Summary
The Huntington's disease (HD) mutation causes polyglutamine expansion in huntingtin (Htt) and neurodegeneration. Htt interacts with a complex containing Rab11GDP and is involved in activation of Rab11, which functions in endosomal recycling and neurite growth and long-term potentiation. Like other Rab proteins, Rab11GDP undergoes nucleotide exchange to Rab11GTP for its activation. Here we show that striatal membranes of HD140Q/140Q knock-in mice are impaired in supporting conversion of Rab11GDP to Rab11GTP. Dominant negative Rab11 expressed in the striatum and cortex of normal mice caused neuropathology and motor dysfunction, suggesting that a deficiency in Rab11 activity is pathogenic in vivo. Primary cortical neurons from HD140Q/140Q mice were delayed in recycling transferrin receptors back to the plasma membrane. Partial rescue from glutamate induced cell death occurred in HD neurons expressing dominant active Rab11. We propose a novel mechanism of HD pathogenesis arising from diminished Rab11 activity at recycling endosomes.
Introduction
Huntington's disease is an autosomal inherited neurodegenerative disorder caused by an expansion of the polyglutamine (polyQ) tract near the NH2-terminus in huntingtin (Htt). Mutant Htt disrupts cellular processes involved with gene transcription (Nucifora et al., 2001; Dunah et al., 2002; Zhai et al., 2005; Cui et al., 2006), energy metabolism (Lin and Beal, 2006), protein clearance (Bence et al., 2001; Zemskov and Nukina, 2003; Bennett et al., 2005; Diaz-Hernandez et al., 2006) and vesicle transport along cytoskeletons (DiFiglia et al., 1995; Gunawardena et al., 2003; Szebenyi et al., 2003; Gauthier et al., 2004; Lee et al., 2004; Trushina et al., 2004). Htt is found on membranes, in coated pits, vesicles, and subcellular organelles (DiFiglia et al., 1995; Velier et al., 1998; Rockabrand et al., 2007; Orr et al., 2008), supporting the idea that Htt functions on membranes. Moreover, Htt interacts with proteins involved in vesicular trafficking (Harjes and Wanker, 2003).
Rabs are small GTPases that regulate processes in vesicular trafficking from vesicle formation to vesicle fusion (Grosshans et al., 2006). Distinct subcellular organelles or membrane domains possess specific Rab proteins, such as Rab5 for early endosomes and Rab11 for recycling endosomes (Zerial and McBride, 2001). Rab proteins undergo cycles that switch Rab activity on and off (Zerial and McBride, 2001; Grosshans et al., 2006). Rab activity is tightly controlled by factors including GEF, GTPase-activating protein (GAP) and RabGDP displacement inhibitor (RabGDI) (Zerial and McBride, 2001; Grosshans et al., 2006). Activation by a guanyl nucleotide exchange factor (GEF) occurs at membranes and converts Rab from the GDP-bound (inactive) to GTP bound (active) state (Zerial and McBride, 2001; Seabra and Wasmeier, 2004; Grosshans et al., 2006). Once activated, the Rab recruits a host of effector proteins that participate in vesicle formation, vesicle delivery and vesicle fusion. After fulfilling functions, RabGTP is inactivated with the aid of a GAP and converts to RabGDP, which is released from membranes by RabGDI (Zerial and McBride, 2001; Grosshans et al., 2006; Jones et al., 2006).
Rab11 has a critical role in controlling post-endocytic recycling of plasma membranes containing important receptors, channels and other membrane constituents (Ullrich et al., 1996; Ren et al., 1998; Zerial and McBride, 2001; Park et al., 2004; Emery et al., 2005; Grosshans et al., 2006; Jones et al., 2006). In flies, proper function of Rab11 is essential for development of the nervous system (Alone et al., 2005; Khodosh et al., 2006; Bhuin and Roy, 2009). Targeted disturbance of Rab11 function causes degeneration of photoreceptor cells (Alone et al., 2005). The impact of Rab11 dysfunction in mammals is not established.
Recently, we found evidence to support a role for normal Htt in the process of activating Rab11 from Rab11GDP to Rab11GTP (Li et al., 2008). Membranes from Htt-null ES cells had reduced levels of Rab11GDP and reduced GEF activity to activate Rab11 compared to membranes from wild-type ES cells. Moreover, endogenous Htt from normal mouse brain interacted with a complex containing inactive Rab11 (Rab11GDP).
In this study we address whether Rab11 dysfunction occurs in the brain and in primary neurons of a knock-in HD mouse model (HD140Q/140Q) and contributes to neurodegeneration. The HD140Q/140Q mice have human exon 1 with expanded CAG repeat inserted into the mouse Htt gene homolog (Menalled et al., 2003). These HD mice express full-length mutant Htt under regulation of the endogenous Htt promoter. We show that striatal membranes of young HD140Q/140Q mice manifest less activity for nucleotide exchange on Rab11. Introducing dominant negative Rab11 into adult normal mouse striatum led to neurodegeneration and motor deficit, indicating a requirement for normal Rab11 activity. HD140Q/140Q primary cortical neurons were delayed in recycling transferrin receptors consistent with Rab11 dysfunction. Glutamate-induced death was ameliorated in HD140Q/140Q neurons that expressed dominant active Rab11. Our results suggest a novel mechanism for HD pathogenesis involving early deficits in the control of Rab11 activity.
Results
HD mouse striatal membranes are deficient in stimulating guanine nucleotide exchange on Rab11
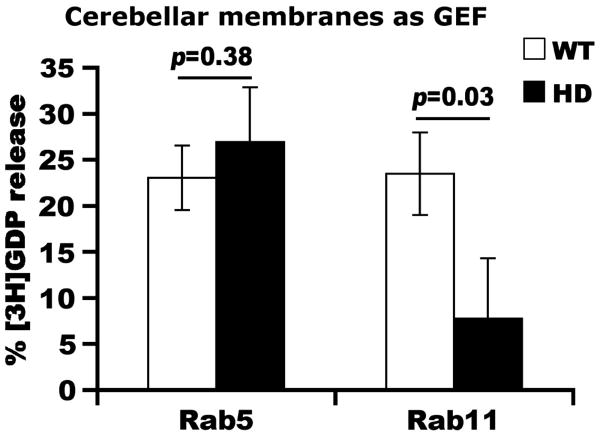
Rab11 activation is reduced in Htt-null mouse ES cells compared to normal mouse ES cells, indicating a requirement of Htt in regulating normal Rab11 activity (Li et al., 2008). To determine if the presence of mutant Htt in the brain affects guanine nucleotide exchange on Rab11, we measured [3H]GDP release from GST-Rab11 in the presence of a GEF activity (Jones et al., 1998; Jones et al., 2000; Shinohara et al., 2002; Hsu et al., 2007). The GEF for Rab11 is not established. We used membrane fractions (high speed pellet) prepared from striatal lysates or Rab11-enriched soluble membrane fractions prepared from whole brain as the sources of GEF (see Methods). When equal amounts of proteins from WT and HD140Q/140Q striatal membranes were added to the GDP release assay, [3H]GDP release from Rab11 catalyzed by HD mouse striatal membranes was significantly lower than that catalyzed by the membranes from WT mouse striata (n=3, Student t-test: p<0.01 at 15min and 30min; Figure 1A). Under the same conditions, [3H]GDP release from Rab5 was similar whether HD or WT striatal membranes were used in the assay (n=3, Student t-test: not significant; Figure 1B). Deficits in activation of Rab11 (n=3, Student t-test: p<0.05 at 15min and 30min; Figure 1C) were also observed when the source of GEF in the [3H]GDP release assay was Rab11-enriched soluble membrane fractions prepared from whole brain. We also observed deficits in Rab11 activation in total membranes from the cerebellum of HD mice (Supplementary Figure 1). Altogether, these findings suggest that the conversion of Rab11GDP to Rab11GTP is impaired in the HD mouse brain, including the striatum and cerebellum.
