Abstract
How TGF-β-type ligands achieve signaling specificity during development is only partially understood. Here we show that Dawdle, one of four Activin-type ligands in Drosophila, preferentially signals through Baboc, one of three isoforms of the Activin Type-I receptor that are expressed during development. In cell culture, Dawdle signaling is active in the presence of the Type-II receptor Punt but not Wit, demonstrating that the Type-II receptor also contributes to the specificity of the signaling complex. During development, different larval tissues express unique combinations of these receptors, and ectopic expression of Baboc in a tissue were it is not normally expressed at high levels can make that tissue sensitive to Dawdle signaling. These results reveal a mechanism by which distinct cell types can discriminate between different Activin-type signals during development as a result of differential expression of Type-I receptor isoforms.
Keywords: TGF-β, Activin, Baboon, Isoform, Drosophila, Specificity
Introduction
The TGF-β superfamily of secreted signaling factors represents an ancient communication pathway that is conserved in all animals. TGF-β factors are dimeric ligands that bind to a complex of Type-II and Type-I receptors. After ligand binding, the Type-II receptor phosphorylates and activates the Type-I receptor, enabling it to recognize and phosphorylate an R- (Receptor-) Smad. Phosphorylated R-Smads bind a Co-Smad to form a trimeric complex that accumulates in the nucleus and regulates transcription in association with other cell type-specific factors (Shi and Massague, 2003).
The TGF-β signaling cascade is conserved throughout the animal kingdom and plays myriad roles within diverse tissues in both development and disease. In the fruit fly Drosophila melanogaster, the TGF-β superfamily is divided into the Bone Morphogenic Protein (BMP) and Activin branches (Parker et al., 2004). The Type-II receptors Punt and Wishful thinking (Wit) function in both pathways (Zheng et al., 2003), while the Type-I receptors Thickveins and Saxophone are BMP-specific receptors that phosphorylate the R-Smad Mad in response to the binding of one of the three BMP ligands: Decapentaplegic (Dpp), Screw (Scw), or Glass bottom boat (Gbb) (Bangi and Wharton, 2006; Shimmi and O’Connor, 2003). Hetero and homodimers of the BMP ligands signal through different combinations of these receptors, producing different levels of Mad phosphorylation. In contrast, The Activin pathway has only one Type-I receptor – Baboon (Babo) – for its four potential ligands: dActivin (dAct), Dawdle (Daw), Myoglianin (Myg), and Maverick (Mav) (Lo and Frasch, 1999; Nguyen et al., 2000; Parker et al., 2007; Serpe and O’Connor, 2006; Zhu et al., 2008). Several of these ligands have been shown to interact with Baboon and either Punt or Wit, leading to phosphorylation of Smox (dSmad2), the Activin R-Smad (Lee-Hoeflich et al., 2005; Parker et al., 2006; Serpe and O’Connor, 2006; Zhu et al., 2008).
Depending on the tissue, the Drosophila Activin ligands Dawdle and dActivin act either redundantly or independently to regulate proliferation, axon guidance and axon remodeling (Parker et al., 2006; Serpe and O’Connor, 2006; Zheng et al., 2003; Zhu et al., 2008). Both ligands are thought to signal via the Type-I receptor Baboon, yet the two ligands are not fully interchangeable. For example, overexpressing daw in the Drosophila wing disc does not phenocopy the large wings that result after similar overexpression of dAct (Gesualdi and Haerry, 2007) or an activated form of Baboon (Brummel et al., 1999). Why is the wing tissue sensitive to one Activin-like signal but not to another, even though both molecules apparently signal upstream of the same receptor in other contexts? How does the wing tissue discriminate between the signals? This issue is especially important in vertebrates, where many of the 33 different ligands circulate systemically via the blood but signal using only 7 Type-I receptors. How do different tissues discriminate between the many different ligands that they likely see during development?
