Abstract
HIV-1 protease processes the Gag and Gag-Pol polyproteins into mature structural and functional proteins, including itself, and is therefore indispensable for viral maturation1,2. The mature protease is active only as a dimer3–5 with each subunit contributing catalytic residues6. The full-length transframe region protease precursor appears to be monomeric yet undergoes maturation via intramolecular cleavage of a putative precursor dimer5,7–11, concomitant with the appearance of mature-like catalytic activity7,9. How such intramolecular cleavage can occur when the amino and carboxy termini of the mature protease are part of an intersubunit β-sheet located distal from the active site is unclear. Here we visualize the early events in N-terminal autoprocessing using an inactive mini-precursor with a four-residue N-terminal extension that mimics the transframe region protease precursor5,12. Using paramagnetic relaxation enhancement, a technique that is exquisitely sensitive to the presence of minor species13–16, we show that the mini-precursor forms highly transient, lowly populated (3–5%) dimeric encounter complexes that involve the mature dimer interface but occupy a wide range of subunit orientations relative to the mature dimer. Furthermore, the occupancy of the mature dimer configuration constitutes a very small fraction of the self-associated species (accounting for the very low enzymatic activity of the protease precursor), and the N-terminal extension makes transient intra- and intersubunit contacts with the substrate binding site and is therefore available for autocleavage when the correct dimer orientation is sampled within the encounter complex ensemble.
The regulation of HIV-1 protease autoprocessing is modulated by the N-terminal flanking transframe region (TFR) sequence (Fig. 1a)2. The catalytic activity of the monomeric protease precursor is approximately three orders of magnitude less than that of the mature protease dimer (which has a monomer–dimer equilibrium dissociation constant Kd < 10 nM)2,5. The appearance of mature-like catalytic activity and stable dimer formation is directly correlated with a single rate-limiting step comprising intramolecular (first order) cleavage of a putative transient dimeric precursor species at the p6pol–protease (PR) junction7,9,10. Mutations within the latter that prevent cleavage lead to the production of an N-terminally extended 17-kDa protease precursor species, and cause a severe defect in Gag polyprotein processing and the complete loss of viral infectivity in vivo17,18. Subsequent cleavage at the C terminus of protease at the PR–reverse transcriptase (RT) junction (Fig. 1a) occurs via an intermolecular (second order) reaction catalysed by a fully active protease dimer19. Mutations within the PR–RT junction that block C-terminal cleavage do not significantly affect either enzymatic activity and dimerization of the protease in vitro19,20 or processing of HIV-1 precursor proteins, virus maturation, viability and morphology in vivo20, indicating that the presence of the C-terminal reverse transcriptase sequence has negligible influence on the protease precursor19,20. Thus, only autoprocessing at the N terminus of protease at the p6pol–PR junction is an absolute prerequisite for stable protease dimer formation, the appearance of mature catalytic activity and complete processing of viral precursors. Before cleavage at the p6pol–PR junction, intermediate precursor forms may be liberated by intramolecular cleavage at competing sites (for example, p2–NC and TFP–p6pol; see Fig. 1a) that become available for productive binding and hydrolysis11, but these precursors will show the same low catalytic activity as that of the p6pol–PR precursor9,10.
