Abstract
In the past 2 years, new gene-targeting approaches using adeno-associated virus and designer zinc-finger nucleases have been successfully applied to the production of genetically modified ferrets, pigs, mice and zebrafish. Gene targeting using these tools has been combined with somatic cell nuclear transfer and germ cell transplantation to generate gene-targeted animal models. These new technical advances, which do not require the generation of embryonic stem cell-derived chimeras, will greatly accelerate the production of non-mouse animal models for biomedical research.
Keywords: gene targeting, recombinant adeno-associated virus, animal models, zinc-finger nucleases, homologous recombination, somatic cell nuclear transfer
Introduction
Animal models of human disease are critically important to biomedical research. Progress in gene targeting has allowed for genetic modification of new animal species, which has greatly facilitated the study of disease processes that are poorly modeled in mice. Targeted mutagenesis can be used to incorporate mutations or deletions into the coding region of a chosen gene—either to disrupt its expression or to produce a mutant protein that is associated with a particular human disease phenotype. Gene targeting can also be used to disrupt or alter cis-regulatory elements that control the expression of a given gene, or to study gene regulation through the targeted insertion of reporter genes. Owing to the technical feasibility of manipulating genes in cultured mouse embryonic stem (ES) cells, transgenic mice are now widely used as animal models in biomedical research. However, for many diseases, genetic mouse models have failed to replicate the human phenotype due to species-specific differences in cell biology and physiology. A well-known example is cystic fibrosis (CF); although more than 11 different CF mice have been created, none has reproduced the pathologies characteristic of the human CF lung or pancreas.
Although disease models from non-rodent mammals or other vertebrates are highly desired in biomedical research, the generation of genetically modified animals of this type has been extremely difficult. The biggest hurdle has been that ES cell lines, with the ability to contribute to the germ cell linage, have not yet been isolated from alternative non-rodent species. Thus, for decades, the production of transgenic non-rodent animals has been accomplished predominantly by the inefficient methodology of injecting DNA into the pronucleus of single-cell embryos. However, this technique is not compatible with site-directed mutagenesis because of the extremely low frequency of homologous recombination.
Given the limitations of ES cells in the generation of larger animal models, cloning by somatic cell nuclear transfer (SCNT)—using genetically modified diploid primary somatic cells (usually fetal fibroblasts) as donors—has been undertaken to produce gene-targeted animals. The major limiting factor in this approach has been the low efficiency of gene targeting in somatic cells. In recent years, a powerful promoter-trap screening strategy has been successfully applied; however, this approach is significantly limited to only genes that are actively expressed in primary fibroblasts. This particular limitation has been successfully overcome in the generation of both CF pig and ferret models by large-scale screening efforts using recombinant adeno-associated virus (rAAV) as a more efficient gene-targeting vector in somatic cells.
Progress
Cloning of CF pigs and ferrets from rAAV gene-targeted somatic cells by nuclear transfer has been successful
CF is one of the most common autosomal recessive diseases, and is caused by mutations in the gene that encodes the CFTR protein. The ΔF508 CFTR mutation, which gives rise to a 3-bp codon deletion that eliminates a phenylalanine at position 508 within exon 10, is present in 79% of patients with CF and allele carriers. CF mice have failed to replicate most of the pathological symptoms of human CF, including the lung defect that is the major cause of mortality in patients with CF. Ferrets and pigs are considered good candidates for modeling CF because of the similarities of their lung cell biology to that of humans.1,2
Initial attempts to target the CFTR gene in fibroblasts were based on transfection and homologous recombination (involving the injection of long linear DNA homologous fragments into the nuclei of fibroblast). These attempts failed badly, indicating that the methods routinely adopted for gene targeting in mouse ES cells are not suitable for targeting in somatic fibroblasts. Indeed, the absolute frequency of homologous recombination in this setting is as low as 2 × 10−7, and even when positive–negative selection (PNS) strategies were used to enrich for targeting events, the efficiency remained two orders of magnitude less efficient than in ES cells. Moreover, the level of nonhomologous integration is much higher in fibroblasts as compared to ES cells. The reason for differences in homologous recombination efficiencies between ES cells and somatic cells remains unclear, but it may be due to differences in chromatin architecture, DNA repair pathways and/or proliferative potential, which may influence the ability to expand and identify gene-targeted clones.