One answer to these questions might involve tissue-specific expression of different receptor isoforms that vary in their extracellular ligand-binding domains. Previously, we described two such isoforms (baboa and babob) for the Activin Type-I receptor Baboon (Brummel et al., 1999), although whether they signal with only one or a subset of ligands has not been established. In this study we report the discovery of a third isoform, baboc. We demonstrate that in tissue culture Baboc is the only Type-I receptor capable of transducing a high level signal from the Activin-like ligand Dawdle, and does so only in the presence of a specific Type-II receptor, Punt. Furthermore, the three babo isoforms are differentially expressed in several issues during larval development. In the wing, for example, expression of baboc is not detected, and wing tissues do not respond to overexpression of daw (Gesualdi and Haerry 2007). However, when we ectopically express baboc, but not baboa and babob, together with daw in the wing, we observe a phenotype that mimics high-level signaling produced by expression of a strong activated Babo receptor. To our knowledge these findings are the first example of Type-I isoform-specific activation by a TGF-β ligand. This signaling specificity, coupled with differential, tissue-specific expression of receptor isoforms, exemplifies a new level of spatial regulation of TGF-β signaling during animal development.
Results
Dawdle signals via the Type-II receptor Punt, but not via the Type-II receptor Wit
Drosophila S2 cells respond to stimulation by TGF-β ligands in cell-based signaling assays as measured by increased phosphorylation of R-Smads (Ross et al., 2001; Serpe and O’Connor, 2006; Shimmi et al., 2005). Exposure to the Activin-like ligand Dawdle (Daw) results in increased levels of phosphorylated Smox (P-Smox), the Activin R-Smad, while addition of the BMP 5/6/7 homologue Glass bottom boat (Gbb) results in increased levels of phosphorylated Mad (P-Mad), the BMP R-Smad (Figure 1A; McCabe et al., 2003; Serpe and O’Connor, 2006). We sought to elucidate which receptors transduce Daw’s signal, starting with the Type-II receptors.
Figure 1. Dawdle signals in the presence of Punt, but not Wit.
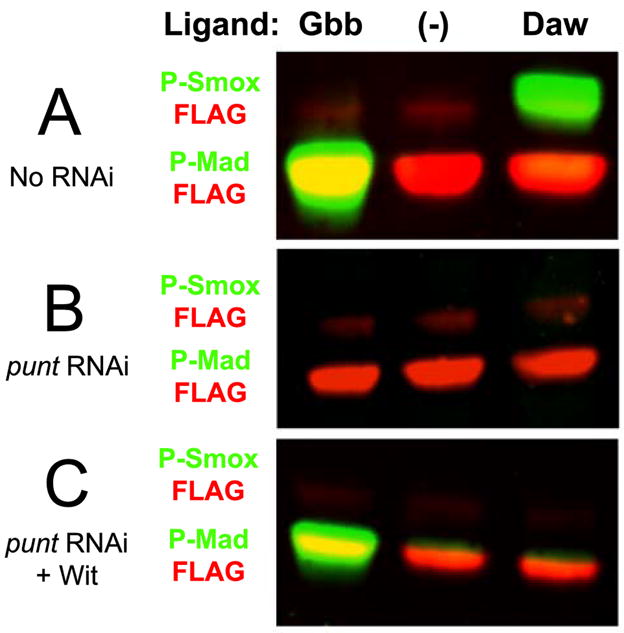
A) S2 cells with endogenous levels of receptors respond to Gbb and Daw by phosphorylating Mad and Smox, respectively, to high levels. B) S2 cells do not respond to Gbb or Daw when the punt transcript is targeted by double-stranded RNA. C) S2 cells respond to Gbb, but not to Daw, when Punt is removed by RNAi and Wit is expressed ectopically. In Figures 1, 3, and 4, phospho-specific antibodies (rabbit) for both species of R-Smad are shown in the green channel, while FLAG antibody (mouse) bound to the Smads’ N-terminal epitope tags is depicted in the red channel. Smad-FLAG levels demonstrate loading consistency; though FLAG-Smox (red) bands may be difficult to visualize in the color images, grayscale representations of the blots’ red channels are shown in Supplementary Figure 1, which demonstrates the presence of Smox in all lanes at comparable levels.
The Drosophila genome encodes two TGF-β Type-II receptors: Punt and Wishful thinking (Wit) (Childs et al., 1993; Marques et al., 2002). Punt is the only Type-II receptor expressed in S2 cells (McCabe et al., 2003), and eliminating its expression by adding double-stranded RNA complementary to a portion of its transcript rendered cells incapable of responding to Gbb or Daw (Figure 1B). This indicates that Punt is sufficient for mediating Daw signaling. To determine if Daw can also signal via Wit, we again targeted the punt transcript by RNAi but also transfected cells with a construct coding for Wit. As shown previously, Gbb is able to signal via Wit (Figure 1C; McCabe et al., 2003), but Daw is not able to signal when Wit is the only available Type-II receptor (Figure 1C). We conclude that in S2 cells, Punt is necessary and sufficient for Daw signaling, but Wit is neither necessary nor sufficient to transduce the same signal.