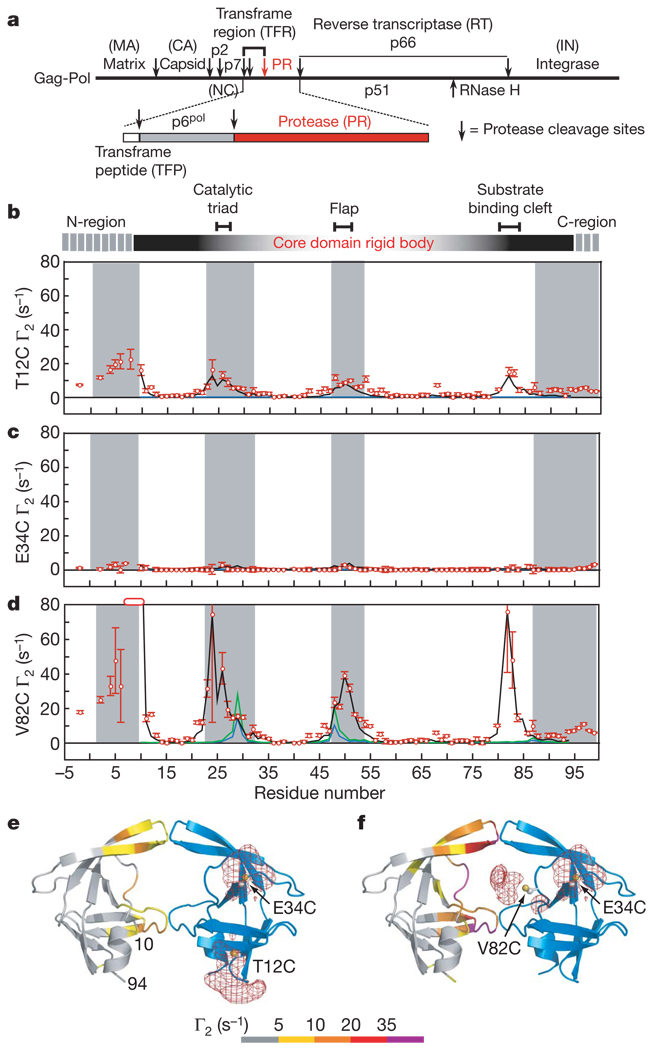
Figure 1. Intermolecular PRE profiles.
a, Organization of the Gag-Pol polyprotein1,2. b–d, Intermolecular PREs observed on U-[2H/13C/ 15N]-labelled SFNFPR(D25N) originating from a spin label conjugated to T12C (b), E34C (c) and V82C (d) of SFNFPR(D25N) at natural isotopic abundance. Residues broadened beyond detection are denoted by open bars. Error bars represent 1 s.d. Γ2 rates back-calculated from the structure of the mature dimer (for the core residues 10–94) at populations of 1% and 2% are shown as blue and green lines, respectively. Average Γ2 rates derived from the top 20 structures of the Ne = 4 simulated annealing calculations at a population of 5% heterodimer are shown as black lines. Grey shaded areas delineate residues that are buried at the dimer interface in the mature protease. e, f, Observed intermolecular PREs originating from the spin label attached to T12C (e) and V82C (f) colour-coded on a ribbon diagram of the mature dimer24 (spin label attached to the blue subunit). Atomic probability density maps25 (plotted at a threshold of 10% of maximum) showing the distribution of the spin-label oxygen radicals are shown as red meshes.
As little as a four-residue extension at the N terminus of protease, corresponding to the C-terminal residues of p6pol, in conjunction with a D25N mutation result in an effectively monomeric species5,12. Disruption of the native protease dimer by N-terminal extension is due to removal of the protons on the secondary amine of the N-terminal proline residue, disrupting the interstrand hydrogen bond between the amine of the N-terminal proline of one subunit and the C-terminal carbonyl oxygen of the second subunit6. C-terminal extension, however, does not have an impact on this interstrand hydrogen bond because the secondary amine of Pro 1 is preserved. Therefore, we made use of the mini-precursor, bearing only the N-terminal cleavage site, to visualize the early transient events involved in autoprocessing of the protease at the p6pol–PR junction that is required for the formation of a fully active, stable protease dimer.
The optimized mini-precursor protease construct SFNFPR(D25N) comprises a four-residue N-terminal extension (Ser-Phe-Asn-Phe) derived from the TFR(Fig. 1a), a D25N mutation to abolish all residual catalytic activity, and C67A and C95A mutations to remove surface cysteines (Supplementary Fig. 1a)9,10,12. The corresponding active SFNFPR(D25) mini-precursor construct undergoes autoprocessing during expression to release the mature protease (see Methods). NMR analysis of SFNFPR(D25N) shows that it is monomeric (with an upper limit of ~10% dimer from translational diffusion measurements); the secondary and tertiary structures of the mature protease are preserved with the exception of the N- and C-terminal strands which form an intersubunit four-stranded anti-parallel β-sheet in the mature dimer; and residues −4 to 9 and 95–99 are disordered and highly mobile (see Methods and Supplementary Fig. 1b–e).