Gene targeting through adeno-associated virus (AAV) has, to some extent, overcome many of the limitations of linear DNA fragments for facilitating homologous recombination. AAV is a small, single-stranded DNA, helper-dependent parvovirus that has been widely used as a gene transfer vector for human gene therapy. The use of rAAV in gene targeting was first reported 10 years ago by David Russell and colleagues. rAAV was shown to yield a remarkably high frequency of site-directed mutagenesis in both cultured human tumor cell lines and primary fibroblasts. We have also observed high frequencies of AAV-mediated homologous recombination—up to 0.1% in human cell lines—even in the absence of selective enrichment. This frequency is at least 3–4 orders of magnitude higher than that achieved by conventional targeting approaches with linear DNA fragments. The mechanism of AAV-mediated homologous recombination remains poorly understood, but may involve unique intermediates of its single-stranded viral genome that are flanked by inverted terminal repeats (ITRs) with a highly ordered secondary structure. These characteristics of the AAV genome may allow it to more efficiently bind DNA repair enzymes. Furthermore, the ITRs that flank the ends of the genome may also prevent rapid degradation, allowing genomes to persist for long periods in the nucleus that may be required for target annealing. The ability of the AAV genome to facilitate homologous recombination appears to be very specific to this type of virus; lentiviral and adenoviral vectors have not worked well for applications of gene targeting. rAAV-mediated gene targeting was recently used to produce CFTR gene-targeted ferret and pig fibroblasts with either complete disruption of exon 10 or the ΔF508 deletion.3–5 These cells were then used as nuclear donors for SCNT-mediated cloning of pigs and ferrets. Only the pig CFTR-deficient model has thus far been bred to homozygosity. Importantly, newborn piglets demonstrate pathologies seen in human CF, including abnormalities of the nasal airways, pancreas, intestine, liver and other organs.4 In the context of lung abnormalities, nasal chloride permeability has been shown to be defective in CF newborn piglets; however, the characterization of bacterial colonization in the adult CF pig lung has not yet been completed.
rAAV has been used to efficiently target the CFTR gene in pig and ferret primary fibroblast
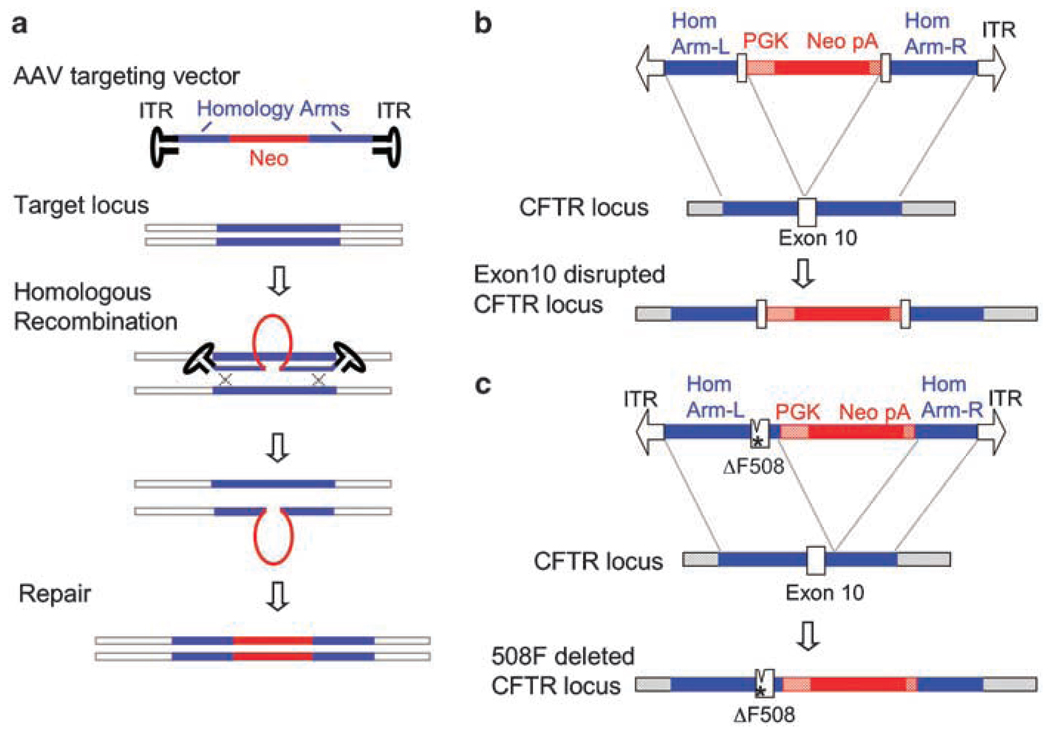
Much has been learned from the rAAV-mediated CFTR gene-targeting experience in the generation of the pig and ferret models. Careful consideration must be given when designing a rAAV-targeting vector, as the design differs significantly from that of the more conventional targeting vectors that have been used in ES cells. First, the optimal insert size for rAAV-targeting vectors (including regions of homology with the genome and a selectable marker expression cassette) is 4.4 kb, and it should never exceed 4.5 kb. For optimal efficiency of homologous recombination, a region with at least 1 kb of homology to the target sequence should be present on both sides of the selectable marker cassette (Figure 1a). When this type of vector construction is used, efficiencies of rAAV-mediated gene targeting are comparable to those achieved using conventional approaches, involving a PNS strategy in mouse ES cells or a promoter-trap strategy in somatic cells. The AAV vectors that were used for targeting the CFTR exon 10 disruption to the ferret and pig genomes of fetal fibroblasts included isogenic homologous sequence from the target cell line, as well as a phosphoglycerate kinase (PGK) promoter that drives expression of a neomycin gene in between the two homology arms (Figure 1b). However, it is currently unclear if it is necessary to use isogenic sequence for the homology arms to obtain high-efficiency targeting. Estimates of targeting efficiencies following first-round PCR screens of neomycin-resistant clones ranged from 0.07 to 10.93% for pig3 and 0.05 to 2% for ferret.5 The higher frequency of repetitive elements in the ferret genome near the CFTR target locus may be responsible for the lower efficiency in ferret CFTR targeting. Additionally, it remains unclear why, in both the ferret and the pig, different attempts at targeting in the same recipient cell line have resulted in large variability in efficiency.