Isolation of baboc
The Drosophila Activin receptor Babo has two known isoforms, Baboa and Babob, which differ only in the extracellular domain that is critical for ligand binding (Brummel et al., 1999; Wrana et al., 1994). These isoforms appear to be products of differential splicing in which each isoform’s mRNA transcript contains a unique fourth exon. We discovered another isoform of baboon, baboc, with its own unique fourth exon that codes for a novel ligand-binding domain (Figure 2). Notably, all three of these isoform-specific exons encode an Asp-Phe-Cys-Asn motif that is typical of sequences found in the previously reported cysteine box region (Wrana et al., 1994) (Figure 2C). All Type-I receptors across the animal kingdom appear to possess an identical or similar motif that is closely preceded by two adjacent cysteine residues (as seen in Figure 2C). Because this D-F-C-N motif is not found in any other reading frame between Exons 3 and 5 in the baboon genomic locus, it appears that there are only three babo isoforms.
Figure 2. Baboonc is an isoform with a novel ligand-binding domain.
A) Schematic representation showing which exons encode which portions of the Baboon protein. Isoforms of Babo are identical except for an extracellular region encoded by the fourth exon (gray), which contains the cysteine box domain that is critical for ligand-binding. The transmembrane and intracellular kinase domains of the protein are also shown. Drawing is to scale and represents the Baboa isoform. B) Schematic representation of the region surrounding the fourth exon(s) of the baboon genomic locus: baboa, top; babob, middle; and baboc, bottom. Alternative splicing leads to inclusion of one isoform-specific fourth exon in the mRNA transcript; unspliced, coding exons are black, and exons that are spliced out of the transcript are gray. C) Divergent amino-acid sequences encoded by the unique fourth exons of each babo splice variant. Because each fourth exon encodes a diverged sequence interspersed between four conserved cysteine residues that are involved in disulfide bonds necessary for ligand binding (Kitisin et al., 2007), each isoform likely exhibits a unique Activin ligand-binding spectrum. Cysteine residues are shown in gray, and the horizontal solid bar denotes the cysteine box motif.
Baboonc is a necessary Type-I receptor for Dawdle signaling
We examined if different isoforms of Babo had different abilities to transduce the Daw signal. First, we measured expression of each isoform in S2 cells by RT-PCR. baboa was expressed at a low level compared to baboc, and the babob transcript was not clearly detected (Supplementary Figure 2A). To determine if Daw signaling utilizes a specific Babo isoform, we eliminated expression of individual isoforms by adding isoform (fourth exon) -specific double-stranded RNA to S2 cells. As illustrated in Supplementary Figure 2B these isoform-specific RNAis are able to knockdown specific isoform protein levels by >95%. We then exposed the cells to Dawdle and measured phosphorylation of Smox. To make sure that a cell’s inability to respond to Daw was due to the loss of the Babo isoform by RNA interference (RNAi) and was not due to the cells’ general inability to respond to a TGF-β signal, an aliquot from each batch of cells was exposed to Gbb, which signals through separate BMP Type-I receptors. As shown in Figures 3A–H, all batches of cells are able to respond to Gbb as measured by an increase in the levels of P-Mad.
Figure 3. Baboc is indispensible for Daw signaling.
A-H) Cells were treated with double-stranded RNA (dsRNA) that targeted different combinations of babo isoforms and were exposed to Gbb and Daw. Cells exposed to no dsRNA (A) or to dsRNA targeting isoforms A (C), B (D), or A and B (F) responded to Daw. In cells where baboc was targeted by RNAi, either alone (E) or in combination with other isoforms (B, G, and H), cells could no longer respond to Daw stimulation. No combination of dsRNA eliminated a response to Gbb (A-H).