Figure 1. Rab11 but not Rab5 activation is inhibited in HD brains.
A and B, 200μg of striatal total membranes prepared from freshly sacrificed WT and homozygous HD mice (see Methods) were used as the source of GEFs for catalyzing [3H]GDP release from [3H]GDP-GST-Rab11 (A) or [3H]GDP-6×His-Rab5 (B) for indicated times. Data were represented as the percentage of [3H]GDP released from GST-Rab11 or 6×His-Rab5 after incubation with striatal total membranes (n=3 for each time point in both A and B, Mean±SD, 2-tailed Student t-test: * p<0.01). C, Rab11-enriched membrane fractions (fraction-7 for HD and fractions-6 and 7 for WT) were prepared as described in Methods. 50μg of HD and WT mouse brain Rab11-enriched membranes were used to catalyze [3H]GDP release from GST-Rab11 for indicated times. Data are represented as the percentage of [3H]GDP released from GST-Rab11 after incubation with Rab11-enriched membrane fractions (n=3 for each time point, Mean±SD, 2-tailed Student t-test: * p<0.05).
Levels of Rab11 are elevated in striatal membranes of HD mouse
Rab11 like other Rab proteins requires secure association with membranes to execute its functions. Recently we found that Htt-null ES cells have impaired nucleotide exchange on Rab11 accompanied by reduced levels of Rab11 in membrane fractions (Li et al., 2008). Therefore, we next determined if the loss of Rab11 activity in HD brain membranes is correlated with reduced levels of membrane-associated Rab11. We examined total membranes from striata of young (10 - 14 weeks old) HD mice and age-matched WT mice. To our surprise, the levels of Rab11 were significantly elevated in striatal membrane fractions of HD mice compared to those of WTs (Mean±SD, 3.4±0.08 fold; Figure 2, A and B). In contrast, the levels of Rab5, a marker of early endosomes, were similar in WT and HD striatal membrane fractions (Figure 2A). Levels of Rab11 and Rab5 in total lysates from striata were not different in WT and HD mice (Figure 2A). These data suggested that the presence of mutant Htt increased levels of Rab11 selectively on membranes.
Figure 2. Impaired extraction of Rab11 by RabGDI contributes to elevation of Rab11 on membranes in HD.
A, Post-debris supernatants and total membranes prepared from striata of young (10 – 14 weeks old) WT and HD mice as described in Methods were analyzed by SDS-PAGE and Western blot analysis with indicated antibodies. Shown are a series of blot analyses. B, Quantitative data for A. Films were scanned and the band density was measured using NIH ImageJ. The density of calnexin was used for normalization of loading variations. Data are represented as ratio of Rab11 on HD striatal membranes to Rab11 on WT striatal membranes (n=6 mice for each of WT and HD, Mean±SD, Student t-test: * p<0.01). C, RabGDI-mediated extraction of Rab11 was performed as described in Methods. In this assay we used 20μg HD membranes and 60μg WT membranes to keep the same level of Rab11GDP to be retrieved by RabGDI. The extracted Rab11 (released) in complex with GST-RabGDI on glutathione and the un-extracted Rab11 (unreleased) in the supernatant was precipitated with chloroform/methanol and was boiled in SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE and Western blot with anti-Rab11 and anti-GDI antibodies. Shown are blot analyses from one of the three experiments. D, Densitometry data obtained in C were used to calculate the percentage of extracted Rab11 by GST-RabGDI by dividing the extracted amount from the total input (released + unreleased). Average percentage of released Rab11 by GST-RabGDI was plotted (n=3, Mean±SD, Student t-test: * p<0.001).
The combination of reduced Rab11 activation and increased levels of Rab11GDP in HD posed an enigma. Usually, non-activated RabGDP is rapidly removed from membranes by RabGDI. We speculated that mutant Htt might affect another process in Rab11 activation, the removal of inactive Rab11 from membranes by RabGDI. To test this idea, we performed in vitro extraction of Rab11GDP by GST-RabGDI. This assay has been used to study the extraction of other Rab proteins by RabGDI (Soldati et al., 1993; Peter et al., 1994). The source of Rab11 for the in vitro extraction experiments was soluble Rab11-enriched whole brain membrane fractions. These soluble membrane fractions were deficient in supporting nucleotide exchange on Rab11 (Figure 1C). In this assay we used 20μg HD membranes and 60μg WT membranes. The rationale was to keep the same level of Rab11GDP to be retrieved by exogenous RabGDI. After 45 min incubation with GST-RabGDI, 75% of Rab11 was extracted from Rab11-enriched brain membranes from WT mice. Under the same conditions, only 32% of Rab11 was extracted from HD mouse brain membranes (Mean±SD, WT vs HD: 75.8±5.4% vs 32±5.8%, Student t-test: p<0.001; Figure 2C, 2D). These data suggested that there is a defect in RabGDI-dependent extraction of Rab11GDP from membranes in HD. The process for RabGDI to remove Rab11GDP from membranes requires hydrolysis of adenosine triphosphate (ATP) (Sakisaka et al., 2002). Addition of excess ATP to the assay recovered the capacity of RabGDI to extract Rab11 from HD brain membranes (Mean±SD, HD vs HD+ATP: 32±5.8% vs 70.2±6.6%, Student t-test: p<0.001; Figure 2C, 2D). ATP production measured by a standard luciferase assay was not different in WT and HD mouse brain lysates (Supplementary Figure 2). Orr et al found that overall levels of ATP production in the cortex of another HD knock-in model (HD150Q/150Q) unchanged compared to WT mice, but significantly reduced in HD cortical synaptosomes (Orr et al., 2008), suggesting that subcellular reductions in ATP levels do occur in HD neurons. Our findings suggest that in HD there is insufficient ATP for the process of extracting Rab11 from HD membranes by RabGDI. An ATP deficit in RabGDI dependent extraction cannot fully account for the excess Rab11GDP on HD membranes since Rab5GDP does not accumulate on HD membranes.
