Abstract
Early diagnosis of lung cancer followed by surgery presently is the most effective treatment for non-small-cell lung cancer (NSCLC). An accurate, minimally invasive test that could detect early disease would permit timely intervention and potentially reduce mortality. Recent studies have shown that the peripheral blood can carry information related to the presence of disease, including prognostic information and information on therapeutic response. We have analyzed gene expression in peripheral blood mononuclear cell (PBMC) samples including 137 patients with NSCLC tumors and 91 patient controls with non-malignant lung conditions, including histologically diagnosed benign nodules. Subjects were primarily smokers and former smokers. We have identified a 29-gene signature that separates these two patient classes with 86% accuracy (91% sensitivity, 80% specificity). Accuracy in an independent validation set, including samples from a new location, was 78% (sensitivity of 76% and specificity of 82%). An analysis of this NSCLC-gene signature in 18 NSCLCs taken pre-surgery, with matched samples from 2-5 months post-surgery, showed that in 78% of cases, the signature was reduced post-surgery and disappeared entirely in 33%. Our results demonstrate the feasibility of using peripheral blood gene expression signatures to identify early-stage NSCLC in at-risk populations.
INTRODUCTION
Lung cancer is the second most-prevalent cancer occurring in both men and women in the United States, accounting for 162,000 deaths in 2008 (1), more than any other cancer. High-risk populations include smokers and former smokers, as well as individuals exposed to second-hand smoke, asbestos, and radon. Presently, there is no easily applied screening protocol for lung cancer similar to those used for breast, prostate, and colon cancers. Screening high-risk patients with low-dose spiral CT (LDCT) (2-5) identifies small, non-calcified pulmonary nodules in approximately 30-70% of high-risk individuals, but only a small proportion (0.4 to 2.7%) of detected nodules ultimately are diagnosed as lung cancers (6-8). Even using the best clinical algorithms, 20-55% of patients selected to undergo surgical lung biopsy for indeterminate lung nodules are found to have benign disease (4), and those that do not undergo immediate biopsy or surgery require sequential imaging studies resulting in continued radiation exposure.
Accordingly, efforts are in progress to develop complementary non-invasive diagnostics using techniques such as detection of methylated tumor DNA in sputum (9), serum proteomics (10-12), detection of auto-antibodies (13, 14), and gene expression profiling in sputum (15) and airway epithelial brushings (16). Although each of these approaches has its own merits, none has yet passed the exploratory stage. Biomarkers that could be identified from a simple blood test, a routine event associated with regular clinical office visits, would be ideal.
Given previous studies that have analyzed gene expression from peripheral blood mononuclear cells for cancer diagnosis or prognosis (17-21), the goals of this study were to determine whether we could identify a gene expression signature in PBMCs that would accurately distinguish patients with early-stage lung cancer from non-cancer controls with similar risk factors (i.e. matched for age, gender, race, and smoking history) and whether such a signature had value in predicting whether lung nodules detected by diagnostic X-ray or CT scans were malignant or benign.
METHODS
Study Populations
Study participants (Supplementary Tables 1A-1B) for the initial training sets were recruited from the University of Pennsylvania Medical Center (Penn) during the period 2003 through 2007: 91 subjects with a history of tobacco use without lung cancer, including 41 subjects that had one non-calcified lung nodule diagnosed as benign after biopsy, and 137 patients with newly diagnosed, histopathologically confirmed, non-small-cell lung cancer. All participants had blood collection in conjunction with a clinical visit or just prior to surgery. None of the case subjects had received any cancer therapy prior to blood collection. Subjects with any prior history of cancer, except non-melanoma skin cancer, were excluded. Obstructive lung disease was defined as an FEV1/FVC < 70%. We recruited a total of 298 cases and controls from Penn. We excluded 10 NSCLC patients that were diagnosed to have a second cancer, and arrays for 6 samples were removed as technical outliers (see Methods). The Penn samples were specifically recruited for this study. PBMC were purified at Penn and RNA extracted at Wistar. The study was approved by the Penn Institutional Review Board. We also received 90 RNA samples processed at the New York University Medical Center (NYUMC), 27 had acceptable RNA quality based on gel electrophoresis and Bioanalyzer analysis and only these 27 were further processed for array analysis. Samples from NYUMC were all collected under IRB approval, and are listed in Supplementary Table 1C.
PBMC Collection and Processing
Blood samples from Penn were drawn in two “CPT” tubes (BD). PBMC were isolated within 90 minutes of blood draw, washed in PBS, transferred into RNAlater (Ambion) and then stored at 4 °C overnight before transfer to −80 °C. A subset of patient PBMCs was analyzed by flow cytometry, with anti-CD3, CD4, CD8, CD14, CD16, CD19, or CD-56 antibodies or isotype controls (BD Biosciences), and analyzed using FlowJo software. Samples collected at NYUMC were processed within 2 hours from collection; PBMC were transferred to Trizol (Invitrogen) and stored at −80 °C. Extracted RNA was transferred to the Wistar Institute for further processing.
Sample Processing
RNA purification of the Penn samples was carried out at Wistar using TriReagent (Molecular Research), as recommended and controlled for quality using the Bioanalyzer. Only samples with 28S/16S ratios >0.75 were used for further studies. A constant amount (400ng) of total RNA was amplified, as recommended by Illumina. The NYU samples required DNAse-treatment before hybridization. Samples were processed as mixed batches of cases and controls and hybridized to the Illumina WG-6v2 human whole genome bead arrays (http://www.illumina.com/pages.ilmn?ID=197)
Array Quality Control and Pre-processing
All arrays were processed in the Wistar Institute Genomics Facility. Arrays were checked for outliers by computing the gene-wise, between-array, median correlation for all the arrays and comparing it with correlation for each array. An array was declared an outlier if the difference between its median correlation with other arrays versus the overall between-array median correlation was greater than 8 median absolute deviations. Non-outlier arrays were quantile normalized and background was subtracted from expression values. Non-informative probes were removed if their intensity was low relative to background in the majority of samples or if maximum ratio between any 2 samples was not at least 1.2. (See Supplementary Methods for details).