Because enzymatically active protease is dimeric, and the rate-limiting step in autoprocessing is unimolecular7,9, transient self-association of the precursor must occur to initiate autoprocessing. To visualize this phenomenon we measured intermolecular paramagnetic relaxation enhancements (PREs) by introducing a spin label via conjugation to three engineered surface-exposed cysteine residues: T12C, E34C and V82C (one at a time). These sites are frequently mutated in viable HIV-1 variants2. T12C and V82C are located at the periphery of the substrate-binding cleft in the mature dimer, whereas E34C is relatively far removed from the dimer interface (Fig. 1e, f). In a rapidly exchanging system, the PRE 1HN-Γ2 rates21 are population-weighted averages of the PRE rates of the species present13,14. Because the PRE rate for a paramagnetic centre-proton pair is proportional to the <r−6> average of the distance between them, and the PRE effect is large owing to the high magnetic moment of an unpaired electron, the PRE in the fast exchange regime is very sensitive to the presence of lowly populated (<5%), highly transient species in solution providing there are paramagnetic centre-proton distances in the minor species that are shorter than in the predominant species13–15.
PREs were measured on a 1:1 mixture of 0.2 mM U-[2H/13C/15N]-labelled SFNFPR(D25N) and spin-labelled SFNFPR(D25N) at natural isotopic abundance. Because 1HN-Γ2 rates are measured using 1H–15N correlation-based experiments21, the observed 1HN-Γ2 rates arise solely from intermolecular interactions between the spin-labelled protein and the isotopically labelled protein (Fig. 1b–d). For the E34C spin label, no 1HN-Γ2 rates greater than 5 s−1 are observed (Fig. 1c); this sample therefore provides a negative control, excluding the existence of solvent PRE effects arising from diffusion and random elastic collisions, or from direct intermolecular interactions between the spin label and the U-[2H/13C/15N]-labelled protein. The PRE profiles for the T12C (Fig. 1b) and V82C (Fig. 1d) spin labels are similar but the magnitude for the latter is 4- to 8- fold greater than for the former. Within the ordered core of the precursor (residues 10–94), large intermolecular PREs are observed for residues 21–30, 46–55 and 80–85 located at or close to the dimer interface. Residues 21–30 encompass the catalytic triad, residues 46–66 correspond to the flap region which gates the active site, and residues 80–81 and 83–84 are located in the substrate binding cleft (Fig. 1e, f). In addition, the N-terminal region experiences sizeable PREs from the T12C (Fig. 1b) and V82C (Fig. 1d) spin labels. These data demonstrate that transient self-association of the precursor involves residues located at the dimer interface in the mature dimer. A similar intermolecular PRE profile is observed from V82C spin-labelled, full-length TFR–PR(D25N) precursor to U-[2H/13C/15N]–SFNFPR(D25N), indicating that the transient dimmerization interface is preserved on further N-terminal extension of the protease precursor (Supplementary Fig. 2a).
Back-calculation of the PREs from the structure of the mature dimer shows that almost zero PRE values are expected for the T12C and E34C spin labels at a population of 1–2% mature heterodimer (Fig. 1b, c). For the V82C label, small PRE values at a population of 1–2% mature heterodimer are predicted for residues 27–30 and 48–50 (Fig. 1d, blue line).The mature dimer does not predict the large observed PRE values observed for residues 20–26, 30–35 and 80–83. Furthermore, in the mature dimer residues 80–83 of one subunit are located on the opposite side of the dimer interface from residues 80–83 of the other subunit, and thus the large intermolecular PREs observed from the V82C spin label to residues 80–83 would require a ~180° rotation of one subunit relative to its position in the mature dimer. Thus, the upper limit of the total population of mature dimer (heterodimer and homodimer) cannot exceed 2–4%.