Expression of dominant negative Rab11 in the brain is sufficient to cause neuropathological changes
If Rab11 dysfunction directly contributes to HD pathogenesis, we would expect that disturbing Rab11 activity in vivo would cause neuronal dysfunction. Dominant negative GTPase mutants are widely used to impair activity of their corresponding GTPases by inhibiting activation of endogenous GTPases (Feig, 1999). We took advantage of the known effects of the dominant negative Rab11S25N mutant (Ullrich et al., 1996; Ren et al., 1998; Wilcke et al., 2000) to look at the consequences of Rab11 dysfunction on neuronal survival in vivo. We unilaterally injected adenoassociated virus (AAV) expressing HA-tagged Rab11S25N or enhanced green fluorescence protein (EGFP) into the striatum of normal adult mice. By 12 and 19 days post-injection, mice injected with HA-Rab11S25N were much slower in crossing a beam compared to mice injected with AAV expressing EGFP (Mean±SD in time increase by fold compared to pre-injection, Rab11S25N vs EGFP, Student t-test: 2.48±0.28 vs 0.7±0.18, p=0.0008, day-12; 5.7±0.7 vs 1.24±0.21, p=0.0004, day-19; Figure 3A). Brain slices stained by the immunoperoxidase method with antibodies against the HA tag showed widespread labeling throughout anterior regions of striatum and overlying cortex on the injected side but no labeling on the non-injected side (results not shown). Labeled neurons showed a somato-dendritic cytoplasmic distribution of HA-Rab11S25N. The labeling was similar to that reported for endogenous immunoreactive Rab11 in brain (Sheehan et al., 1996). No gross atrophy of the injected striatum was evident but there was shrinkage of some labeled striatal neurons (Figure 3B upper panel). Dendrites of some labeled cortical neurons were retracted (Figure 3B lower panel). These pathological features were not evident in brains of mice injected with AAV expressing EGFP for the same time period (Supplementary Figure 3). The findings demonstrate that hindering Rab11 function in the brain is sufficient to cause neuronal and movement abnormalities.
Figure 3. Interference with Rab11 function causes behavioral and neuronal anomalies in vivo.
A, AAV expressing EGFP or HA-tagged Rab11S25N were unilaterally injected into the striatum and overlying cortex. Beam walking was tested at indicated dates post-injection as described in Materials and Methods. The time for mice to cross the beam was measured pre- and post-injection of virus. Data were graphed as the average increase in time at each post-injection period compared to pre-injection time (n=6, Mean±SD, Student t-test: * p<0.01). B, Shown are representative vibratome cut brain sections at the level of the striatum from two of the mice reported in (A). Brain slices were stained with anti-HA antibody or anti-GFP antibody using the immunoperoxidase method. Arrow points to the side injected with AAV-HARab11S25N or AAV-EGFP. C, Expression of HA Rab11S25N led to neuropathological changes. Shown are representative examples of neurons expressing HA-tagged Rab11S25N in the striatum and cortex that appear shrunken (arrowhead) or have a retracted proximal dendrite (arrow). Scale bar: 5μm.
Transferrin receptor recycling is slowed in HD neurons
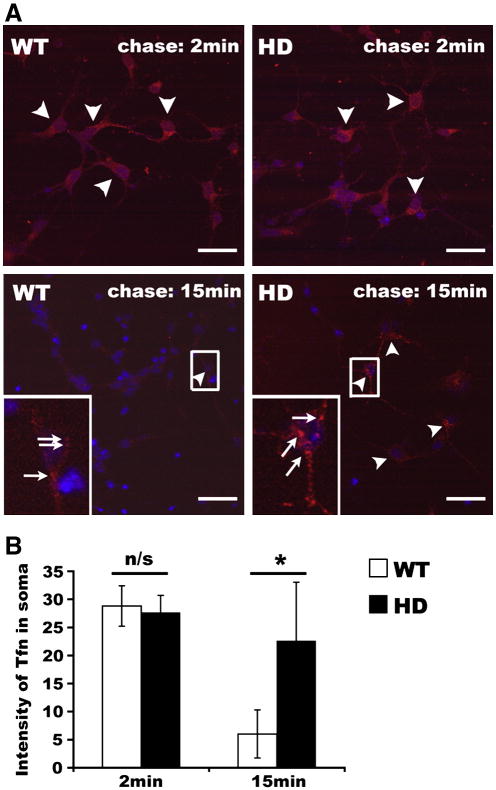
Having established that Rab11 activity is impaired in HD brains, we next investigated if an effect downstream of Rab11 activation is altered in HD. Rab11 is well known as a regulator for endocytic recycling of transferrin receptors (Ullrich et al., 1996). In previous studies, neurons expressing WT and mutant Htt had similar total cumulative uptake of Alexa568-transferrin (Trushina et al., 2006), indicating that transferrin receptor functions normally in neurons with mutant Htt. To explore if transferrin receptor recycling is affected in the presence of mutant Htt, we conducted a pulse-chase uptake experiment, in which cell surface transferrin receptor was labeled with Alexa568-transferrin on ice. After washes to remove un-bound Alexa568-transferrin, primary neurons were chased in the presence of unlabeled transferrin at 37°C. Transferrin receptor recycling can be determined by detecting the decline in fluorescence intensity of Alexa568-transferrin. When primary neurons were chased for 2min, neurons expressing mutant Htt were not different from those of WT in taking up Alexa568-transferrin (n=42 WT and 42 HD neurons, Mean±SD fluorescence intensity of Alexa568-transferrin in the soma region; WT vs HD: 28.8±3.6 vs 27.6±3.1; Student's t-test: p=0.108; Figure 4, A and B), suggesting that transferrin receptor functions normally in HD neurons. After chase for 15min, the fluorescence intensity of Alexa568-transferrin in HD cortical neurons was significantly stronger than that in WT cortical neurons (n=42 WT and 42 HD neurons, Mean±SD fluorescence intensity of Alexa568-transferrin in the soma region; WT vs HD: 6.1±4.3 vs 22.6±10.5; Student's t-test: p<0.0001; Figure 4, A and B), indicating a delay in the return of transferrin receptor back to the cell surface. An analysis was performed to determine the number of neurons with fluorescence intensity of Alexa568-transferrin at least 1.5 fold of adjacent background fluorescent intensity. Results showed that after chase for 15min, 54.8% of HD neurons had intensity of Alexa568-transferrin in the soma region at least 1.5-fold greater than background, whereas no WT neurons met this criteria (n=42 WT and 42 HD neurons), indicating that the majority of HD neurons were impaired in recycling transferrin receptors to the cell surface. These data support insufficient Rab11 activation in HD neurons.