Analysis
Classification was performed using a Support Vector Machine with recursive feature elimination (SVM-RFE)(22) using random, tenfold, cross-validation repeated 10 times. Classification scores for each tested sample were recorded at each reduction step, down to a single gene. Average accuracy for each reduction step was calculated and all the genes at the points of maximal accuracy formed the initial discriminator, which then underwent additional reduction to form the final discriminator (see Supplementary Methods for details). Pathway analysis was carried out using Ingenuity Pathways Analysis software (http://www.ingenuity.com/). Significance of the changes in the SVM score before and after surgery was determined with a one-sided t-test.
Validation of the Classifier on Independent Samples
Each of the genes in the signature from SVM analysis of the microarray data identified in the training set is assigned a coefficient that defines its importance in the classifier. In validating or testing the accuracy of the signature on new samples that are not identified by class association, the analysis is carried out essentially as follows: The signature is applied as an equation of the form:
where A, B, C, etc. are the microarray expression levels of each of the signature genes, and a,b,c, etc. are the coefficients by which each expression level is multiplied to give a value for X (the classification score). The expression levels of the 29 genes [A, B, C...Z] determined by microarray for a new patient are each multiplied by the appropriate coefficient (a, b, c...z) to determine a classification score, “X.” If the threshold value of X is set to be zero, then patients with positive scores will be declared to have malignant disease and those with negative scores will be called non-malignant. The higher the positive score, the greater is the confidence of malignancy, and the more negative the score, the greater is the confidence of no malignancy (Supplementary Figure 2).
RESULTS
Characteristics of the Case and Control Populations
Clinical and demographic variables for 137 non-small-cell lung cancer (NSCLC) cases and 91 controls with non-malignant lung disease, including those with pathologically diagnosed benign nodules collected at the Penn, are summarized in Table 1 and detailed in Supplementary Tables 1A and 1B. The case and control groups were similar in terms of age, race, gender, and smoking history. Fifty-five percent of the cancer patients were Stage 1, 13%, Stage 2, and 32% Stages 3 and 4. Eighty-four percent of the control group and 93% of the NSCLC group were current or previous smokers. Samples used for independent validation included an additional 12 cases and 15 controls collected at the NYUMC and 26 additional cases and 2 controls collected at Penn (Supplementary Table 1C). These samples were not included in the studies to develop a general classifier.
Table 1.
Demographics of Patients
| Category | Cases (n=137) | Controls (n=91) |
|---|---|---|
| Age (yrs) | ||
| Average | 66 | 63 |
| Median | 68 | 64 |
| Max | 84 | 88 |
| Min | 39 | 38 |
| Gender | ||
| Male | 69 | 55 |
| Female | 68 | 36 |
| Race | ||
| Caucasian | 125 | 78 |
| African-American | 11 | 11 |
| Other | 1 | 1 |
| Tobacco Use | ||
| Current | 26 | 8 |
| Former | 102 | 68 |
| Never | 9 | 15 |
| Histology | ||
| Adenocarcinoma | 85 | NA |
| Squamous Cell Carcinoma | 42 | |
| NSCLC, NOS | 10 | |
| Cancer Stage | ||
| Stage I | 75 | |
| Stage II | 18 | |
| Stage III | 39 | NA |
| Stage IV | 5 | |
| Obstructive Lung Disease | ||
| Yes | 63 | 65 |
| No | 65 | 17 |
| Unknown | 9 | 9 |
| Benign Lung Nodule | ||
| Yes | NA | 41 |
| No | 50 |
Flow cytometry was performed on peripheral blood mononuclear cells (PBMC) from 35 cases and 14 controls collected at Penn. As shown in Supplementary Table 2, there were no significant differences in the percentages of T-cells, CD4 cells, B-cells, monocytes, or NK cells. The tumor group had a slightly lower percentage of CD8 cells (18.9%) than the controls (24.5%), which did reach significance (p=0.03).
Gene Expression in PBMC Can Identify Individuals with NSCLC
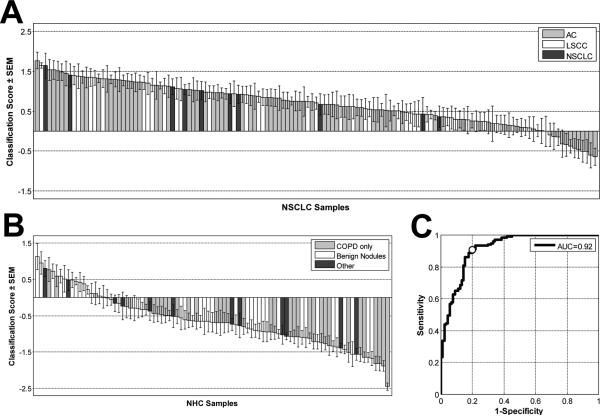
We compared gene expression profiles in PBMC samples from the 137 NSCLC cases to 91 controls with non-malignant lung disease. We applied a support vector machine with recursive feature elimination (SVM-RFE) and tenfold cross-validation (22) to the data to find the minimal number of genes that could most accurately distinguish the case and control groups by their PBMC gene expression (see Supplementary Methods and Supplementary Figure 1). We identified a 29-gene signature that distinguished the cases from controls with an overall classification accuracy of 86%, a sensitivity of 91%, and a specificity of 80%. The distribution of SVM scores, which measure how well a particular sample is classified, is shown in Figure 1A for each NSCLC patient and in Figure 1B for each control. The numerical classification score of each sample, together with its clinical annotation, is listed in Supplementary Table 3. The 29 genes used for classification are listed in Table 2 ordered by their SVM score, which is a measure of each gene's contribution to the classifier.
