:217-227. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, H. J., D. J. Schibli, W. Jing, E. M. Lohmeier-Vogel, R. F. Epand, and R. M. Epand. 2002. Towards a structure-function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides. Biochem. Cell Biol. 80:49-63. [DOI] [PubMed] [Google Scholar]
- 34.Vorland, L. H. 1999. Lactoferrin: a multifunctional glycoprotein. APMIS 107:971-981. [DOI] [PubMed] [Google Scholar]
- 35.Vorland, L. H., H. Ulvatne, J. Andersen, H. H. Haukland, O. Rekdal, J. S. Svendsen, and T. J. Gutteberg. 1999. Antibacterial effects of lactoferricin B. Scand. J. Infect. Dis. 31:179-184. [DOI] [PubMed] [Google Scholar]
- 36.Wessolowski, A., M. Bienert, and M. Dathe. 2004. Antimicrobial activity of arginine- and tryptophan-rich hexapeptides: the effects of aromatic clusters, D-amino acid substitution and cyclization. J. Pept. Res. 64:159-169. [DOI] [PubMed] [Google Scholar]
- 37.Yamauchi, K., M. Tomita, T. J. Giehl, and R. T. Ellison, 3rd. 1993. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect. Immun. 61:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, G. H., D. M. Mann, and C. M. Tsai. 1999. Neutralization of endotoxin in vitro and in vivo by a human lactoferrin-derived peptide. Infect. Immun. 67:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]