Figure 1.
Recombinant adeno-associated viral vectors (rAAV)-mediating gene-targeting approaches used to generate new cystic fibrosis ferret and pig models. (a) Schematic representation of the mechanism of gene targeting using rAAV. Target sequence homology (blue) and a selectable marker (red) are packaged between two AAV inverted terminal repeats (ITRs). Following infection, homologous recombination between the rAAV vector and the target genomic locus mediates insertion of the selectable marker. (b) Design of rAAV vectors used to disrupt the coding sequence of the CFTR gene through targeted insertion of a neomycin expression cassette in exon 10. (c) Design of rAAV vectors used to delete the F508 codon in exon 10 of the CFTR gene. In this approach, the neomycin expression cassette is inserted into intron 11 of the CFTR gene.
As mentioned above, AAV-mediated gene targeting has also been used to introduce the disease-associated 3-bp deletion that results in expression of the ΔF508 CFTR mutant protein into the CFTR coding sequence. Generating this mutant required the introduction of two noncontiguous changes in the genome—deletion of the phenylalanine 508-encoding codon from exon 10 and the introduction of the neomycin selection cassette into intron 11 (Figure 1c). The distance between the desired mutation and the neomycin insertion was critically important in the design of this vector, as it ensured that the mutation was introduced primarily in neomycin-resistant cells. If this distance is too large, homologous recombination between the mutation and the selectable marker can occur more frequently and this reduces the rate of appearance of the mutation in neomycin-resistant clones. This approach has been successfully applied to generate a ΔF508 CFTR pig,3 and has also proven successful in the generation of targeted ferret fibroblasts (personal observation, ZY and JFE), although ferrets have not yet been cloned from these cells.
A regenerative cloning approach has been used to expand gene-targeted fibroblasts for SCNT
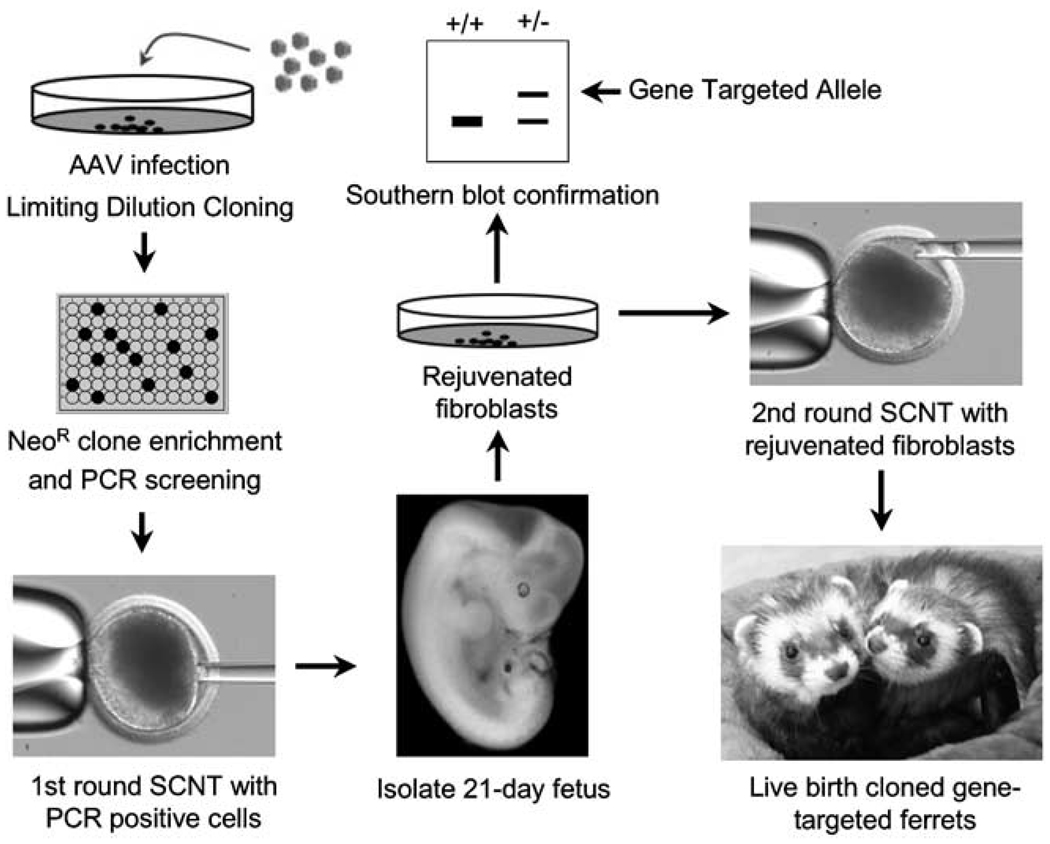
A significant obstacle to cloning animals from gene-targeted primary fibroblasts is the limited life span and rapid senescence of primary fibroblasts following selection. In addition, long-term culturing following selection can lead to chromosome aberrations, and consequently to heterogeneity in the populations derived from a single cell. Each of these obstacles can reduce the efficiency of SCNT. The life span of neomycin-resistant fetal fibroblasts appears to differ across species, and this may reflect species-specific toxicity of G418. In porcine