Figure 1. Classification scores assigned by the NSCLC- classifier to 137 NSCLC patients and 91 patients with non-malignant lung disease.
A positive score indicates classification as a cancer, a negative score as a non-malignant disease. The column heights are a measure of how well the sample is classified by the SVM algorithm for the 29 genes and the error bars are a measure of the classification variance across the 100 resamplings. SEM: standard error of the mean. A) NSCLC patients: AC-adenocarcinoma, LSCC-lung squamous cell carcinoma, NSCLC-samples not further characterized. B) Non-healthy control samples (NHC) include patients with non-malignant lung disease: COPD: only chronic obstructive pulmonary disease, Benign Nodules: (determined by biopsy), Other: various types of lung diseases C) Receiver-Operator Characteristic curve for classification of samples shown in A and B. AUC: area under the curve. White circle indicates sensitivity-specificity value corresponding to classification score threshold of 0.
Table 2.
Twenty-nine genes that distinguish patients with NSCLC from controls with non-malignant lung disease ordered by their contribution to the final classification score. Fold change = average change of NSCLC/NHC.
| # | Accession | Symbol | Description | Fold change |
|---|---|---|---|---|
| 1 | NM_016578 | RSF1 | Remodeling and spacing factor 1 | 1.27 |
| 2 | NM_003583 | DYRK2 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | −1.34 |
| 3 | NM_003403 | YY1 | YY1 transcription factor | −1.08 |
| 4 | NM_001031726 | C19orf12 | Chromosome 19 open reading frame 12 | 1.36 |
| 5 | NM_018473 | THEM2 | Thioesterase superfamily member 2 | −1.13 |
| 6 | NM_007118 | TRIO | Triple functional domain (PTPRF interacting) | −1.16 |
| 7 | NM_001020820 | MYADM | Myeloid-associated differentiation marker | −1.34 |
| 8 | NM_017450 | BAIAP2 | BAI1-associated protein 2 | −1.34 |
| 9 | NM_024589 | ROGDI | Rogdi homolog (Drosophila) | −1.18 |
| 10 | NM_024920 | DNAJB14 | DnaJ (Hsp40) homolog, subfamily B, member 14 | −1.14 |
| 11 | NM_199191 | BRE | TNFRSF1A modulator | 1.04 |
| 12 | NM_080652 | TMEM41A | Transmembrane protein 41A | 1.15 |
| 13 | NM_032307 | C9orf64 | Chromosome 9 open reading frame 64 | −1.14 |
| 14 | NM_031424 | FAM110A | Family with sequence similarity 110, member A | −1.14 |
| 15 | NM_014801 | PCNXL2 | Pecanex-like 2 (Drosophila) | 1.21 |
| 16 | NM_005612 | REST | RE1-silencing transcription factor | 1.29 |
| 17 | NM_014173 | C19orf62 | Chromosome 19 open reading frame 62 | 1.10 |
| 18 | NM_138779 | C13orf27 | Chromosome 13 open reading frame 27 | −1.18 |
| 19 | NM_022091 | ASCC3 | Activating signal cointegrator 1 complex subunit 3 | 1.83 |
| 20 | NM_005628 | SLC1A5 | Solute carrier family 1 (neutral amino acid transporter), member 5 | −1.16 |
| 21 | NM_016395 | PTPLAD1 | Protein tyrosine phosphatase-like A domain containing 1 | −1.22 |
| 22 | NM_005590 | MRE11A | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | −1.18 |
| 23 | NM_033107 | GTPBP10 | GTP-binding protein 10 (putative) (GTPBP10), transcript variant 2 | −1.27 |
| 24 | BX118737 | N/A | BX118737 Soares fetal liver spleen 1NFLS | −1.40 |
| 25 | NM_006217 | SERPINI2 | Serpin peptidase inhibitor, clade I (pancpin), member 2 | −1.41 |
| 26 | AK126342 | CREB1 | CAMP responsive element binding protein 1 | −1.45 |
| 27 | NM_016053 | CCDC53 | Coiled-coil domain containing 53 | −1.07 |
| 28 | NM_032236 | USP48 | Ubiquitin specific peptidase 48 | −1.17 |
| 29 | NM_001007072 | ZSCAN2 | Zinc finger and SCAN domain containing 2 | 1.18 |
Although an SVM score of 0 achieved the greatest degree of accuracy in separating case and control classes, additional clinical utility can be derived from this data by taking advantage of the value of the assigned SVM predictive score in the class assignments. For example, individuals with an SVM score of less than −0.65 are classified as controls with 100% specificity. Similarly, an SVM threshold of +0.65 or above would eliminate 12 of 17 false positives and could identify a lung cancer case with 95% sensitivity. The scores have confidence levels which are proportionate to the score itself as shown in Supplementary Figure 2. The ROC curve (Figure 1C) demonstrates the full spectrum of performance characteristics for various cutoffs of the SVM scores. The overall area under the curve (AUC) achieved by the classifier was 0.92.