While cells with endogenous levels of receptors respond robustly to Daw (Figure 3A), simultaneous RNAi against all three babo isoforms eliminated Smox phosphorylation (Figure 3B). RNAi against baboa or babob had no effect on the stimulation of Smox phosphorylation by Daw (Figures 3C and 3D), and cells with baboa and babob simultaneously targeted by RNAi, leaving only Baboc, also respond to Daw (Figure 3F). However, cells whose baboc was targeted by RNAi, regardless if other isoforms were similarly targeted, never transduce Daw’s signal (Figures 3E, 3G, and 3H). These data demonstrate that Baboc is a necessary Type-I receptor for Daw signaling in S2 cells.
Other isoforms of Baboon are neither necessary nor sufficient for Dawdle signaling
baboa and babob are expressed at lower levels than baboc in S2 cells (Supplementary Figure 2A). This observation raises the possibility that Baboa and Babob might indeed be able to transduce the Daw signal, but do not appear to do so in S2 cells because they are expressed at low levels. We increased the levels of Baboa and Babob in S2 cells by transfection with constructs encoding each individual isoform. This expression was combined with the RNAi strategy utilized in Figure 3 to enable us to test each isoform individually for its ability to transduce the Daw signal. As before, Gbb was able to stimulate each batch of cells, suggesting that any loss of Daw signaling was due to the loss of Babo and not to the cells’ general inability to respond to TGF-β ligands (Supplementary Figure 3).
As previously shown in Figure 3, cells expressing endogenous receptors respond robustly to Daw, and RNAi against Baboa and Babob, leaving only Baboc, had no effect on this response (Fig. 4A). Cells ectopically expressing Baboa had a baseline level of P-Smox even when exposed only to conditioned medium, indicating that the overexpressed receptor is able to signal to some extent in S2 cells in the absence of added ligand (Figure 4B). However, these cells still show a significant increase in Smox phosphorylation in response to Daw when endogenous Baboc was not targeted by RNAi (Figure 4B). In contrast, when Babob and Baboc were eliminated by RNAi, leaving only overexpressed Baboa, cells no longer respond to Daw (Fig. 4B). The same is true for Babob: cells overexpressing Babob could respond to Daw at levels above background stimulation, but only if endogenous Baboc was not targeted by RNAi (Figure 4C). These results suggest that neither Baboa nor Babob is capable on its own of responding to Daw, even under overexpressed conditions.
Figure 4. Daw cannot signal using other isoforms of Babo.
Daw is able to signal in the presence of endogenous receptors or when baboa and babob are targeted by RNAi, leaving only Baboc (A). Expression of baboa did not change cells’ ability to respond to Daw, but addition of dsRNA targeting babob and baboc, leaving only Baboa, left cells unable to respond to Daw stimulation (B). Similarly, cells expressing Babob could respond to Daw in the presence of endogenous Baboc, but could not respond if baboa and baboc were targeted by RNAi (C).
Isoforms of baboon are expressed in different tissues during development
We sought to determine whether individual babo isoforms are differentially expressed in various tissues during larval development. Previous in situ hybridization experiments have shown that all larval tissues express baboon mRNA (Brummel et al., 1999), but the probe that was used in these studies was complementary to a nucleotide sequence shared by all three babo isoforms. To determine if the isoforms of baboon have distinct spatial expression patterns during larval development, we isolated RNA from tissues that were dissected from third-instar larvae and performed semi-quantitative RT-PCR analysis using isoform-specific primers.
Consistent with the previous study of baboon expression, all larval tissues express at least one baboon isoform (Figure 5). The third-instar brain expresses predominantly baboa, while the wing disc expresses both baboa and babob but not high levels of baboc. In contrast baboc and not baboa or babob is expressed principally in the gut and fat body. Punt, the necessary and sufficient Type-II receptor for Daw signaling in S2 cells (Figure 1), is expressed ubiquitously during development (Childs et al., 1993).
Figure 5. Expression of isoforms of babo in third-instar larval tissues.

Primers for actin (A) and each isoform of babo (B-D) amplified PCR products from cDNA collected from tissues that were dissected from third-instar larvae. actin transcript was present in all tissues tested (A). baboa is expressed in the brain and the wing (B), and babob was detected primarily in the wing disc (C). baboc is expressed in the gut and the fat body, but not in the brain or the wing disc at measurable levels (D).
Dawdle induces wing patterning defects only in the presence of ectopically-expressed baboc
Since we found that signaling by Daw in tissue culture requires Baboc, the inability of Daw to produce a phenotype when ectopically expressed in the wing (Gesualdi, S.C. and Haerry, 2007) might be attributable to the low endogenous level of Baboc in the wing. To examine this possibility, we ectopically expressed Daw in the wing either with or without coexpression of Baboc.