Figure 4. Slowed recycling of transferrin receptor in HD neurons.
A, Primary neurons on coverslips were cultured in PBS for 1hr. After two washes in cold PBS and incubation in cold PBS on ice for 15min, primary neurons on coverslips were incubated with 5μg ml-1 Alexa568-transferrin on ice for 30min. After two washes in cold PBS, coverslips were incubated in pre-warmed culture medium containing 10μg ml-1 non-labeled transferrin at 37°C for 2min or 15min and fixed for analysis. Shown are confocal images. Nuclear stain with Hoechst 33258 is in blue. Alexa568-transferrin is in red. Fewer neurons appear in the HD cultures than in the WT cultures. Arrowheads show neurons with strong Alexa568-transferrin signals. Boxed regions are shown at higher magnification at lower left of each panel. Arrows identify endosomes or transport intermediates labeled with Alexa568-transferrin. Scale bar: 20 μm. B, Signal intensity for Alexa568-transferrin in soma of neurons and the corresponding background levels were measured using NIH ImageJ. Data are represented as mean signal intensity of Alexa568-transferrin in the soma region per neuron (Mean±SD, Student t-test; 2min chase, n=42 neurons for each of WT and HD140Q/140Q, n/s: no significance; 15min chase, n=42 neurons for each of WT and HD140Q/140Q, * p<0.001).
Dominant active Rab11 protects HD neurons from glutamate-induced cell death
Compared to neurons expressing normal Htt, neurons expressing mutant Htt are more sensitive to various stressors such as a glutamate challenge (Estrada Sanchez et al., 2008). We addressed if introducing dominant active Rab11 into primary neurons could improve the tolerance of primary HD neurons to glutamate exposure. Primary cortical neurons were prepared from WT and HD140Q/140Q mouse embryos. Lentivirus expressing EGFP or lentivirus expressing dominant active Rab11 (Rab11Q70L) and EGFP were added to primary neuronal cultures for one week. After verifying the presence of EGFP expression in the cultures, primary neurons were challenged with 0.2mM glutamate for 20min and extensively washed to remove glutamate. Neuronal death was detected 24hr after glutamate by determining the number of trypan blue positive neurons. In agreement with previous studies (Shin et al., 2005), cortical neurons expressing endogenous mutant Htt were significantly more vulnerable to glutamate compared to cortical neurons expressing endogenous wild-type Htt (percent of trypan blue positive neurons, WT vs HD: 45.2% vs 76.1%; Chi square: p<0.001; Figure 5). Viral delivery of Rab11Q70L ameliorated the susceptibility of WT neurons and HD140Q/140Q neurons to glutamate-induced cell death and restored HD neurons to survival levels of glutamate treated WT neurons without viral infection or infection with virus expressing EGFP alone. Taken together, these data suggest that elevating Rab11 activity can decrease the sensitivity of HD neurons to glutamate induced cell death.
Figure 5. Dominant active Rab11 improves the tolerance of primary HD neurons to glutamate.
Glutamate-induced cell death in primary cortical neurons. Primary WT and HD cortical neurons were infected with lentivirus or left alone for seven days and challenged with 0.2mM glutamate for 20min. After washout of glutamate, primary neurons were cultured in fresh media for 24hr. Cell death was determined by trypan blue uptake. Data in bar graph are represented as the percentage of trypan blue positive neurons from four independent preparations of neurons (n=462 for WT and 285 HD neurons for no-virus; n= 403 for WT and 349 for HD neurons infected with virus-EGFP; n= 432 for WT and 409 for HD neurons infected with virus-Rab11Q70L; Chi square: # p<0.0001; * p<0.000001).
Discussion
Rab11 is a key regulator for endocytic recycling. We show that mutant Htt hampers the activity of Rab11 in the brain of HD140Q/140Q mice. Using a standard GEF activity assay, we found that HD striatal membranes were deficient compared to WT striatal membranes in supporting the conversion of inactive Rab11 into active Rab11 (Figure 6). On the other hand, HD striatal membranes did not impair activation of Rab5, which functions at early endosomes. The deficiency in supporting Rab11 activation was detected using whole brain, striatal, or cerebellar membranes of 10 to 14 week old HD mice. In this HD mouse model, a reduction in climbing behavior is detected at about 6 weeks but other measurements of motor activity (rotarod, pole task, running wheel) and neuropathological changes start between 16-24 weeks (Menalled et al., 2003; Hickey et al., 2008). Thus, the inhibitory effects of mutant Htt on Rab11 activity are widespread in the brain and present at an early stage of the disease in HD mice. Total protein levels of Rab11 were not changed in HD striatum compared to WT striatum, suggesting that neuronal vulnerability in HD is not related to a change in levels of Rab11. Our data support a model for HD pathology in which deficient GEF activity on Rab11 slows the recycling of cargo proteins critical to the function of vulnerable neurons.
Figure 6. Schematic representations of Rab11 cycles in WT and HD conditions.
Left panel depicts the two cycles of Rab11 that are required for its proper function (normal). Rab11 associates and dissociates with membranes and alternates between GDP-bound inactive and GTP-bound active states. In the cytoplasm, GDP-bound Rab11 partners with Rab-GDP dissociation inhibitor (GDI). To fulfill functions, Rab11-GDP is recruited onto endosomal membranes for activation by a guanine nucleotide exchange factor (GEF) and conversion into GTP-bound form. After fulfilling the function by recruiting a cohort of effectors, Rab11-GTP is inactivated by a GTPase-activating protein (GAP) and returns back to GDP-bound form that is extracted by RabGDI. Right panel shows a proposed model for Rab11 dysfunction in HD. The key event causing neurodegeneration is a deficient nucleotide conversion from Rab11GDP to Rab11GTP or deficient activation of Rab11 (indicated by long dashed arrow), which occurs on endosomal membranes. Another problem in HD endosomal membranes is the accumulation of inactive GDP-bound form of Rab11 (enlarged oval) caused in part by reduced extraction of RabGDP by RabGDI (indicated by short dashed arrow). The role of the accumulation of Rab11GDP to neuronal dysfunction is unclear.