To address the issue of data over-fitting and to test the generality of the classification model, we also performed the analysis using only 80% of the samples for training and set aside 20% of the samples for validation. We repeated that process for 5, non-overlapping, 20% set-asides. Similar average accuracies were found over the 5 training sets (81.8%) and the 5 validation sets (81.1%) (Supplementary Table 4) demonstrating the ability of the algorithm to classify new samples with the predicted accuracy. The overall accuracy is slightly reduced when using the smaller training sets (81% vs. 86%). The average accuracy of the analysis with randomly permuted sample labels was 58% across 10 permutation runs.
Classification Accuracy for Tumor Subclasses and by Smoking Status with the NSCLC Classifier
We also determined the accuracy of the NSCLC- classifier on histological subtypes and clinical tumor stages (Supplementary Table 6). The sensitivity for adenocarcinoma (AC) samples was 86%, while the squamous cell carcinomas (LSCC) were classified significantly better with 98% sensitivity (p=0.04, chi-squared test). We also determined whether classification sensitivity varied with increasing pathological stages. As shown in Supplementary Table 6, we find a significant increase in sensitivity from Stage 1A (83%) to Stages 3 and 4 (100%) (p=0.005, chi-squared test), suggesting the PBMC cancer signature becomes more pronounced with disease burden.
The accuracy of the NSCLC- classifier varied slightly based on the smoking status of the participants (although there are a limited number of non-smokers in the study population). The overall accuracy was 79%, 87%, and 88% for current, former, and never smokers, respectively (non-significant difference, p=0.28 by Fisher exact test). (The accuracy data based on smoking status and case/control status are shown in Supplementary Table 7).
The NSCLC signature was generated with controls from two different at-risk populations. About half (50) were “high risk” based on underlying lung disease and smoking history, while an additional 41 had been further diagnosed by CT or chest X-ray with lung nodules and were to undergo surgical evaluation. When we calculated classification accuracy for the two control populations separately, the NSCLC- classifier had a specificity of 89%, if only the “high risk” controls without lung nodules are considered, whereas the specificity was 71% for the controls with confirmed benign nodules. Although the difference in specificity appears to be large for these 2 control groups, it does not quite reach statistical significance (p=0.051, Fisher Exact Test), limited in part by sample numbers. However, we further explored this difference in accuracy by analyzing patients with confirmed benign nodules separately. We were able to obtain a 24-gene nodule classifier by cross-validation (Supplementary table 5) using only the 41 benign nodule samples as the control group and data from a randomly selected group of 54 NSCLC case samples. This classifier had a somewhat better apparent specificity of 80% as determined by SVM, but the difference in accuracy between the NSCLC and nodule classifiers did not reach significance (p=0.44, Fisher Exact Test). Because of its higher accuracy and potentially broader applicability, the following analyses were carried out with the 29 gene NSCLC- classifier
Validation of the NSCLC- Classifier on Independent Samples
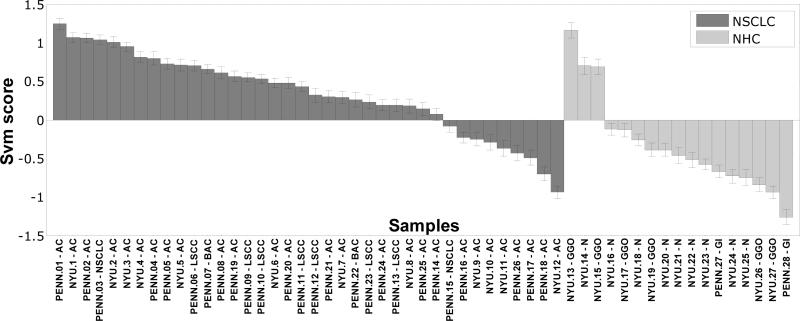
Although we had used cross-validation to establish our NSCLC- classifier, to further validate the utility of the classifier for analyzing new samples we assessed the classification accuracy using samples not included in the 29-gene selection process. The validation set included 38 NSCLC samples and 17 controls. Twenty-seven of the validation samples (Supplementary Table 1C) were collected at the NYU Lung Cancer Biomarker Center, an Early Detection Research Network (EDRN) Clinical and Epidemiologic Validation Center. The dataset included 12 Stage 1 NSCLC (5 of whom were never smokers) and 15 smoker and ex-smoker controls. Six of the controls were diagnosed by serial CT scans as having non-malignant Ground Glass Opacities (GGO) (23). No GGO patient samples were included in our original training set. The RNA for these samples was prepared at NYU. An additional 26 patients and 2 control samples were collected at Penn and had not been analyzed previously. The NSCLC classification algorithm is applied to these samples with no knowledge of whether a sample is a case or control (see Methods). The classification for the validation set is shown in Figure 2 and in more detail in Supplementary Table 8. The overall accuracy for the validation set was 78%, with 76% sensitivity and 82% specificity. This small decrease in accuracy and sensitivity (although with an increase in specificity) was not unexpected since the NYU samples were not specifically collected for these studies and, as a result, the sample collection and RNA purification were not standardized for these samples.
Figure 2. Application of the NSCLC classifier to independent validation sets.
PBMC-derived RNA of lung cancer patients and controls collected at the New York University Lung Cancer Biomarker Center have labels prefaced by NYU. Lung cancer and control RNAs collected at Penn are prefaced by Penn. IDs that end in GGO= ground glass opacities, GI= granulomatous inflammation, AC=adenocarcinoma, LSCC=lung squamous cell carcinoma, NSCLC=non-small cell lung cancer, and NHC=non-healthy control.
