Abstract
Salmonella genomic island 2 (SGI2) is an independently derived genomic island related to SGI1 with the integron in a different position. The integron in SGI2 was found to include an additional 2.1 kb derived from the tni module of Tn5058, Tn502, or Tn512 that was not detected previously. Independent evolution of the backbone was confirmed with 21 single base differences found in over 11.5 kb, representing 40% of the 27.4-kb SGI2 backbone.
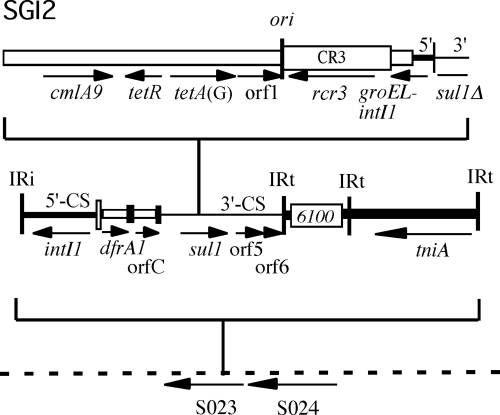
Antibiotic resistance genes carried in complex class 1 integrons that are located in Salmonella genomic island 1 (SGI1) play an important role in resistance development in Salmonella enterica (1-4, 10, 13, 14, 18), and SGI1 or variants of it are found in many different serovars (2, 6, 7, 11-13). We recently reported the structure and partial sequence of a second genomic island, SGI2, carrying multiple antibiotic resistance genes in a complex In4-type class 1 integron, which was found in S. enterica serovar Emek (9). While made up of components similar to those of SGI1, SGI2 clearly has an independent origin, with the integron in a different position in the backbone (9). In the course of that work, we noticed that the sequence of a 2.5-kb PCR product amplified from the region between IS6100, found near the right end of In4-type integrons (17), and S024, which lies to the right of the integron in SGI2, appeared to consist of two superimposed sequences over a short segment that corresponds to 152 bp from the inverted repeat IRt end of In4-type integrons (Fig. 1). The two sequences corresponded to closely related sequences derived from Tn402 (as expected, adjacent to IS6100) and the equivalent part of Tn5058 (GenBank accession no. Y17897). This finding indicated that two copies of this region were present, but the possibility that there were two copies of SGI2 was eliminated (9). Here, we have further examined the S. enterica serovar Emek strains used in our original study in order to resolve this ambiguity.
FIG. 1.
Sequence of the 152-bp IRt end of the tni modules of Tn402 and Tn5058 and the hybrids recovered by PCR amplification. The Tn402 and Tn5058 sequences are at the top and bottom, respectively, with the hybrid sequences detected in clones from the PCR amplicon between them. Numbering above the alignment refers to positions in the 152-bp region, and the flanking sequences in the clones are in lower case. Bases in the sequence of Tn402 that differ from those in Tn5058 are highlighted in black. The 25 bases of IRt are indicated by bold type.
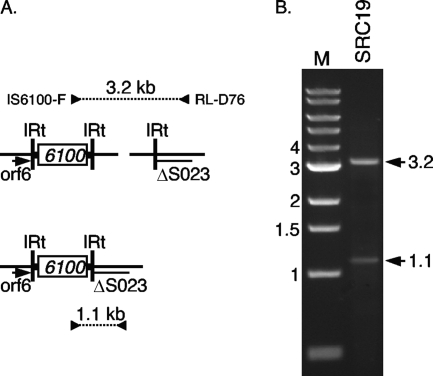
Whole-cell DNA was extracted from two independent cultures of strain SRC19 and used as a template for PCR with primers IS6100-F (5′-AAGGGATTCGAAGTCATG-3′) in IS6100 and RL-D76 (5′-AAACTGGGTAGTAAGCC-3′) in the part of S023 located to the right of the integron. The products were cloned into pGEM-T Easy by using the instructions of the manufacturer (Promega, Madison, WI). Several clones from each amplification were sequenced as described previously (4) using the amplification primers. In most clones, only the 152-bp tni region separated IS6100 from S023, and it was a hybrid. Four hybrid types were found (Fig. 1), but in all of them the first part was derived from the Tn402-type IRt end and the following part was derived from the Tn5058 IRt end. These hybrid products presumably arose either prior to amplification via homologous recombination occurring within the duplication described below or during amplification via template switching in the PCR. However, a few clones containing longer products were also obtained, and these included a further region of 2,113 bp derived from the tni module of Tn5058, Tn502 (GenBank accession no. EU306743), or Tn512 (GenBank accession no. EU306744) and located immediately adjacent to the 152-bp tni segment which, in these clones, was entirely of the Tn402 sequence type (Fig. 2A). This additional tni segment can be viewed as part of the integron, as it is located between the 5-bp duplication created by insertion of the integron and identified previously (9). The same integron arrangement has recently been found in Salmonella serovar Virchow (5), where it is also located in the same position in S023.