The present work in HD mice and our previous study using Htt-null ES cells (Li et al., 2008) demonstrate that the presence of mutant Htt or the absence of wild-type Htt diminishes nucleotide exchange activity on Rab11. Our previous study suggested that Htt participates in a complex that activates Rab11, indicating that the GEF is in the complex. The GEF for Rab11 in mammalian cells is not known. Knowledge of this factor is needed to know if Htt interacts directly with this protein and modulates its function. These data will be important to fully understand the mechanism for deficient Rab11 activity in HD membranes. Nevertheless, our study identifies a novel effect of mutant Htt on the activity of a Rab protein. Interfering with Rab11 activity by the expression of dominant negative Rab11 in the cortex and striata of normal adult mice was sufficient to cause motor deficits and neuropathology. Our data suggest a novel pathway for HD pathogenesis involving the dysfunction of Rab11.
Although total levels of Rab11 were not changed in HD, we found redistribution of Rab11 onto HD membranes. This change was not observed for Rab5. There are no previous conditions described in which a Rab protein accumulates on membranes so it is unclear if more Rab11GDP on HD membranes per se affects neuronal function. There was a deficiency in the ATP dependent removal of inactive Rab11 from membranes by RabGDI. The extraction process by RabGDI is shared by all Rab proteins (Soldati et al., 1993; Peter et al., 1994) and therefore in HD would be expected to increase levels of other Rab proteins on membranes. This does not appear to be the case, since Rab5 levels on membranes were normal in HD membranes. Thus, impaired retrieval by RabGDI does not fully account for Rab11 membrane accumulation. The processes of Rab recruitment to membranes, activation, and retrieval from membranes are interdependent and efficient such that only those RabGDP molecules in the process of nucleotide exchange are on membranes. The Rab11GDP molecules that accumulate on HD membranes may arise from a combination of not being activated and retrieved by RabGDI (Figure 6).
It is well known that transferrin receptors constitutively recycle back to the plasma membrane in a Rab11-dependent manner (Ullrich et al., 1996; Ren et al., 1998; Wilcke et al., 2000). Consistent with a deficit in Rab11 activity, we found that HD primary cortical neurons were delayed in recycling of transferrin receptor back to the plasma membrane. Besides regulation of transferrin receptor recycling, Rab11 regulates the delivery of other important proteins to cell surfaces. These include AMPA receptor (Park et al., 2004; Correia et al., 2008) and the neuronal cysteine/glutamate transporter EAAC1 (Gonzalez et al., 2007). Based on the function of these proteins, a deficiency in their recycling back to the plasma membrane might alter cellular response to glutamate or increase oxidative stress, which have been reported to occur in brains of HD patients (Dure et al., 1991; Wagster et al., 1994; Browne et al., 1997). Thus, Rab11 dysfunction in HD has the potential to affect functions related to a broad range of proteins in neurons.
In flies, disturbance of Rab11 activity by expressing dominant negative Rab11 is lethal (Alone et al., 2005; Khodosh et al., 2006). Targeted expression of dominant negative Rab11 causes degeneration of photoreceptor cells (Alone et al., 2005) and deficits in development of the nervous system (Bhuin and Roy, 2009). Consistent with these studies in flies, we found that introducing dominant negative Rab11 into the striatal region and underlying cortex of adult normal mice led to neuropathological changes. Motor deficits were detectable as early as twelve days post-injection of virus expressing dominant negative Rab11 and were progressive. These in vivo data demonstrate that disturbance of endogenous Rab11 activity is sufficient for neuronal dysfunction.
We found that expression of dominant active Rab11 partially rescued the vulnerability of primary cortical neurons to glutamate induced cell death. The precise mechanism for how a dominant active Rab11Q70L provides neuroprotection from glutamate insult is not clear. Expression of the dominant active Rab11Q70L in mouse atrial myocytes increased the steady state cell surface levels of the voltage-gated potassium channel Kv1.5, a protein constitutively recycling through recycling endosomes (McEwen et al., 2007). In neuronal cells, neuronal glutamate transporter EAAC1 constitutively recycles in a Rab11-dependent manner (Gonzalez et al., 2007). EAAC1 is the primary route for neuronal uptake of glutathione precursor cysteine, thus critical for defense against oxidative stress (Aoyama et al., 2006). Glutathione depletion contributes to glutamate induced cell death (Murphy et al., 1989). It is possible that a neuroprotective effect of dominant active Rab11Q70L against glutamate insult was mediated by increased steady state cell surface levels of EAAC1 in a manner similar to increasing levels of Kv1.5 in atrial myocytes.
The deficit we observed in activation of a Rab protein (Rab11) in the presence of mutant Htt is novel. Previous studies suggest that mutant Htt may affect other Rab dependent pathways but downstream of Rab activation-that is, on Rab effectors and the functions related to those effectors (Hattula and Peranen, 2000; Pal et al., 2006). For example, Pal et al. found that the huntingtin interactor HAP40 interacted with Rab5GTP and was at higher levels in HD fibroblasts. The increase in HAP40 inhibited the recruitment of Rab5-positive early endosomes onto microtubules (Pal et al., 2006). Del Toro et al. found that immortalized neurons expressing mutant Htt had impaired localization of optineurin-Rab8 complex at Golgi membranes and this resulted in impaired transport of secretory vesicles to lysosomes (Del Toro et al., 2009). The mechanisms for these deficits are not known but do not involve impaired Rab activation. The inhibition of Rab11 activation by mutant Htt found in our study is the basis for slowed recycling of cargo (transferrin receptor) from recycling endosomes back to the plasma membrane. It will be of interest to know if the deficits in effector functions of Rab5, Rab8 and Rab11 occur in the same or different HD neurons in the brain and if cargoes that recycle in a Rab11-dependent manner are more vulnerable to Rab11 dysfunction in some types of HD neurons.
In summary, our study establishes that Rab11 activity is impaired in the brain of young HD knock-in mice. A modest loss of activation of Rab11 by a GEF in HD neurons early in the disease may have cumulative effects on neuronal function over time that correlate with slow progression characteristic of HD. One challenge in future studies will be to determine how Rab11 dysfunction increases vulnerability for cell death in select populations of neurons of the cortex and striatum. Knowledge of the cargoes that are recycled in a Rab11 dependent manner in these brain regions will be critical to understanding selective neuronal degeneration. Nevertheless, an increase in Rab11 activity protected HD primary cortical neurons from the toxic effects of glutamate challenge. Thus, our data suggest that approaches to improve the activity of Rab11 may be beneficial for HD.
Materials and Methods
Monoclonal antibodies against Rab11 and the HA tag were purchased from BD biosciences-CHEMICON and Covance, respectively. Polyclonal antibodies were commercially available (calnexin c-terminus: Stressgen; Rab5: Santa Cruz Biotech and GDI1: ProteinTech Group Inc). Conjugated secondary antibodies were obtained from Jackson Laboratory.