cells, neither limited life span nor senescence has been an overwhelming problem and, although some variability in the life span of different gene-targeted clones has been observed, it has been possible to expand clones sufficiently for Southern blot confirmation of gene-targeting events before SCNT. In fetal fibroblasts from ferret, however, it has never been possible to expand gene-targeted fibroblast clones to this extent. This problem was solved using a sequential two-step cloning approach. The first round of SCNT used gene-targeted (PCR positive) senescent fibroblast clones as the nuclear donor to generate reconstructed oocytes. These reconstructed oocytes were transplanted into pseudopregnant female ferrets and cloned embryos were harvested at 21 days gestation and used to rederive highly proliferative gene-targeted fetal fibroblasts.5 These rejuvenate fibroblasts were then used for Southern blot genotyping and confirmation of normal karyotypes before a second round of SCNT cloning to produce live ferrets (Figure 2). This two-step recloning method has also been shown to be much more efficient than direct cloning of gene-targeted cells in the generation of α1,3-galactosyl-transferase gene knockout pigs by SCNT.6
Figure 2.
Summary of the somatic cell nuclear transfer (SCNT) approach used to produce CFTR gene-targeted ferrets. Primary fetal ferret fibroblasts are infected with rAAV-targeting virus and subsequently cloned by limiting dilution under G418 selection. Following replica plating and PCR screening in 96-well plates, cells are minimally expanded and then used for SCNT. Embryo fibroblasts (21 days) are then derived under G418 selection and screened by Southern blotting for the CFTR gene-targeting event. Rejuvenated fibroblasts containing the CFTR-targeted allele are then used for a second round of SCNT to clone live ferrets.
Cultured spermatogonial stem cells have been used to generate gene-targeted knockout mice through germ cell transplantation
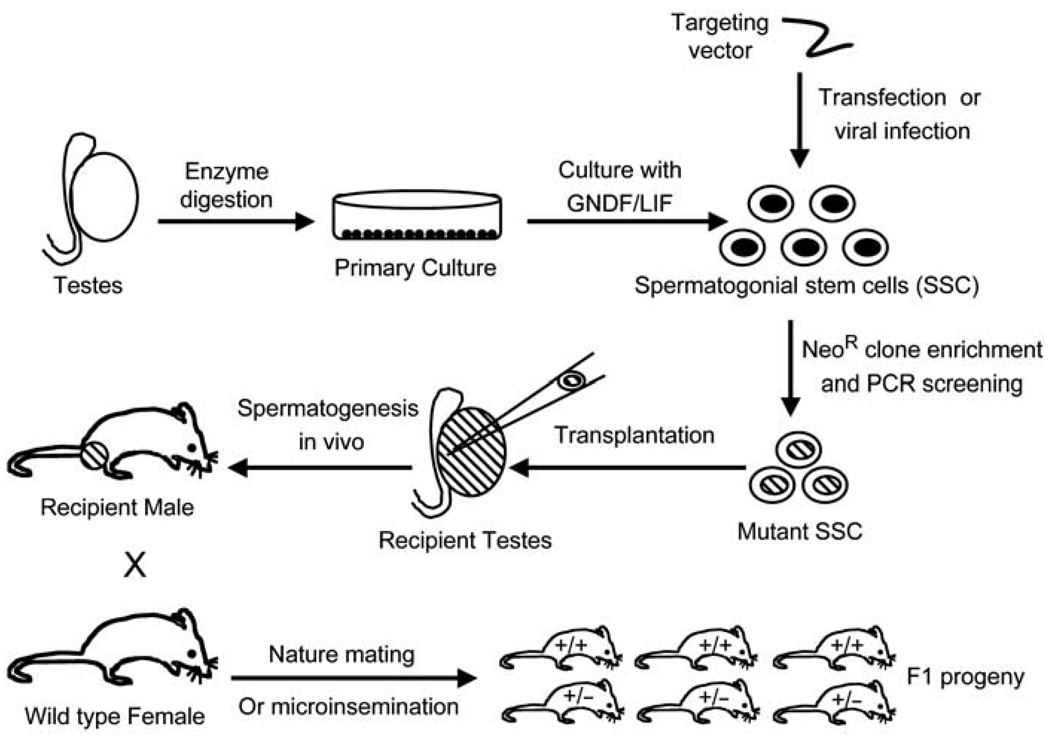
Recently, great technological progress in transgenesis has been made using mouse male germ line stem cells as a vehicle for genetic manipulation. This approach allows for the production of heterozygous mice with gene-targeted mutations (as opposed to chimeras generated by blastocyst injection of gene-targeted ES cells) through the mating of wild-type females and recipient males transplanted with genetically modified spermatogonial stem cells (SSCs) (Figure 3). This approach may streamline the generation of knockout models in the mouse, and may potentially also be applicable to the generation of large animal models.