Effect of Tumor Removal on Individual Classification Scores
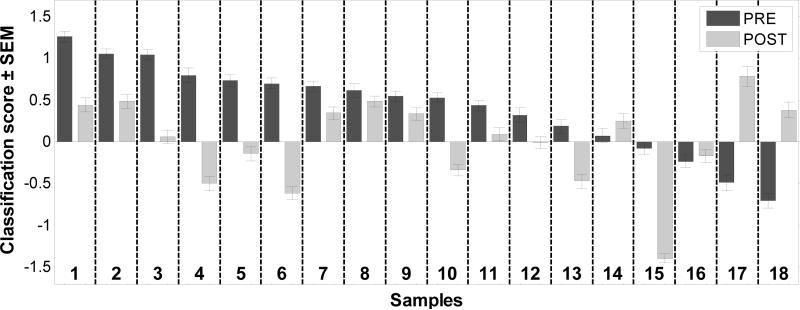
Eighteen of the NSCLC patients in the validation set shown in Figure 2 also had post-resection blood samples that were collected 2-5 months after surgery (Supplementary Table 9). To assess how the removal of the tumor affected the NSCLC- SVM score we had determined for the pre-surgery samples, we also determined the scores for the post-resection samples from each pair (Figure 3) Of the 14 patients that classified as cancer in the validation set (i.e. had positive SVM scores), 13 (93%) showed a decrease in their SVM scores in the post-resection samples. Five of these post-surgery samples (4, 5, 6, 10, and 13) had clearly negative SVM scores and would be classified as non-cancer samples in the analysis. Of the 4 misclassified, pre-surgery patients, 1 showed a highly decreased score and 3 showed increases in their scores. Although the time intervals between the first and second samples ranged between 2 and 5 months (Supplementary Table 9), there was no obvious relationship between the change in the scores and the time to post-resection sample collection. In the large majority of the patients (14 out of 18), tumor removal was associated with a decrease in the cancer signature score.
Figure 3. Classification scores are altered by tumor removal.
The samples are arranged as paired pre- and post-surgery samples to allow a comparison of the classification scores with the 29-gene diagnostic panel.
Effect Of Tumor Presence on Expression of Genes Associated with Immune Functions
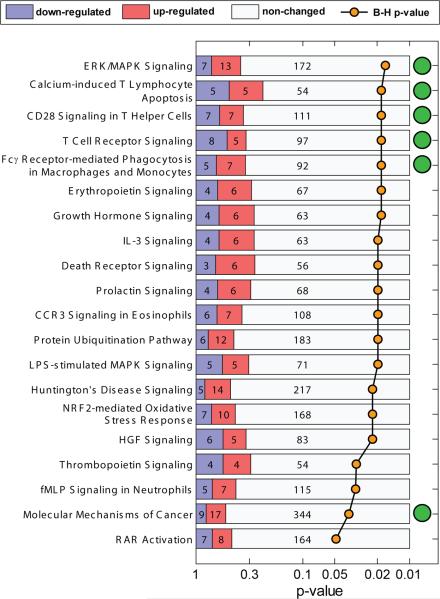
Although 29 genes were sufficient to distinguish cancer and control classes, many more statistically significant genes were differentially expressed providing some indication of the nature of the changes we are detecting. We used Ingenuity Core Analysis to determine the functions significantly and preferentially represented after correction for multiple testing in the top 1,000 significant genes from the NSCLC vs NHC and NSCLC vs. benign nodule comparisons (from a total of 2386 and 3276 differentially expressed genes respectively, p<0.05 by t-test). We did both analyses to further assess the similarities and differences between the genes identified in the 2 comparisons. Details are in Supplementary Methods. A list of statistically significantly enriched pathways is shown in Figure 4. As expected, pathways associated with specific immune functions are well represented, and highly significant, including pathways for CD28 and T-cell receptor signaling, calcium induced T-cell apoptosis. and macrophage and monocytes phagocytosis The top 5 pathways by p value in the NSCLC/NHC comparison are also found to be significant for the NSCLC vs. benign nodule comparison and rank among the top 6 pathways for that analysis. There were, in addition, 3 significantly enriched pathways that were unique to the latter comparison, SAPK/JNK Signaling, p38 MAPK Signaling and Lymphotoxin β Receptor Signaling.
Figure 4. Significantly enriched canonical pathways from Ingenuity Pathway Analysis of the genes differentially regulated between NSCLC and NHC samples.
Numbers in the bars show the number of genes in the pathway significantly higher in cancer (red) or lower in cancer (blue). B-H = Benjamini-Hochberg multiple testing correction. Green circles indicate pathways that were also enriched in NSCLC vs benign nodule comparison.
In addition to identifying significant canonical pathways, we looked at genes associated with functional categories. We focused on those functional categories associated with the innate and humoral immune response, in particular, those functions associated with inflammation and infection. The overlap of genes associated with these 2 processes is significant. Under the functional categories of cell mediated and humoral immunity, we found that 13/13 (p=9.2E-06) differentially expressed anti-pathogen response genes and 8/9 genes (p=5.04E-04) associated with the generation of reactive oxidative species, an end product of Toll Receptor (TLR) activation, are downregulated in the NSCLCs as compared to controls with benign nodules. In parallel we found that 7/7 antibacterial response genes are downregulated in the NSCLCs compared to all NHC (p=4.15E-02). Five genes are common to the 2 comparisons including Toll receptor 5 (TLR5), the surface receptor for bacterial lipopolysaccahrides. TLRs 1, 7 and 8 are down in NSCLCs compared to either control class. We also find that genes associated with activation of the NFκb pathway, through which the TLR signals are transmitted (24), are down while pathway inhibitory genes like IkB are up in NSCLC PBMC. Recently an important role for Toll receptor functions in respiratory diseases has emerged, in particular for COPD a condition affecting the majority of both our case and control subjects (24-26) suggesting that innate response pathways are suppressed in our cancer samples despite the presence of the activating condition of COPD.