Preparation of plasmid DNA and viral vectors
The cDNA encoding Rab11 (BC010722) was amplified by polymerase chain reaction (PCR), purified from agarose gel using QIAEX II kit (Qiagen) and digested with BamH I/Xho I. After purification from agarose gel, the digested Rab11 cDNA fragment was cloned into the BamH I/Xho I sites of the HA-pcDNA3 plasmid. Rab11 mutants (Rab11S25N and Rab11Q70L) were generated by PCR-based site-specific mutagenesis. All Rab11 sequences were verified by DNA sequencing. HA-tagged Rab11S25N was obtained by digesting HA-Rab11S25N-pcDNA3 with Hind III and Xho I, and subcloned into the Hind III/Xho I sites of the AAV2-CBA-W plasmid.
HA-Rab11Q70L-pcDNA3 was digested with BamH I and Xho I to obtain Rab11Q70L, which was subcloned into the BamH I/Xho I sites of the pENTR1A plasmid. HA-Rab11Q70L was then released from HA-Rab11Q70L-pENTR1A after treatment with Nhe I and Xho I. This Nhe I/Xho I fragment was cloned into the Nhe I/Xho I sites of lentivirus vector CSCW2-pgk-EGFP. This vector has a bi-cistronic promoter enabling the independent expression of HA-Rab11Q70L and enhanced green fluorescent protein (EGFP).
Viral packaging and titration
Production of AAV2 and determination of AAV2 viral titers were performed exactly as previously described (Broekman et al., 2006). Viral titers used for in vivo striatal injection of AAV-HARab11S25N and AAV-EGFP were up to 2 × 1012 g.c. (genome copies) ml-1 and 9 × 1012 g.c. ml-1, respectively, and 6 × 109 g.c. of AAV- HARab11S25N (3μl) or AAV-EGFP (0.7μl diluted in 2.3μl PBS).
293T producer cells were used to generate lentivirus expressing enhanced green fluorescent protein (EGFP) (CSCW2-pgk-EGFP plasmid) or bi-cistronically expressing Rab11Q70L and EGFP (Rab11Q70L-CSCW2-pgk-EGFP plasmid). Transfection was performed according to the manufacturer's instructions (Roche). In brief, 7×106 of 293T cells were plated in a 15cm plate and cultured in 30ml DMEM media supplemented with 10% fetal bovine serum, L-glutamine and sodium pyruvate at 37°C in a cell culture incubator (Thermo-Fisher) over night. Prior to preparation of packaging mixtures, 293T cells were changed into fresh media cultured as above. 6.75μg of CSCW2-pgk or Rab11Q70L-CSCW2-pgk plasmid DNA, 6μg of pCMV-dR8.2 dvpr, 0.75μg of pCMV-VSV-G and 243μl of FuGene 6 (Roche) were used to prepare packaging mixtures. After applying packaging mixtures, 293T cells were cultured 37°C in the cell culture incubator for 24hr and changed into fresh culture media. 48hr later, culture media were collected and passed through a 0.45μm filter. The filtrated culture media were centrifuged at 4°C 24, 000rpm in a T50.2 rotor (Beckman) for 90min. Viral pellet was resuspended in 0.2ml PBS containing 0.5% BSA. Titers of lentivirus were determined using a HIV-1 p24 Antigen ELISA kit (ZeptoMetrix Corp.). The viral titer for Rab11Q70L/EGFP was 1.8×108 pg ml-1 and for EGFP was 1.1×108 pg ml-1. Lentivirus preparations were used for infection of primary neurons.
Membrane preparations
Striata were dissected from brains of 10-14 week old WT and HD mice and minced into pieces in homogenization buffer (50mM HEPES-Na, pH7.0, 200mM NaCl, 5mM MgCl2, 1mM DTT, 1mM EDTA and 0.25M sucrose containing protease inhibitors). Homogenates were prepared by passing striatal pieces through a dounce homogenizer for 12 strokes on ice and centrifuged at 4°C 14, 000rpm for 10 min. The post nuclear supernatant (S1) was further centrifuged at 4°C 55, 000 rpm for 1hr in a TLA120.2 rotor (Beckman-Coulter). The supernatant (S2, cytosol) was discarded and the pellet (P2, total membrane) was re-suspended in homogenization buffer supplemented with 1% Triton X-100 or directly in sample buffer. These striatal membrane preparations were used for nucleotide exchange activity assays and for determination of Rab11 and Rab5 levels by Western blot analysis.
To obtain sufficient amounts of soluble Rab11-enriched membrane from one mouse, we used whole brain preparations. Whole fresh WT and HD mouse brains were homogenized in 50mM Tris/Cl, pH7.4, 150mM NaCl, 1mM EDTA containing 0.5mM GTPγS and protease inhibitors as above. Post-nuclear supernatants were supplemented with sucrose to a final concentration of 40% (w/v), and overlayed with a 10 - 40% (w/v) continuous sucrose gradient and centrifuged at 4°C 100, 000×g for 24hr in a SW41 rotor (Beckman-Coulter). Twelve fractions, 1ml each, from top to bottom were collected. Equal volumes of each fraction were analyzed by SDS-PAGE and Western blotting. Rab11-positive membrane fractions were determined by analyzing the enrichment of Rab11 by Western blot. Rab11 from HD mouse brains is enriched in fraction-7, whereas Rab11 from WT mouse brains is equally distributed in fraction-6 and fraction-7. Rab11 in these fractions (HD fraction-7; WT fractions 6 and 7) was recoverable by a centrifugation at 4°C 100, 000×g for 1hr after diluting the sucrose concentration to 0.25M, indicating that these fractions have membranes. These soluble Rab11-enriched membrane preparations were used for GEF activity assay and for GDI-extraction assay (see below).
Guanine nucleotide exchange assay
[3H]GDP release was used to determine the GEF activity as performed previously (Jones et al., 1998) with minor modifications (Li et al., 2008). In brief, 0.5μg of purified GST-Rab11 on glutathione beads or 0.3μg of hexahistine (6×His)-Rab5 on Ni-NTA beads was loaded with 20 picomol of [3H]GDP (11.9Ci/mmol, Amersham) in 20mM HEPES, pH7.2, 20mM potassium acetate, 1mM DTT, 5mM EDTA at 30°C for 30min. Loaded GST-Rab11 on glutathione beads or 6×His-Rab5 on Ni-NTA beads was washed twice in loading buffer containing 10mM MgCl2 to remove unbound [3H]GDP. Samples (striatal total membranes or whole brain Rab11-enriched membranes) were diluted in assay buffer (20mM HEPES, pH7.2, 5mM Mg(OAc)2, 1mM DTT and 0.75mM GTP/GDP). After adding the [3H]GDP-loaded GST-Rab11 or 6×His-Rab5, the GEF reaction was initiated by incubating at 30°C for indicated times and stopped by cooling the samples on ice. After each reaction, GST-Rab11 on glutathione beads or 6×His-Rab5 on Ni-NTA beads was washed twice in cold wash buffer (20mM Tris/Cl, pH7.4, 20mM NaCl, 5mM MgCl2, 1mM DTT) and counted with Aquasol-2 (Du Pont) Scintillation fluid.