Figure 3.
Summary of methods for producing gene knockout mice by gene targeting in male spermatogonial stem cells (SSCs) and germ cell transplantation. See text for details.
Germ-line cells in the adult reproductive organs of both sexes have the ability to transmit genetic information from parent to offspring. However, unique to the male germ line are stem cells (that is, SSCs) with a capacity for both self-renewal and differentiation into spermatozoa through spermatogenesis.7,8 Hence, genetic modification of SSCs can be carried out to create permanent mutations in the germ line. SSC transplantation into germ-cell-depleted recipient testes also has the advantage that it is a well-established technique, with demonstrated utility in species as diverse as mouse, rat, pig, goat and monkey. However, the use of germ cell transplantation as a method for transgenesis in animal models will be attractive only if SSCs can be expanded in culture for genetic manipulation. Thus, a significant amount of research has been carried out to understand how to propagate SSCs from multiple species.
One breakthrough in the propagation of SSCs came from the finding that glial cell line-derived neurotrophic factor (GDNF) promotes the survival and/or proliferation of these cells. Subsequently, it was discovered that the addition of both GDNF and leukemia inhibitory factor (LIF) to male germ-line stem cells collected from either the neonatal mouse testis or the adult testis allows the cells to be cultured and expanded. By calculating shortening of telomere length, it was estimated that SSCs have a limited proliferative potential of 34 months in vitro, hence allowing for an ~10120-fold expansion. Importantly, SSCs cultured for 2 years retain a normal karyotype and the ability to produce normal fertile offspring following transplantation into the seminiferous tubule.9–11 This feature allows for the genetic manipulation and selection of cultured germ-line stem cells. Transgenic animals have been generated from cultured SSCs, into which exogenous DNA sequences have been introduced, using both transfection and viral transduction (Figure 3). The selected transgenic cells are transplanted into the seminiferous tubules of infertile mice by microinjection. After transplantation, normal spermatogenesis occurs in the testis, and the recipients become fertile within months. Heterozygous offspring are then generated by mating or micro-insemination.
The transplantation of SSCs has led to the generation of not only transgenic mice, but also knockout mice for the occludin gene.12 Cultured mouse germ-line cells were transfected, by electroporation, with linear DNA containing a sequence targeting the occludin gene. From the total 2.4 × 108 transfected cells, 120 neomycin-resistant colonies (~2 × 106 cells per clone) were obtained after being cultured under G418 selection for 12–14 weeks. The population was enriched for recombinant clones using a promoter-trap vector that selectively activates the expression of a neomycin-resistant gene following homologous recombination, and the use of a diphtheria toxin A-chain gene at the terminal end of the targeting DNA sequence to select negatively against random integrants. Of these G418-resistant clones, 1.7% clones contained the correct targeting event and 85–90% of the gene-targeted clones retained a normal karyotype. Following transplantation of the targeted cells into the seminiferous tubules of sterile recipient males, 37.5% (18 of 48) of recipients became fertile within 4 months. Seventy-one percent (10 of 14) of the F1 offspring were heterozygous for the targeted occludin gene, and homozygous mutants were obtained in F2 progeny. These mice did not express occludin mRNA or protein at detectable levels.
This germ cell-based approach for generating genetic knockout mice has several advantages and disadvantages with respect to traditional ES cell-based procedures. For example, heterozygous mutants can be obtained in F1 offspring using SSC transplantation, whereas germ-line transmission using ES cell gene targeting requires breeding to the F2 generation to generate heterozygotes, because F1 generations are chimeras. However, due to the slow rate of SSC proliferation, clonal selection and expansion to obtain sufficient cells for both targeted colony screening and germ cell transplantation takes 12–14 weeks. Thus, the time required to generate gene knockout mice by these two procedures is comparable. However, the SSC-based approach may be more flexible with respect to generating mice on pure inbred backgrounds. In contrast, ES cell targeting is typically carried out on a strain 129 background to ensure optimal efficiency of germ-line transmission, and moving the targeted locus onto the appropriate inbred genetic background requires extensive breeding. Although ES cell exist for certain inbred strains of mice (namely, C57BL6), they are much less efficient than 129 ES cells and hence more costly to use.