DISCUSSION
We previously suggested that chemokines and cytokines released by malignant cells could impose a tumor-specific signature on normal immune cells of patients with non-hematopoietic cancers (27). Gene expression profiles from PBMC that identify blood signatures associated with a variety of cancers, including metastatic melanoma (18), breast (20), renal (17, 21), and bladder cancers (19) have now been reported. However, most of these studies have focused on later-stage cancers or response to therapy and used healthy control groups for comparison. We now have identified gene expression signatures in PBMC that can distinguish patients with early-stage NSCLC from appropriate at-risk controls with non-malignant lung diseases common to both patient and control classes.
The observed classification is not likely to be influenced by circulating tumor cells since 1) our classifiers do not contain genes characteristic of lung tumors such as SFTBP(28) or lung specific keratins (29); and 2) any tumor cells would be diluted to an extraordinary degree by the PBMC without efforts to enrich for such cells. This classifier appears not to be smoking dependent. Lung cancer in individuals who have never smoked has been shown to have several important differences from tobacco-associated lung tumors, and some molecular changes have been suggested to be unique to non-smokers (30, 31). There were 14 NSCLC patients in our study that had no prior history of smoking. Despite this, 11 of the 14 “never” smokers in our dataset were correctly classified as cancer by our NSCLC- panel.
The mechanism(s) for the effect we have detected remains to be determined. Interactions between the tumor and immune cells could be direct or mediated by cytokines or other tumor-released factors. The effects are enhanced with tumor progression, as evidenced by the increased accuracy of our gene panel in classifying late-stage NSCLC. Our ability to build a classifier from peripheral immune cells is consistent with recent findings from both mouse models and studies of immune suppression by tumors in humans. For example, Redente et al (32) showed, in a mouse lung-cancer model, that soluble factors produced in lung pre-malignant lesions influenced expression of specific macrophage activation markers in bone marrow macrophages and that the effect on gene expression was enhanced with tumor progression. The ability of tumors to induce myeloid-derived suppressor cells in lymph nodes, spleens, and peripheral blood in mouse models is now well established (33-35). The observation that tumor-resection results in disappearance of these myeloid-derived suppressor cells (36) supports our observations that the PBMC tumor signature diminished after tumor removal in the majority of the patients we examined. Similar tumor-induced suppressor cells in the PBMC fraction of blood also have been identified in human cancer patients (37, 38). Evidence from recent studies, comparing gene expression in PBMC and tumor-infiltrating lymphocytes from patients with either liver cirrhosis alone or in conjunction with liver cancer, suggests that the tumor presence can be communicated to the peripheral immune system and that the signal can be detected in the PBMC gene expression patterns (39). These observations support our finding that the NSCLC signature detected in PBMC diminishes in a majority of post-surgery patients.
The 5 pathways most significantly represented among the top 1,000 differentially expressed genes between cases and controls were significant for both the comparison of NSCLC and all controls and for the comparison of NSCLC and nodule controls. There is significant, but not complete overlap in the genes associated with these 5 pathways for the 2 comparisons. For 3 of the pathways (1, 2 and 5) <50% of the genes are common to both comparisons. Clearly there are significant similarities as well as some differences in the 2 comparisons we have carried out to identify our NSCLC general classifier. Recent studies have suggested that while diagnostic genes detected in various pathways may vary, the pathways themselves are better classifiers (40, 41).
We also identified some interesting differences between cases and controls in relation to immune response functional categories. The reduction in TLR expression in NSCLC was somewhat surprising as a high proportion of our patients and controls have COPD which would normally be expected to have activated TLR pathways (25). TLR function has been studied primarily in response to pathogens but a more expansive role in immune regulation has been emerging for recognition of self-antigens associated with auto-immunity (42-45). In addition endogenous ligands for Toll receptors have been identified including MUC1 a tumor expressed antigen that has been shown to be a negative regulator of TLR signaling (46) and heat shock proteins (47-51).
Our study follows the paradigm for biomarker development described by Pepe et al. (52) and adopted by the NCI Early Detection Research Network (EDRN). This paradigm first outlines the use of cross-sectional studies of patients with cancer versus appropriately chosen controls without disease to document initial estimates of sensitivity and specificity. Biomarkers meeting appropriate thresholds are then to be tested in external populations and finally in prospective studies. Following this model, our first analysis showed that a 29-gene panel could differentiate between a lung cancer population and an appropriate at-risk control population. Additional validation studies were then carried out on an external, independent dataset. Plans for prospective studies are in progress.
Although the NSCLC- signature could be developed as a screening tool for high-risk patients, the initial clinical use of our biomarkers is more likely to provide additional data to a clinician trying to evaluate a pulmonary nodule diagnosed by CT scan or chest X-ray. Based on prevalence data from a large CT screening study (3), the 29-gene NSCLC classifier has a PPV of 0.06 and an NPV of 1.0. (3) (Supplementary Table 10). This is comparable to the PPV and NPV values calculated using the same prevalence values for the 80-gene classifier derived from lung epithelial cells obtained from bronchial brushing recently described by Spira et al (16).