Strongly activating the Activin pathway in the Drosophila wing disc results in significant vein-patterning defects, as shown by overexpression of a constitutively active form of Babo (Babo*) (Figures 6A vs. 6B). A milder ectopic vein phenotype is seen using weaker Babo* transgenes (Brummel et al., 1999) and is also seen upon overexpression of dActivin (Figure 6C). Ectopic wing expression of neither Daw nor Baboc alone induces a patterning phenotype (Figures 6D and 6E), but when Daw is co-overexpressed with Baboc we observe a striking defect in wing (Figure 6F) and disc (Supplemental Figure 4) patterning that is identical to that seen upon overexpression of a strong activated Babo receptor line. These patterning defects are not seen when Daw is co-overexpressed together with Baboa and Babob. (Figure 6G), nor are they seen when dActivin is co overexpressed with Baboc (Figure 6H). Taken together, these data suggest that Baboc is necessary in vivo to transduce a high level of Daw signal. Furthermore, overexpression of Baboc does not appear to enhance signaling by dActivin. Taken together, these data demonstrate that tissue-specific isoform expression can dictate which tissues respond to different Activin-family signal(s) during development.
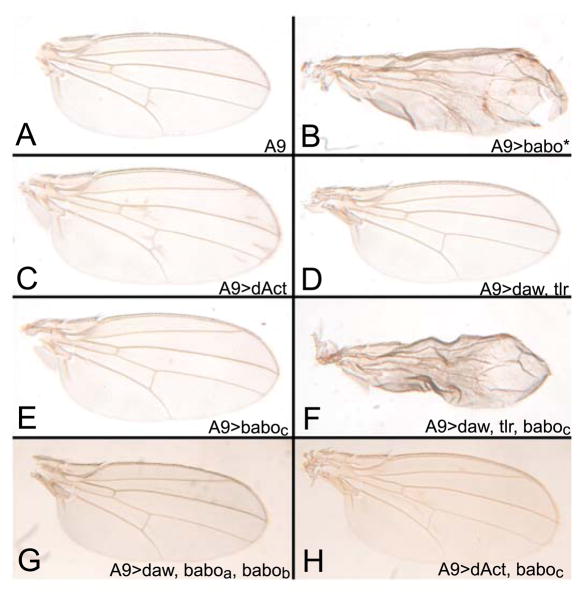
Figure 6. Dawdle overexpression induces wing patterning defects only in the presence of Baboc.
Patterning defects result from ectopic expression of a constitutively active form of Baboon (Babo*) using the A9 Gal4 driver (A and B). Overexpression of the ligand dActivin produces a moderate vein-patterning defect (C), but A9-driven Dawdle does not induce either of these phenotypes, even when it is co-overexpressed with its activating protease Tlr (Serpe and O’Connor, 2006) (D). However, patterning defects occur when Daw alone (not shown) or Daw and Tlr are co-overexpressed with Baboc (F), even though Baboc has no effect when it is expressed alone (E). Co-overexpression of Daw and both Baboa and Babob does not result in patterning defects (G). We know this recombinant line produces protein since it is able to rescue the neuroblast defects of babo mutants (Zhu et al. 2008). Simultaneous expression of dAct and Baboc does not result in a phenotype (H).
Discussion
The data presented in this study demonstrate that the gene for the Drosophila Activin receptor, Baboon, encodes three isoforms that differ only in their extracellular ligand-binding domains and that one of these isoforms, Baboc, is uniquely required for signaling by the Activin-like ligand Dawdle. The ability to express ligand-specific receptor isoforms is likely to have important implications for cells during development. For example, different profiles of receptor expression, combined with different ligand/receptor affinities, would allow neighboring cells to receive different levels of Activin signaling, even if they are exposed to the same suite of Activin ligands. Restricted receptor expression patterns also offer a means to enable systemically delivered ligands such as Daw to still exhibit tissue-specific effects. One example of this regulation may occur at the Drosophila neuromuscular junction, where multiple Activin-like ligands are expressed either pre- or post-synaptically [Zhu et al., 2008; Serpe and O’Connor, 2006; Parker et al., 2006; M.B.O., unpublished]. Differential expression of Baboon isoforms may be an important way for the muscle and neuron to distinguish between different Activin inputs.