Perhaps the greatest expected advantage of SSC-based gene targeting is its potential application in species for which ES cells are not available. Indeed, the feasibility of transgenesis by genetic modification of SSCs has been demonstrated in the rat using retroviral and lentiviral vectors,13 and also in the goat using rAAV2-transduced SSCs.14 Although live birth of transgenic goats was not evaluated (these experiments were terminated at the embryo stage), this same report demonstrated that in the case of mouse SSCs, rAAV2-mediated transduction led to transmission of the transgene through the male germ line and gave rise to the live birth of transgenic F1 progeny.14 It should be noted that the use of SSC transplantation to create gene knockouts has thus far only been successfully applied in mice. However, there is great potential for this technology to create gene knockout models in other species if certain hurdles can be overcome.
Challenges remain in the use of SSC transplantation for transgenesis in larger species
In spite of the above-discussed potential advantages of SSC transplantation and the significant progress that has already been made in resolving some of the hurdles, much remains to be done before it can be used effectively for transgenesis in larger species. The difficulties of long-term culture and expansion of SSCs due to the slow proliferation rates in vitro are significant. Moreover, SSC gene targeting would benefit from eliminating the requirement for both promoter-trap positive selection and negative selection against random integrants. Although the ability of rAAV to transduce mammalian SSCs14 suggests that this virus may be a useful tool for gene targeting of transcriptionally silent genes in SSCs, it is currently unclear if the challenges of cloning SSCs by limiting dilution or manual harvesting are compatible with the large-scale screening efforts needed for successful rAAV gene-targeting in fetal fibroblasts, as shown for the porcine and ferret CFTR gene.3,5
Zinc-finger nucleases have been successfully used to produce gene-targeted zebrafish
Over the last three decades, zebrafish have become an important and flexible model species for the study of human disease.15,16 Unlike most vertebrate model systems, zebrafish are small in size and develop externally—features of significant advantage with regard to genetic manipulation and economic large-scale forward and reverse genetic screens.17 The ability to create specific mutations at predetermined gene loci in the zebrafish genome would further increase the potential utility of this impressive model organism. Although zebrafish embryonic cells remain pluripotent and germ-lime competent for multiple passages in culture,18 and targeted insertion of exogenous DNA by homologous recombination in zebrafish ES-like cells has been accomplished, the ability of gene-targeted embryonic cells to produce viable germ cells following transplantation back into an embryo has thus far not been successful.19
Recently, a new approach using designer zinc-finger nucleases (ZFNs) has succeeded in introducing targeted mutations to the zebrafish genome.20 ZFNs are chimeric molecules consisting of a nonspecific FokI endonuclease cleavage domain and three or four engineered zinc-finger DNA-binding domains. Each of the zinc-finger DNA-binding domains interacts with a particular triplet base pair of DNA; their combination allows specific binding to 9- or 12-bp DNA motifs in the chromosome.21 Nuclease activity and efficient DNA cleavage requires that dimerization of two FokI subunits, each linked to independent ZFN sets, assemble on the DNA with appropriate geometry. The linkage of three or four ZFNs to each FokI subunit provides high specificity for DNA recognition at 18- or 24-bp motifs, respectively, ensuring that these enzymes produce double-stranded DNA breaks (DSBs) at only the site of interest in the genome.22,23
ZFN-induced DSBs can lead to targeted gene disruption, creating a knockout phenotype by altering the reading frame of the target gene. This occurs by error-prone, nonhomologous end joining (NHEJ) repair, which leads to base addition or deletion, at the ZFN cleavage site. Two groups have reported similar approaches—based on the injection of mRNAs encoding ZFNs into single-cell embryos—to direct targeted mutagenesis in the zebrafish germ line.24,25 ZFN-induced mutations in the zebrafish kdr gene are one example of the successful application of this approach; ZFN-induced mutagenesis at the target site occurred in 10% of embryos, and these mutations were transmitted through the germ line in approximately one-third of screened founders carrying the kdr gene lesion.25 Similar results were obtained in the case of ZFN-mediated mutagenesis of the zebrafish gol and ntl genes, with over half of the founders transmitting the disrupted ntl allele and the observed phenotypic changes at frequencies averaging 20%.24 Sequencing of these mutated genes revealed that this approach produced both deletions and insertions that are typical of the NHEJ repair pathway. Off-target cleavage rates of ~1–5% were observed when the three zinc-fingers were used to target the kdr allele, and this was reduced by increasing the number of zinc-fingers to four.25 The toxicity of injected ZFN mRNAs in zebrafish embryos was quite different in these two studies, ranging from pg to ng quantities of mRNA.24,25 These observed differences may reflect variation in the specificity of the ZNF cassettes in the two studies.
It is noteworthy that ZFN-induced DSBs can also enhance homologous recombination in the presence of mutagenic donor DNA with matching homologous sequences near the target cleavage site.23 Given the high efficiency of DNA cleavage by ZFNs in zebrafish embryos, one can envision modifications to this approach that allow for more sophisticated manipulation of the genome through ZFN-enhanced homologous recombination. In addition, this procedure may be applicable to other species in which embryo manipulation is already established.