RabGDI dependent extraction of Rab11 from membranes
We used whole brain Rab11-enriched membranes for this assay. 0.1 μg of GST-RabGDI (a generous gift of Dr. William E Balch, the Scrippes Institute) was used for each reaction. GST-RabGDI was bound onto glutathione beads. After washes in extraction buffer (50mM HEPES-K, pH6.9, 100mM KCl, 10mM MgCl2, 1mM DTT and 0.5mM GDP), GST-RabGDI on glutathione beads was incubated with Rab11-enriched membranes in the absence (WT and HD) or presence (HD) of 5mM ATP at 37°C for 45min. Since there is more Rab11GDP on HD membranes and we wanted to maintain similar levels of Rab11GDP for extraction by RabGDI, we used 20μg HD membranes and 60μg WT membranes. The extracted Rab11 was in complex with GST-RabGDI on glutathione beads (released) and the un-extracted Rab11 remained in the solution. A centrifugation was applied to separate the extracted Rab11 on glutathione beads from the un-extracted Rab11 in the supernatant (unreleased). The extracted Rab11 on glutathione beads was washed once in extraction buffer and eluted into sample buffer. The un-extracted Rab11 in the supernatant was precipitated with chloroform/methanol and re-suspended in sample buffer. Samples were analyzed by SDS-PAGE and Western blot with anti-Rab11 and anti-GDI1 antibodies.
Isolation and culture of embryonic primary cortical neurons and glutamate challenge
Cortices were dissected from litters of E18 mice (WT and HD140Q/140Q), dissected free of meninges and other tissue, and incubated in HEPES-buffered Hanks' balanced salt solution (HBSS) without calcium and magnesium containing antibiotics (penicillin, streptomycin and neomycin) and 0.25% trypsin for 10 min at 37°C (all reagents were purchased from Invitrogen except where indicated). After incubation, cells were washed twice in HBSS supplemented with calcium and magnesium, then dissociated in Neurobasal DMEM medium containing B27 supplement, 25mM β-mercaptoethanol and L-glutamine, and seeded onto coverslips (Warner Instruments) or coated plates (BD biosciences) coated with poly-D-lysine and gelatin (CHEMICON). 24 to 48 hrs post plating, primary cultures were treated with cytosine arabinoside for 24 hrs and then changed into fresh media for culturing as above. Primary neurons were infected with lentivirus (MOI=30:1) expressing EGFP or lentivirus bi-cistronically expressing EGFP and Rab11Q70L or left alone for one week. Upon detecting EGFP expression under microscopy, primary neurons were challenged with 0.2mM glutamate for 20min and washed three times in pre-warmed culture medium to remove glutamate. The challenged primary neurons were cultured in fresh media for 24hr. After three washes in PBS, primary neurons were stained with trypan blue for counting the percentage of trypan blue-stained neurons under a microscope. Chi square test was used to determine the statistical significance.
Transferrin uptake
Synchronized uptake of transferrin was used to study the recycling of transferrin receptor in primary cortical neurons. Primary cortical neurons were seeded on coverslips and cultured as described above. At DIV7-9, primary cortical neurons on coverslips were washed and cultured in pre-warmed serum-free DMEM medium at 37°C for 1hr. Then, primary neurons were washed in ice-cold serum-free DMEM and incubated in cold DMEM on ice for 15min. Neurons were changed into fresh cold serum-free DMEM containing 5μg ml-1 Alexa568-transferrin (Invitrogen Inc.) and incubated on ice for 30min. After aspirating Alexa568-transferrin, neurons on coverslips were washed three times in cold serum-free DMEM to completely remove unbound Alexa568-transferrin. Neurons were chased in pre-warmed serum-free DMEM containing 10μg ml-1 unlabeled transferrin (Sigma-Aldrich) at 37°C for 2min or 15min. After two washes in cold PBS, neurons were fixed with 4% paraformaldehyde in PBS at room temperature for 15min, washed in PBS, incubated in 50mM NH4Cl to quench free aldehyde and stained with Hoechst 33258 (Invitrogen Inc.). The coverlips were mounted onto a slide containing drops of Prolong (Pierce), and examined using a fluorescence/confocal microscope (BioRad2100) after 24 hours.
Intensities of Alexa568-tranferrin signals in soma of neurons and the corresponding background signals were captured by confocal microscopy and the images were measured using NIH ImageJ. The average area of all examined neurons (n=42 WT and 42 HD neurons: 84 neurons) was used for normalizing the Alexa568-tranferrin fluorescence intensity in the soma region of each neuron. The mean intensity of Alexa568-tranferrin signals per neuron was graphed. Two-tailed Student t-test was performed to determine the statistical significance.
Immunohistochemistry and Western blot
For brain tissue immunohistochemistry, AAV-injected mice were perfused with cold 4% paraformaldehyde in PBS for 20 min. Brains were removed, stored overnight in PBS, and sectioned on a Vibratome at 40 μm. Brain slices were labeled with anti-HA (1:100) and detected with the immunoperoxidase method as described previously (DiFiglia et al., 1995). Western blot analysis was performed as described previously (Kegel et al., 2000). In brief, proteins in sample buffer were separated on a tris-glycine SDS gel by electrophoresis and transferred onto a nitrocellulose blot. The blot was blocked with 5% non-fat milk in PBS containing 0.05% Tween-20 and incubated with primary antibodies accompanied by secondary antibodies. Concentrations for primary antibodies are: monoclonal anti-Rab11 (1:1, 000), polyclonal anti-Rab5 (1:1, 000), polyclonal anti-calnexin (1:1, 000) and polyclonal anti-GDI (1:500). Peroxidate-conjugated secondary antibodies were diluted at 1:5, 000. Films were developed using enhanced ECL and scanned for densitometry analysis. Two-tailed student t-test was performed to determine the statistical significance.
Virus injection and Beam walking test
Mice were bred and tested according to the Animal protocol at the University of Massachusetts Medical School (A-978). The right striata of mice were injected with 6 × 109 g.c. of AAV-HARab11S25N or AAV-EGFP (0.7μl diluted in 2.3μl PBS for total of 3μl) with a 30G needle by means of an Ultra Micro Pump 3 (World Precision Instruments) at a speed of 0.1μl min-1. To test beam walking, mice were first familiarized with the testing situation by allowing them to cross a beam four times. During testing, each mouse was placed at one end of the beam and allowed to cross to the other end in four consecutive trials. The trials for each mouse were videotaped. The time to cross the beam was determined from the videotapes. For each testing date, time to cross was the average of four trials. Mice were sacrificed at 3 weeks post-injection for immunohistochemical studies.