Since higher SVM score increases the likelihood of a sample being cancer the specific SVM value may be useful for clinical decision making in patients with suspected lung cancer or a non-calcified nodule and thus could help determine which patients require immediate interventions such as biopsy or surgical resection. This could potentially decrease the number of patients with benign lung nodules that would otherwise undergo biopsy or surgery (i.e. false positives).
Our results represent an encouraging first step, but several tasks remain to be addressed. Additional external validation sets are required to establish a standard collection protocol and to confirm the gene signatures and their accuracy. A larger prospective cohort study in patients with lung nodules is needed to more fully determine the role of smoking or other potentially confounding effects or diseases and to evaluate the overall clinical feasibility and utility of this approach. In addition, the observed reduction of the NSCLC cancer signature in the post-surgery samples suggests the possibility that post-surgery gene expression profiles might contain information predictive of recurrence. Ongoing follow-up studies are being conducted to determine the applicability of our approach to recurrence and response to therapy.
In summary, we have found gene expression signatures in PBMC that can distinguish individuals with early-stage NSCLC from individuals with non-malignant lung disease. The changes in PBMC gene expression with tumor removal suggest some specific functional effects of the tumor on the immune system that can be detected in the gene expression profiles. Although we have only examined NSCLC in this study, other types of lung cancer also may be detectible by gene expression in the peripheral immune cells.
*Gene expression data is available in the gene expression omnibus (GEO). The index code is GE12355.
Supplementary Material
ACKNOWLEDGEMENTS
We thank WenHwai Horng, Linda Alila, and Shere Billouin for technical assistance and support from the Genomics and Bioinformatics Cores. This project was supported by PA DOH Tobacco Settlement grants SAP 4100020718 and 4100038714, the PA DOH Commonwealth Universal Research Enhancement Program, EDRN Set-Aside funds. Wistar Cancer Center Support Grant P30 CA010815. A.V. was supported by NCI K07 CA111952. There were no conflicts of interests.
REFERENCES
- 1.ACS . Cancer Facts and Figures. American Cancer Society; Atlanta: 2007. 2008. [Google Scholar]
- 2.Diederich S, Wormanns D. Impact of low-dose CT on lung cancer screening. Lung Cancer. 2004;45(Suppl 2):S13–9. doi: 10.1016/j.lungcan.2004.07.997. [DOI] [PubMed] [Google Scholar]
- 3.Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, Miettinen OS. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–71. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
- 4.Jett JR. Limitations of screening for lung cancer with low-dose spiral computed tomography. Clin Cancer Res. 2005;11:4988s–92s. doi: 10.1158/1078-0432.CCR-05-9000. [DOI] [PubMed] [Google Scholar]
- 5.Mulshine JL. Current issues in lung cancer screening. Oncology (Williston Park) 2005;19:1724–30. discussion 30-1. [PubMed] [Google Scholar]
- 6.Bach PB, Jett JR, Pastorino U, Tockman MS, Swensen SJ, Begg CB. Computed Tomography Screening and Lung Cancer Outcomes. JAMA. 2007;297:953–61. doi: 10.1001/jama.297.9.953. [DOI] [PubMed] [Google Scholar]
- 7.Deppermann KM. Lung cancer screening--where we are in 2004 (take home messages). Lung Cancer. 2004;45(Suppl 2):S39–42. doi: 10.1016/j.lungcan.2004.07.994. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda K, Awai K, Mori T, Kawanaka K, Yamashita Y, Nomori H. Differential diagnosis of ground-glass opacity nodules: CT number analysis by three-dimensional computerized quantification. Chest. 2007;132:984–90. doi: 10.1378/chest.07-0793. [DOI] [PubMed] [Google Scholar]
- 9.Machida EO, Brock MV, Hooker CM, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res. 2006;66:6210–8. doi: 10.1158/0008-5472.CAN-05-4447. [DOI] [PubMed] [Google Scholar]
- 10.Gao WM, Kuick R, Orchekowski RP, et al. Distinctive serum protein profiles involving abundant proteins in lung cancer patients based upon antibody microarray analysis. BMC Cancer. 2005;5:110. doi: 10.1186/1471-2407-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patz EF, Jr., Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE., 2nd Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol. 2007;25:5578–83. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- 12.Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 13.Brichory FM, Misek DE, Yim AM, et al. An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proc Natl Acad Sci U S A. 2001;98:9824–9. doi: 10.1073/pnas.171320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pontes ER, Matos LC, da Silva EA, et al. Auto-antibodies in prostate cancer: humoral immune response to antigenic determinants coded by the differentially expressed transcripts FLJ23438 and VAMP3. Prostate. 2006;66:1463–73. doi: 10.1002/pros.20439. [DOI] [PubMed] [Google Scholar]
- 15.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 16.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–6. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 17.Burczynski ME, Twine NC, Dukart G, et al. Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res. 2005;11:1181–9. [PubMed] [Google Scholar]
- 18.Critchley-Thorne RJ, Yan N, Nacu S, Weber J, Holmes SP, Lee PP. Down-regulation of the interferon signaling pathway in T lymphocytes from patients with metastatic melanoma. PLoS Med. 2007;4:e176. doi: 10.1371/journal.pmed.0040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osman I, Bajorin DF, Sun TT, et al. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–80. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 20.Sharma P, Sahni NS, Tibshirani R, et al. Early detection of breast cancer based on gene-expression patterns in peripheral blood cells. Breast Cancer Res. 2005;7:R634–44. doi: 10.1186/bcr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twine N, Stover J, Marshall B. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. . Cancer Res. 2003;6:6069–75. al. e. [PubMed] [Google Scholar]