Prospects
Although genetic manipulation in mice has contributed greatly to advances in biomedical research, there is an increasing need for flexible approaches for the genetic manipulation of the genomes of alternative species. Although SCNT cloning and germ cell transplantation have led to significant advances in the genetic engineering of alternative species over the past 2 years, the pace of advancements is still slow due to the inefficiencies of current gene-targeting techniques. However, the field appears to be poised for major breakthroughs in the near future. In this regard, the integration and combination of several new techniques for gene targeting and germ-line transmission may greatly enhance the field’s ability to precisely manipulate the genome of many species in years to come.
ZFNs have demonstrated enormous potential to improve the specificity of gene targeting. Not only have they been shown to selectively and efficiently target gene disruption as discussed above,26 but they have also been used to significantly enhance homologous recombination and the insertion of DNA sequences at specific sites in the genome.27 Highly effective ZFN delivery methods that take advantage of defective adenoviral vectors28 and integrase-defective lentiviral vector29 are currently under development. The application of ZFNs to gene-targeting technologies will likely also be greatly facilitated by the development of screening methods that will make it possible to rapidly identify zinc-finger combinations specific to new gene targets.
As discussed above, numerous new techniques for transgenesis in animal models are emerging. Although the vast majority of these techniques rely on expression of the target locus within the cell type that is to be manipulated, rAAV-mediated gene targeting has provided new opportunities for targeting transcriptionally silent genes. It is noteworthy that the efficiency of Raav-mediated homologous recombination, which is high to begin with, can be further enhanced by the introduction of DSBs near the target locus. The combination of rAAV and ZFNs in targeting transcriptionally silent genes thus should have a significant future impact on the development of new animal models. Indeed, the recent report of rAAV transduction in stem cells of not only the mouse male germ line, but also that of the goat, suggests that there is ample opportunity to adapt combinatorial rAAV and ZFN techniques for the genetic manipulation of cultured SSCs. The current success in producing gene knockout mice from genetically modified SSCs12 and from multipotent germ-line stem cells30 may also become applicable to other species in the near future. A better understanding of the basic biology of male germ cells and the processes that control spermatogenesis may also provide critical clues for overcoming the remaining technical challenges to applying this technology. In this regard, the recent establishment of human adult pluripotent germ-line stem cells from spermatogonial cells of adult human testis is an important breakthrough. 31 Future progress in the propagation of male germ-line stem cells from different species, together with enhancements in gene-targeting technologies, will provide tremendous new opportunities for directed genetic engineering of new animal models of human diseases.
Genetic engineering in nonhuman primates (NHP) also offers great potential for understanding the molecular basis of genetic disease and the development of effective therapies. Although gene-targeted NHP models have not yet been generated, a transgenic model of Huntington’s disease (HD) was recently generated in rhesus macaques.32 A previously developed method for transgenesis in NHPs was used to generate the HD model. This approach involved recombinant lentiviral infection of oocytes by injection of virus into the perivitelline space. The lentiviral vector encoded exon 1 of the human huntingtin (HTT) gene with an expanded polyglutamine CAG codon repeat, which is associated with HD. This technique of lentiviral-mediated gene delivery, in combination with intracytoplasmic sperm injection to fertilize the oocytes, successfully generated transgenic animals expressing the HTT minigene. Importantly, these animals developed morphological symptoms of HD, including nuclear inclusions and neuropil aggregates in the brain, and clinical symptoms of dystonia and chorea. Other progress toward genetic engineering of rhesus macaques has included the generation of NHP ES cell lines by SCNT.33,34 However, a major hurdle that remains in the use of ES cells for modeling in this species includes the fact that cloning of NHPs by SCNT has thus far been unsuccessful. Other techniques discussed in this review may provide new opportunities for the development of gene-targeted models of diseases in NHP.
In brief
Progress
Cloning of cystic fibrosis pigs and ferrets from recombinant adeno-associated virus (rAAV) gene-targeted somatic cells by nuclear transfer has been successful.
rAAV has been used to efficiently target the CFTR gene in pig and ferret primary fibroblast.
A regenerative cloning approach has been used to expand gene-targeted fibroblasts for somatic cell nuclear transfer.
Cultured spermatogonial stem cells have been used to generate gene-targeted knockout mice through germ cell transplantation.
Challenges remain in the use of spermatogonial stem cell transplantation for transgenesis in larger species.
Zinc-finger nucleases have been successfully used to produce gene-targeted zebrafish.
Prospects
More flexible and effective gene-targeting techniques for genome engineering in somatic cells are under development.
The development of new approaches for the production of genetically defined animal models from somatic cells and adult stem cells will be important in the application of this technology.
Disease modeling in nonhuman primates is beginning to show promise.