Supplementary Material
Acknowledgments
Xueyi Li was supported by a John J. Wasmuth Postdoctoral Fellowship from the Hereditary Disease Foundation. Other support included grants from High Q/CHDI Foundation to M. D, Huntington's Disease Society of America to M. D. and from NIH (NS35711 to M. D. and NS38194 to N. A.). We are indebted to Dr. William E. Balch (The Scripps Research Institute, La Jolla USA) for his generous gift of GST-RabGDI.
Footnotes
Conflict interests claims: authors declared no conflict of interests.
References
- Alone DP, Tiwari AK, Mandal L, Li M, Mechler BM, Roy JK. Rab11 is required during Drosophila eye development. Int J Dev Biol. 2005;49:873–879. doi: 10.1387/ijdb.051986da. [DOI] [PubMed] [Google Scholar]
- Aoyama K, Suh SW, Hamby AM, Liu J, Chan WY, Chen Y, Swanson RA. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat Neurosci. 2006;9:119–126. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Bhuin T, Roy JK. Rab11 is required for embryonic nervous system development in Drosophila. Cell Tissue Res. 2009;335:349–356. doi: 10.1007/s00441-008-0711-8. [DOI] [PubMed] [Google Scholar]
- Broekman ML, Comer LA, Hyman BT, Sena-Esteves M. Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or -2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience. 2006;138:501–510. doi: 10.1016/j.neuroscience.2005.11.057. [DOI] [PubMed] [Google Scholar]
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- Correia SS, Bassani S, Brown TC, Lise MF, Backos DS, El-Husseini A, Passafaro M, Esteban JA. Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat Neurosci. 2008;11:457–466. doi: 10.1038/nn2063. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, Canals JM. Mutant Huntingtin Impairs Post-Golgi Trafficking to Lysosomes by Delocalizing Optineurin/Rab8 Complex from the Golgi Apparatus. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Hernandez M, Valera AG, Moran MA, Gomez-Ramos P, Alvarez-Castelao B, Castano JG, Hernandez F, Lucas JJ. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J Neurochem. 2006;98:1585–1596. doi: 10.1111/j.1471-4159.2006.03968.x. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Dure LSt, Young AB, Penney JB. Excitatory amino acid binding sites in the caudate nucleus and frontal cortex of Huntington's disease. Ann Neurol. 1991;30:785–793. doi: 10.1002/ana.410300607. [DOI] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Estrada Sanchez AM, Mejia-Toiber J, Massieu L. Excitotoxic neuronal death and the pathogenesis of Huntington's disease. Arch Med Res. 2008;39:265–276. doi: 10.1016/j.arcmed.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gonzalez MI, Susarla BT, Fournier KM, Sheldon AL, Robinson MB. Constitutive endocytosis and recycling of the neuronal glutamate transporter, excitatory amino acid carrier 1. J Neurochem. 2007;103:1917–1931. doi: 10.1111/j.1471-4159.2007.04881.x. [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Hickey MA, Kosmalska A, Enayati J, Cohen R, Zeitlin S, Levine MS, Chesselet MF. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–788. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- Jones MC, Caswell PT, Norman JC. Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol. 2006;18:549–557. doi: 10.1016/j.ceb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Jones S, Richardson CJ, Litt RJ, Segev N. Identification of regulators for Ypt1 GTPase nucleotide cycling. Mol Biol Cell. 1998;9:2819–2837. doi: 10.1091/mbc.9.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosh R, Augsburger A, Schwarz TL, Garrity PA. Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development. 2006;133:4655–4665. doi: 10.1242/dev.02650. [DOI] [PubMed] [Google Scholar]
- Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sapp E, Valencia A, Kegel KB, Qin ZH, Alexander J, Masso N, Reeves P, Ritch JJ, Zeitlin S, Aronin N, Difiglia M. A function of huntingtin in guanine nucleotide exchange on Rab11. Neuroreport. 2008;19:1643–1647. doi: 10.1097/WNR.0b013e328315cd4c. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- McEwen DP, Schumacher SM, Li Q, Benson MD, Iniguez-Lluhi JA, Van Genderen KM, Martens JR. Rab-GTPase-dependent endocytic recycling of Kv1.5 in atrial myocytes. J Biol Chem. 2007;282:29612–29620. doi: 10.1074/jbc.M704402200. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- Murphy TH, Miyamoto M, Sastre A, Schnaar RL, Coyle JT. Glutamate toxicity in a neuronal cell line involves inhibition of cystine transport leading to oxidative stress. Neuron. 1989;2:1547–1558. doi: 10.1016/0896-6273(89)90043-3. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J Cell Biol. 2006;172:605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Peter F, Nuoffer C, Pind SN, Balch WE. Guanine nucleotide dissociation inhibitor is essential for Rab1 function in budding from the endoplasmic reticulum and transport through the Golgi stack. J Cell Biol. 1994;126:1393–1406. doi: 10.1083/jcb.126.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci U S A. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Sakisaka T, Meerlo T, Matteson J, Plutner H, Balch WE. Rab-alphaGDI activity is regulated by a Hsp90 chaperone complex. Embo J. 2002;21:6125–6135. doi: 10.1093/emboj/cdf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Sheehan D, Ray GS, Calhoun BC, Goldenring JR. A somatodendritic distribution of Rab11 in rabbit brain neurons. Neuroreport. 1996;7:1297–1300. doi: 10.1097/00001756-199605170-00016. [DOI] [PubMed] [Google Scholar]
- Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171:1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, Gotoh Y, Ihara S, Nagata S, Itoh H, Fukui Y, Jessberger R. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature. 2002;416:759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]
- Soldati T, Riederer MA, Pfeffer SR. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, Gould M, Selkoe K, Stenoien DL, Young M, Faber PW, MacDonald ME, McPhaul MJ, Brady ST. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Trushina E, Singh RD, Dyer RB, Cao S, Shah VH, Parton RG, Pagano RE, McMurray CT. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum Mol Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- Wagster MV, Hedreen JC, Peyser CE, Folstein SE, Ross CA. Selective loss of [3H]kainic acid and [3H]AMPA binding in layer VI of frontal cortex in Huntington's disease. Exp Neurol. 1994;127:70–75. doi: 10.1006/exnr.1994.1081. [DOI] [PubMed] [Google Scholar]
- Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemskov EA, Nukina N. Impaired degradation of PKCalpha by proteasome in a cellular model of Huntington's disease. Neuroreport. 2003;14:1435–1438. doi: 10.1097/00001756-200308060-00006. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.