FIG. 2.
Revised structure of the integron of SGI2. The InEmek region of SGI2 is drawn to scale. The SGI2 backbone is shown as a dashed line with the backbone of the In4-type integron above and the additional segment found in the integron depicted above that. Different discrete segments such as the gene cassettes are represented by open boxes and lines of different thicknesses, and in the top line, “5′” and “3′” indicate the regions derived from the 5′ conserved sequence (5′-CS) and 3′-CS, respectively. Arrows indicate the positions and orientations of genes and open reading frames. Vertical bars indicate the inverted repeats (IRi and IRt) of class 1 integrons and Tn5058. The attI1 site is represented by a tall open box, and gene cassettes (dfrA1 and orfC) are shown as open boxes with a black bar at one end, indicating the attC sites (59-be). IS6100 and CR3 are represented by open boxes. The SGI1 backbone adjacent to the integron is shown as a dashed line with only relevant open reading frames indicated.
We concluded that the mixed sequence that we observed previously resulted from the use of PCR primers and amplification conditions that failed to detect the longer, correct PCR product because the primers were too far apart and the extension time was too short. The shorter, sequenced product, which consisted of a mixture of the various types of hybrids seen in the clones, was preferentially recovered. By using longer extension times and primers that would yield a smaller product, both the short and long products could be readily detected (Fig. 3). To facilitate the future detection of SGI2 variants with this integron configuration, we have designed primers for the detection of the right boundary of the integron (InEmek) with the backbone. PCR amplification with primers RH837 (5′-TTCATGCCCGACCACATCAA-3′), located in the tniA gene of the Tn5058 segment, and RL-D4 (5′-TTCATGATCTTGTGCCGCTAGC-3′), located in S023, produced a product of 473 bp by using genomic DNA from all three SGI2-containing strains and an SGI2-A variant described previously (9) as the template.
FIG. 3.
PCR products amplified from SGI2. (A) The regions amplified are indicated schematically. Symbols are as in Fig. 2. (B) The products produced by amplification with primers IS6100-F and RL-D4 are indicated by arrows with sizes marked in kilobases. M, molecular size markers.
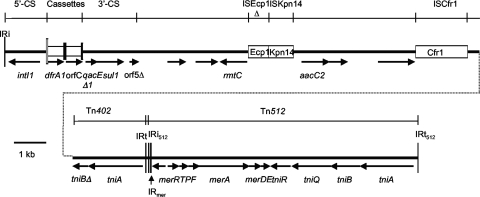
In the case of Salmonella serovar Virchow (5), the authors have argued that SGI2 should be viewed as a variant of SGI1 and named in the SGI1 series. However, all the other variants of SGI1 differ mainly within the boundaries of an integron which is in an invariant location (between tnpR and S044) and hence can be viewed as part of a specific lineage. Two features clearly distinguish SGI2 from this group. First and most importantly, the different location of the class 1 integron in SGI2 indicates that two separate integron acquisition events have occurred. Second, the sequence of the 20% of the 27.4-kb SGI2 backbone that we originally reported (9) differed at 0.3% of positions from the SGI1 backbone sequence, indicating that it had evolved independently for a long time. The 2.7 kb (10%) of backbone sequence reported for serovar Virchow (GenBank accession no. EU924797) also contained differences from SGI1. Here, we have sequenced an additional 5.8 kb of the backbone and have found further differences. A total of 11,558 bp, representing over 40% of the 27.4-kb backbone, has now been sequenced, and 21 single base differences (representing 0.18% of positions), 9 in coding regions and 3 synonymous, were found. These differences are unlikely to be due to errors in the original SGI1 sequence (GenBank accession no. AF261825), as the backbone sequence of a new SGI1 variant, SGI1-S, from Salmonella serovar Virchow SL491 (Fig. 4) which was reported recently (GenBank accession no. ABFH02000001) is identical. We also sequenced 6 kb of the backbone of another SGI1 variant, SGI1-K, and except for an IS1359 insertion and adjacent deletion, this sequence (GenBank accession no. AY463797) was also identical to SGI1. A shorter segment of 344 bp at the int end of the backbone that includes four single base differences between SGI1 and SGI2 (9) has also been reported for SGI1 from S. enterica serovar Patratyphi B d-tartrate-utilizing strains (4) (GenBank accession no. FJ477835) and is identical to the SGI1 backbone. Hence, we conclude that the differences between the SGI1 and SGI2 backbones do not result from sequencing errors.
FIG. 4.