References
- 1.Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB, Jr, et al. The porcine lung as a potential model for cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;295:L240–L263. doi: 10.1152/ajplung.90203.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Luo M, Zhang L, Ding W, Yan Z, Engelhardt JF. Bioelectric properties of chloride channels in human, pig, ferret, and mouse airway epithelia. Am J Respir Cell Mol Biol. 2007;36:313–323. doi: 10.1165/rcmb.2006-0286OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers CS, Hao Y, Rokhlina T, Samuel M, Stoltz DA, Li Y, et al. Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. 2008;118:1571–1577. doi: 10.1172/JCI34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun X, Yan Z, Yi Y, Li Z, Lei D, Rogers CS, et al. Adeno-associated virus-targeted disruption of the CFTR gene in cloned ferrets. J Clin Invest. 2008;118:1578–1583. doi: 10.1172/JCI34599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimura T, Murakami H, Kurome M, Takahagi Y, Shigehisa T, Nagashima H. Effects of recloning on the efficiency of production of alpha 1,3-galactosyltransferase knockout pigs. J Reprod Dev. 2008;54:58–62. doi: 10.1262/jrd.19110. [DOI] [PubMed] [Google Scholar]
- 7.Dobrinski I. Transplantation of germ line stem cells for the study and manipulation of spermatogenesis. Ernst Schering Res Found Workshop. 2006;60:175–193. doi: 10.1007/3-540-31437-7_12. [DOI] [PubMed] [Google Scholar]
- 8.Dobrinski I. Male germ cell transplantation. Reprod Domest Anim. 2008;43 Suppl 2:288–294. doi: 10.1111/j.1439-0531.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 9.Kanatsu-Shinohara M, Inoue K, Lee J, Miki H, Ogonuki N, Toyokuni S, et al. Anchorage-independent growth of mouse male germline stem cells in vitro. Biol Reprod. 2006;74:522–529. doi: 10.1095/biolreprod.105.046441. [DOI] [PubMed] [Google Scholar]
- 10.Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Yoshida S, Toyokuni S, et al. Leukemia inhibitory factor enhances formation of germ cell colonies in neonatal mouse testis culture. Biol Reprod. 2007;76:55–62. doi: 10.1095/biolreprod.106.055863. [DOI] [PubMed] [Google Scholar]
- 11.Kubota H, Brinster RL. Technology insight: in vitro culture of spermatogonial stem cells and their potential therapeutic uses. Nat Clin Pract Endocrinol Metab. 2006;2:99–108. doi: 10.1038/ncpendmet0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanatsu-Shinohara M, Ikawa M, Takehashi M, Ogonuki N, Miki H, Inoue K, et al. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proc Natl Acad Sci USA. 2006;103:8018–8023. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu BY, Orwig KE, Oatley JM, Lin CC, Chang LJ, Avarbock MR, et al. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J Androl. 2007;28:353–360. doi: 10.2164/jandrol.106.001511. [DOI] [PubMed] [Google Scholar]
- 14.Honaramooz A, Megee S, Zeng W, Destrempes MM, Overton SA, Luo J, et al. Adeno-associated virus (AAV)-mediated transduction of male germ line stem cells results in transgene transmission after germ cell transplantation. FASEB J. 2008;22:374–382. doi: 10.1096/fj.07-8935com. [DOI] [PubMed] [Google Scholar]
- 15.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 16.Skromne I, Prince VE. Current perspectives in zebrafish reverse genetics: moving forward. Dev Dyn. 2008;237:861–882. doi: 10.1002/dvdy.21484. [DOI] [PubMed] [Google Scholar]
- 17.Jao LE, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:427–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L, Collodi P. Zebrafish embryonic stem cells. Methods Enzymol. 2006;418:64–77. doi: 10.1016/S0076-6879(06)18004-0. [DOI] [PubMed] [Google Scholar]
- 19.Fan L, Moon J, Crodian J, Collodi P. Homologous recombination in zebrafish ES cells. Transgenic Res. 2006;15:21–30. doi: 10.1007/s11248-005-3225-0. [DOI] [PubMed] [Google Scholar]
- 20.Woods IG, Schier AF. Targeted mutagenesis in zebrafish. Nat Biotechnol. 2008;26:650–651. doi: 10.1038/nbt0608-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 22.Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, et al. Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–D283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porteus M. Design and testing of zinc finger nucleases for use in mammalian cells. Methods Mol Biol. 2008;435:47–61. doi: 10.1007/978-1-59745-232-8_4. [DOI] [PubMed] [Google Scholar]
- 24.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 30.Takehashi M, Kanatsu-Shinohara M, Miki H, Lee J, Kazuki Y, Inoue K, et al. Production of knockout mice by gene targeting in multipotent germline stem cells. Dev Biol. 2007;312:344–352. doi: 10.1016/j.ydbio.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 31.Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, et al. Generation of pluripotent stem cells from adult human testis. Nature. 2008;465:344–349. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- 32.Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, et al. Towards a transgenic model of Huntington's disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 34.Cram DS, Song B, Trounson AO. Genotyping of Rhesus SCNT pluripotent stem cell lines. Nature. 2007;450:E12–E14. doi: 10.1038/nature06456. [DOI] [PubMed] [Google Scholar]