- 22.Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- 23.Ohta Y, Shimizu Y, Kobayashi T, et al. Pathologic and biological assessment of lung tumors showing ground-glass opacity. Ann Thorac Surg. 2006;81:1194–7. doi: 10.1016/j.athoracsur.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 24.Brody JS, Spira A. State of the art. Chronic obstructive pulmonary disease, inflammation, and lung cancer. Proc Am Thorac Soc. 2006;3:535–7. doi: 10.1513/pats.200603-089MS. [DOI] [PubMed] [Google Scholar]
- 25.Pan MM, Sun TY, Zhang HS. [Expression of toll-like receptors on CD14+ monocytes from patients with chronic obstructive pulmonary disease and smokers]. Zhonghua Yi Xue Za Zhi. 2008;88:2103–7. [PubMed] [Google Scholar]
- 26.Sabroe I, Whyte MK. Toll-like receptor (TLR)-based networks regulate neutrophilic inflammation in respiratory disease. Biochem Soc Trans. 2007;35:1492–5. doi: 10.1042/BST0351492. [DOI] [PubMed] [Google Scholar]
- 27.Kari L, Loboda A, Nebozhyn M, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477–88. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vachani A, Nebozhyn M, Singhal S, et al. A 10-Gene Classifier for Distinguishing Head and Neck Squamous Cell Carcinoma and Lung Squamous Cell Carcinoma. Clinical Cancer Research. 2007;13:2905–15. doi: 10.1158/1078-0432.CCR-06-1670. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–70. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 31.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 32.Redente EF, Orlicky DJ, Bouchard RJ, Malkinson AM. Tumor signaling to the bone marrow changes the phenotype of monocytes and pulmonary macrophages during urethane-induced primary lung tumorigenesis in A/J mice. Am J Pathol. 2007;170:693–708. doi: 10.2353/ajpath.2007.060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–79. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- 34.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: Recruitment, phenotype, properties, and mechanisms of immune suppression. Seminars in Cancer Biology. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Sinha P, Clements VK, Bunt SK, Albelda SM, Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol. 2007;179:977–83. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 36.Salvadori S, Martinelli G, Zier K. Resection of solid tumors reverses T cell defects and restores protective immunity. J Immunol. 2000;164:2214–20. doi: 10.4049/jimmunol.164.4.2214. [DOI] [PubMed] [Google Scholar]
- 37.Diaz-Montero C, Salem M, Nishimura M, Garrett-Mayer E, Cole D, Montero A. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. . Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kusmartsev S, Su Z, Heiser A, et al. Reversal of myeloid cell-mediated immunosuppression in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2008;14:8270–8. doi: 10.1158/1078-0432.CCR-08-0165. [DOI] [PubMed] [Google Scholar]
- 39.Sakai Y, Honda M, Fujinaga H, et al. Common transcriptional signature of tumor-infiltrating mononuclear inflammatory cells and peripheral blood mononuclear cells in hepatocellular carcinoma patients. Cancer Res. 2008;68:10267–79. doi: 10.1158/0008-5472.CAN-08-0911. [DOI] [PubMed] [Google Scholar]
- 40.Efroni S, Schaefer CF, Buetow KH. Identification of Key Processes Underlying Cancer Phenotypes Using Biologic Pathway Analysis. PLoS ONE. 2007;2:e425. doi: 10.1371/journal.pone.0000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee E, Chuang H-Y, Kim J-W, Ideker T, Lee D. Inferring Pathway Activity toward Precise Disease Classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Zhou Y, Feng G, Su SB. The critical role of Toll-like receptor signaling pathways in the induction and progression of autoimmune diseases. Curr Mol Med. 2009;9:365–74. doi: 10.2174/156652409787847137. [DOI] [PubMed] [Google Scholar]
- 43.Fischer M, Ehlers M. Toll-like receptors in autoimmunity. Ann N Y Acad Sci. 2008;1143:21–34. doi: 10.1196/annals.1443.012. [DOI] [PubMed] [Google Scholar]
- 44.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–69. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 45.Ehlers M, Ravetch JV. Opposing effects of Toll-like receptor stimulation induce autoimmunity or tolerance. Trends Immunol. 2007;28:74–9. doi: 10.1016/j.it.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Ueno K, Koga T, Kato K, et al. MUC1 mucin is a negative regulator of toll-like receptor signaling. Am J Respir Cell Mol Biol. 2008;38:263–8. doi: 10.1165/rcmb.2007-0336RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen K, Huang J, Gong W, Iribarren P, Dunlop NM, Wang JM. Toll-like receptors in inflammation, infection and cancer. Int Immunopharmacol. 2007;7:1271–85. doi: 10.1016/j.intimp.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Qazi KR, Oehlmann W, Singh M, Lopez MC, Fernandez C. Microbial heat shock protein 70 stimulatory properties have different TLR requirements. Vaccine. 2007;25:1096–103. doi: 10.1016/j.vaccine.2006.09.058. [DOI] [PubMed] [Google Scholar]
- 49.Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. J Leukoc Biol. 2004;76:514–9. doi: 10.1189/jlb.0304127. [DOI] [PubMed] [Google Scholar]
- 50.Tsan MF, Gao B. Heat shock proteins and immune system. J Leukoc Biol. 2009;85:905–10. doi: 10.1189/jlb.0109005. [DOI] [PubMed] [Google Scholar]
- 51.Vabulas RM, Wagner H, Schild H. Heat shock proteins as ligands of toll-like receptors. Curr Top Microbiol Immunol. 2002;270:169–84. doi: 10.1007/978-3-642-59430-4_11. [DOI] [PubMed] [Google Scholar]
- 52.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.