Structure of the integron found between tnpR and S044 in SGI1-S from S. enterica serovar Virchow strain SL491. The structure is drawn to scale from the sequence of a contig in the unfinished genome sequence of S. enterica serovar Virchow strain SL491 (GenBank accession no. ABFH02000001; M. J. Rosovitz, P. McDermott, D. White, J. E. LeClerc, M. K. Mammel, T. A. Cebula, and J. J. Ravel, Craig Venter Institute, Rockville, MD). The integron in SGI1-S is located in the same position in the SGI1 backbone (between tnpR and S044) as those in all other members of the SGI1 group, and the 27.4-kb backbone sequence is identical to that of SGI1 (GenBank accession no. AF261825). Arrows indicate the positions and orientation of genes and open reading frames. Thin vertical bars indicate inverted repeat sequences of class 1 integrons (IRi and IRt) and IRmer from the part of a class II transposon found within Tn512. IS elements are represented by open boxes. The attI1 site is shown as a tall open box, and gene cassettes (dfrA1 and orfC) are depicted as open boxes with a black vertical bar at one end, representing the cassette-associated attC site (59-be). The divisions along the thin horizontal line above the schematic indicate the origins of particular regions. The drfA1 gene confers resistance to trimethoprim, sul1 confers resistance to sufonamides, rmtC confers resistance to gentamicin, kanamycin, tobramycin, and amikacin, and aacC2 confers resistance to gentamicin.
Estimates of the rate of divergence between bacterial nucleotide sequences vary substantially. One is of the order of 0.9% differences representing over 1 million years of separation (15). A more recent estimate of the rate at which single nucleotide polymorphisms accumulate suggests that about one synonymous base substitution arises each year in a bacterial genome of 4 Mb (8). Using each of these estimates, we calculated that the backbones of SGI1 and SGI2 have been diverging for around either 30,000 or 1,000 years. In contrast, the integrons of SGI1 and SGI2 have identical sequences over an equivalent length (11,075 bp), if the gene cassettes and a region before and in the floR or cmlA9 gene that was previously shown to have altered due to a recombinational exchange (9) are excluded. Hence, not only does SGI2 have the integron in a different position from SGI1, but the backbones of the SGI1 and SGI2 families evolved separately for a long time before they acquired very similar complex class 1 integrons. This scenario is similar to that for transposons Tn21 and Tn1696, which carry class 1 integrons located at different positions in related backbone mercuric ion resistance regions (16).
Nucleotide sequence accession numbers.
The additional 7.9 kb of sequence from SGI2 has been added to GenBank under accession no. AY963803. The additional sequence from SGI1-K has been added to GenBank under accession no. AY463797.
Acknowledgments
R.M.H. is supported by NHMRC fellowship grant 358713. N.L.W. and the project were supported by NHMRC project grant 352352.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Amar, C. F., C. Arnold, A. Bankier, P. H. Dear, B. Guerra, K. L. Hopkins, E. Liebana, D. J. Mevius, and E. J. Threlfall. 2008. Real-time PCRs and fingerprinting assays for the detection and characterization of Salmonella Genomic Island-1 encoding multidrug resistance: application to 445 European isolates of Salmonella, Escherichia coli, Shigella, and Proteus. Microb. Drug Resist. 14:79-92. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli, A., E. Filetici, L. Villa, A. M. Dionisi, A. Ricci, and I. Luzzi. 2002. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djordjevic, S. P., A. K. Cain, N. J. Evershed, L. Falconer, R. S. Levings, D. Lightfoot, and R. M. Hall. 2009. Emergence and evolution of multiply antibiotic-resistant Salmonella enterica serovar Paratyphi B d-tartrate-utilizing strains containing SGI1. Antimicrob. Agents Chemother. 53:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doublet, B., C. Chu, C. H. Chiu, Y. C. Fan, and A. Cloeckaert. 2009. Truncated tni module adjacent to the complex integron of Salmonella genomic island 1 in Salmonella enterica serovar Virchow. Antimicrob. Agents Chemother. 53:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublet, B., K. Praud, S. Bertrand, J. M. Collard, F. X. Weill, and A. Cloeckaert. 2008. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52:3745-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublet, B., K. Praud, F. X. Weill, and A. Cloeckaert. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J. Antimicrob. Chemother. 63:282-289. [DOI] [PubMed] [Google Scholar]
- 8.Feng, L., P. R. Reeves, R. Lan, Y. Ren, C. Gao, Z. Zhou, Y. Ren, J. Cheng, W. Wang, J. Wang, W. Qian, D. Li, and L. Wang. 2008. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS One 3:e4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levings, R. S., S. P. Djordjevic, and R. M. Hall. 2008. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levings, R. S., D. Lightfoot, R. M. Hall, and S. P. Djordjevic. 2006. Aquariums as reservoirs for multidrug-resistant Salmonella Paratyphi B. Emerg. Infect. Dis. 12:507-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levings, R. S., S. R. Partridge, S. P. Djordjevic, and R. M. Hall. 2007. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 51:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915-1922. [DOI] [PubMed] [Google Scholar]
- 14.Ng, L. K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26: 74-86. [DOI] [PubMed] [Google Scholar]
- 16.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]