Because Daw appears to signal though a specific Type-I receptor isoform that is not necessary for dAct signaling (Figure 6C), it is curious that the two ligands appear to function redundantly in one case, the regulation of neuroblast proliferation (Zhu et al., 2008). This is especially surprising since baboc is not highly expressed in the brain, at least as measured by low-cycle RT-PCR (Figure 5D). One possible explanation for this discrepancy is that baboc may be expressed only in a small subset of cell types in the brain, like neuroblasts, and our whole-brain RT-PCR using a pan-babo 5′ primer was biased towards the more prevalent baboa transcript. However, our attempts to examine the tissue distribution of individual isoforms by in situ hybridization using isoform-specific mRNA probes has not been successful. Ultimately, isoform-specific antibodies may be necessary to elucidate higher-resolution spatial expression patterns of the three isoforms. Such studies, together with a more careful analysis of each ligand’s expression pattern and the generation of isoform-specific loss-of-function mutants, will help elucidate the extent of potential functional redundancies between ligands and if specific receptor isoforms regulate unique biological processes.
Baboon is not the only Drosophila Type-I receptor with multiple isoforms: the Drosophila BMP receptors Sax and Tkv also have several isoforms that differ in their extracellular regions (Brummel et al., 1994). Similarly, recent work has uncovered Type-I receptor isoforms that are divergent in their extracellular domains in many mammalian species (Konrad et al., 2007). In both of these cases, however, the divergent isoforms do not affect the cysteine box as seen in the Babo isoforms. For this reason it is not clear if, or to what degree, these more subtle changes might affect affinity of ligand-binding or the ability to form effect signaling complexes. Genes encoding mammalian Type-II receptors also produce multiple isoforms that differ in their extracellular domains (Suzuki et al., 1994; Hirai and Fujita, 1996), and some of these isoforms can bind the same ligand with different affinities (Attisano et al., 1992) or bind different ligands with different affinities (Parker et al., 2007; Rotzer et al., 2001). Some of these isoforms are also expressed tissue-specifically (Rotzer et al., 2001). When coupled with these findings, our study uncovers an evolutionarily conserved mechanism by which cells can regulate their response to TGF-β ligands via the type of receptor isoforms that they express. The many potential combinations of Type-I and Type-II isoforms likely enable a cell to fine-tune its response when presented with numerous TGF-β family members, especially in mammals, where 33 ligands appear to signal via a limited set of 5 Type-II and 7 Type-I receptors (Kitisin et al., 2007).
In Drosophila, only one isoform of each Type-II receptor has been found, but because flies express so few ligands, many combinations of Type-I-Type-II signaling complexes may not be needed. For example, the Drosophila genome encodes two Type-II receptors and three isoforms of Babo, giving six combinations of homomeric Type-I/Type-II receptor complexes. The fly genome also encodes four Activin-style ligands (e.g., with nine cysteines versus the seven found in BMPs): dActivin, Dawdle, Myoglianin and Maverick. It is possible, therefore, that each ligand could have a specific combination of high affinity receptors for signaling. We have examined this possibility using S2 signaling assays, but we have been unable to see reproducible signaling in vitro from dActivin, Myoglianin, and Maverick, even in the presence of every combination of receptors (data not shown). Others have had noted similar difficulties (Gesualdi and Haerry, 2007; Lee-Hoeflich, 2005). Perhaps, in addition to expressing different isoforms of Baboon, cells also control other Activin-like signals by regulating expression of a necessary co-receptor not found in S2 cells. Indeed, a co-receptor is required by the ligand Nodal to phosphorylated Smads 2/3 in vertebrates (Yeo and Whitman, 2001).
In summary, our demonstration of Type-I receptor isoform-specific signaling reveals an additional mechanism by which signal specificity can be achieved within the TGF-β pathway. Our findings suggest that, by regulating which complement of receptor isoforms they express, together with different ligand/receptor affinities, distinct cell types within a tissue may be able to discriminate between Activin-family signals during development. This specificity may allow a tissue or cell type to maintain a unique level of Activin signaling, different from its neighbor’s, despite similar exposure to several systemically expressed ligands that can all activate a common intracellular Activin signaling cascade.
Experimental Procedures
S2 Signaling Assay
S2 cell culture and transfection for monitoring ligand signaling have been described previously (Serpe and O’Connor, 2006). Briefly, conditioned medium was collected from cells that were transfected with an expression construct coding for a Drosophila TGF-β ligand. Conditioned medium from mock-transfected cells was used as a negative control. Cells used in the signaling assay were all transfected with constructs coding for N-terminally FLAG-tagged Mad and Smox, and, as noted, were also transfected with constructs coding for Wishful thinking or various isoforms of Baboon. Double-stranded RNA was prepared as described previously (Shimmi and O’Connor, 2003), and 10 μg RNA was added to cultured cells at the time of transfection and then subsequently at 24 hours and 48 hours post-transfection. Separate batches of cells were transfected with ligand or with Smox and receptors. After 72 hours of incubation, 250μL of supernatant from ligand-expressing cells were added to 250μL of Smad/receptor-transfected cells suspended in their own culture media and incubated at room temperature with rolling for 90 minutes. Cells were then pelleted, resuspended in 1X SDS sample buffer, and boiled.
Western Blotting and Antibodies
Samples were loaded onto NuPAGE© 4–12% Bis-Tris Gels (Invitrogen) and transferred after electrophoresis to PVDF membrane (Bio-Rad). Membranes were probed with primary antibodies against phosphorylated Mad (gift from E. Leof, 1:1000), phosphorylated Smox (Cell Signaling, 1:1000), and the FLAG epitope (M2 from Sigma, 1:2000). The α-Alpha-Tubulin antibody (Sigma T-9026) was used 1:10,000. (IRDye secondary antibodies (Rockland) were used 1:2000, and blots were imaged on a Li-Cor Odyssey infrared imaging system. The α-Sal antibody (gift from A. Salzberg) was used 1:1000 on third-instar tissue that was fixed using standard methods.
Isolation of baboonc
The baboc isoform was identified by sequencing baboon PCR products amplified from a cDNA library. As illustrated in Supplementary Figure 2, the baboc isoform is also expressed by the embryonically-derived S2 cell line. The baboc cDNA sequence is available from FlyBase (www.flybase.org; transcript ID: FBtr0300599).
RNA Isolation and Reverse Transcription – Polymerase Chain Reaction (RT-PCR)
RNA was isolated from either S2 cells or from tissues that were dissected from third-instar Drosophila larvae using Trizol® (Invitrogen); 1 μg total RNA was reverse transcribed using the Thermoscript® RT-PCR kit (Invitrogen) and a Oligo dT primer. All products were used per suppliers’ instructions. 1 μL of the RT reaction was amplified by PCR for 25 cycles (S2 cDNA) or 30 cycles (fly-tissue cDNA). Sequences of primers used in PCR reactions are listed in Supplementary Table 1.
Drosophila stocks and husbandry
The A9 Gal4 driver was used to induce overexpression of transgenes in the wing disc (Brummel et al. 1999). A9 females were crossed to males carrying the following transgenes or combinations thereof, as noted: UAS babo*1A2, UAS dAct 2X, UAS baboc, or the UAS daw 9D2 UAS tlr2 recombinant. Flies were reared at 25 degrees C on standard food, and wings from adult females were mounted in standard mounting medium.
Supplementary Material
Acknowledgments
We thank members of the O’Connor lab for helpful discussions and A. Peterson for both technical support and comments on the manuscript. P.A.J. was funded, in part, by NIH predoctoral training grant T32 HD007430-11A1. T.L. and X.Z. were funded by NIH grant NS42049 to T.L. M.B.O. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attisano L, Wrana JL, Cheifetz S, Massague J. Novel activin receptors: distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell. 1992;68:97–108. doi: 10.1016/0092-8674(92)90209-u. [DOI] [PubMed] [Google Scholar]
- Bangi E, Wharton K. Dual function of the Drosophila Alk1/Alk2 ortholog Saxophone shapes the Bmp activity gradient in the wing imaginal disc. Development. 2006;133:3295–303. doi: 10.1242/dev.02513. [DOI] [PubMed] [Google Scholar]
- Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O’Connor MB. The Drosophila activin receptor baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes Dev. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs SR, Wrana JL, Arora K, Attisano L, O’Connor MB, Massague J. Identification of a Drosophila activin receptor. Proc Natl Acad Sci U S A. 1993;90:9475–9. doi: 10.1073/pnas.90.20.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. A gene complex acting downstream of Decapentapleic in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Gesualdi SC, Haerry TE. Distinct signaling of Drosophila Activin/TGF-beta family members. Fly (Austin) 2007;1:212–21. doi: 10.4161/fly.5116. [DOI] [PubMed] [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE 2007. 2007 doi: 10.1126/stke.3992007cm1. cm1. [DOI] [PubMed] [Google Scholar]
- Konrad L, Scheiber JA, Volck-Badouin E, Keilani MM, Laible L, Brandt H, Schmidt A, Aumuller G, Hofmann R. Alternative splicing of TGF-betas and their high-affinity receptors T beta RI, T beta RII and T beta RIII (betaglycan) reveal new variants in human prostatic cells. BMC Genomics. 2007;8:318. doi: 10.1186/1471-2164-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Zhao X, Mehra A, Attisano L. The Drosophila type II receptor, Wishful thinking, binds BMP and myoglianin to activate multiple TGFbeta family signaling pathways. FEBS Lett. 2005;579:4615–21. doi: 10.1016/j.febslet.2005.06.088. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech Dev. 1999;86:171–5. doi: 10.1016/s0925-4773(99)00108-2. [DOI] [PubMed] [Google Scholar]
- Marques G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–43. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, O’Connor MB. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39:241–54. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Parker L, Arora K. Identification of maverick, a novel member of the TGF-beta superfamily in Drosophila. Mech Dev. 2000;95:201–6. doi: 10.1016/s0925-4773(00)00338-5. [DOI] [PubMed] [Google Scholar]
- Parker L, Ellis JE, Nguyen MQ, Arora K. The divergent TGF-beta ligand Dawdle utilizes an activin pathway to influence axon guidance in Drosophila. Development. 2006;133:4981–91. doi: 10.1242/dev.02673. [DOI] [PubMed] [Google Scholar]
- Parker L, Stathakis DG, Arora K. Regulation of BMP and activin signaling in Drosophila. Prog Mol Subcell Biol. 2004;34:73–101. doi: 10.1007/978-3-642-18670-7_4. [DOI] [PubMed] [Google Scholar]
- Parker WL, Finnson KW, Soe-Lin H, Knaus P, Philip A. Expression and function of TbetaRII-B, a variant of the type II TGF-beta receptor, in human chondrocytes. Osteoarthritis Cartilage. 2007;15:442–53. doi: 10.1016/j.joca.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O’Connor MB, Marsh JL. Twisted gastrulation is a conserved extracellular BMP antagonist. Nature. 2001;410:479–83. doi: 10.1038/35068578. [DOI] [PubMed] [Google Scholar]
- Rotzer D, Roth M, Lutz M, Lindemann D, Sebald W, Knaus P. Type III TGF-beta receptor-independent signalling of TGF-beta2 via TbetaRII-B, an alternatively spliced TGF-beta type II receptor. Embo J. 2001;20:480–90. doi: 10.1093/emboj/20.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe M, O’Connor MB. The metalloprotease tolloid-related and its TGF-beta-like substrate Dawdle regulate Drosophila motoneuron axon guidance. Development. 2006;133:4969–79. doi: 10.1242/dev.02711. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shimmi O, O’Connor MB. Physical properties of Tld, Sog, Tsg and Dpp protein interactions are predicted to help create a sharp boundary in Bmp signals during dorsoventral patterning of the Drosophila embryo. Development. 2003;130:4673–82. doi: 10.1242/dev.00684. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O’Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005 doi: 10.1016/j.cell.2005.02.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Tran H, Attisano L, Arora K, Childs SR, Massague J, O’Connor MB. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol. 1994;14:944–50. doi: 10.1128/mcb.14.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7:949–57. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY-H, Martin J, O’Connor MB, C-HJ L, Lee T. Baboon/Smad2-mediated TGF-β signaling activates ecdysone receptor B1 expression during neuronal remodeling in the Drosophila Brain. Cell. 2003 doi: 10.1016/s0092-8674(03)00072-2. in press. [DOI] [PubMed] [Google Scholar]
- Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O’Connor MB. Drosophila Activin- and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development. 2008;135:513–21. doi: 10.1242/dev.010876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.