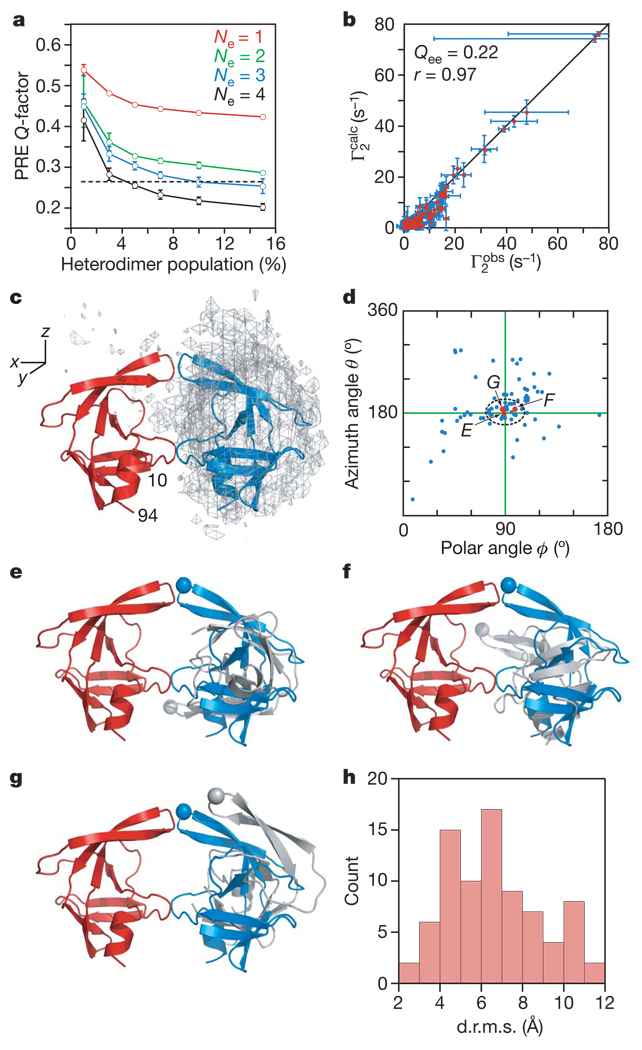
Transient interactions between SFNFPR(D25N) precursor monomers were visualized semi-quantitatively using rigid-body simulated annealing calculations14,16,22 to optimize the agreement between observed and calculated C2 rates arising from the T12C, E34C and V82C spin labels simultaneously (see Methods). The flexible N- and C-terminal regions (residues −4 to 9 and 95–99, respectively) were excluded from the calculations. A single conformer representation (Ne = 1) for the transient dimer does not account for the PRE data and even at a heterodimer population of 15% the PREQ-factor23 (see Methods for definition) has a value of greater than 0.4 (Fig. 2a). Thus, the dimeric SFNFPR(D25N) precursor is an ensemble of multiple encounter complexes. For Ne ≥ 2, the average PRE Q-factor decreases rapidly as the heterodimer population is increased above 1%, levelling off at a population of ~5% (Fig. 2a). The best results are obtained with Ne = 4, and larger ensemble sizes are unjustified and would result in over-fitting the data. For Ne = 4, the PRE Q-factors at a heterodimer population of 3–5% are close to the expected PRE Q-factor based on experimental error (Fig. 2a), consistent with translational diffusion data (Methods and Supplementary Fig. 1d). Given a total protein concentration of 0.4 mM, the apparent Kd for self-association is therefore 3–6 mM. A comparison of the calculated and observed PRE profiles and a correlation plot of observed versus calculated Γ2 rates for Ne = 4 at a heterodimer population of 5% are shown in Fig. 1b–d and Fig. 2b, respectively.
Figure 2. Ensemble simulated annealing and the protease mini-precursor encounter complex ensemble.
a, PRE Q-factor as a function of ensemble size and population of heterodimer. Dashed line denotes the expected Q-factor when agreement between observed and calculated Γ2 rates is comparable to the experimental error in the measurements. b, Correlation between observed and calculated Γ2 rates for Ne = 4 and a heterodimer population of 5%. Qee is the ensemble of ensembles average PRE Q-factor for the 20 calculated Ne = 4 ensembles and r the correlation coefficient. Error bars in a and b represent 1 s.d. c, Atomic probability density map25 (grey mesh, plotted at a threshold of 20% of maximum) showing the distribution of the spin-labelled subunit relative to the isotopically labelled subunit (red ribbon) in the SFNFPR(D25N) encounter complexes. The location of the second subunit in the mature dimer is shown as a blue ribbon. d, Orientations in spherical coordinates of the vector joining the centre of masses of the two interacting molecules in the encounter complexes relative to the coordinate system shown in c with the z axis corresponding to the C2 symmetry axis of the mature dimer. The ϕ,θ angles for the mature dimer are located at the crosshair. e–g, Representative encounter complexes (labelled and denoted by red dots in d) corresponding to the structures with the closest spherical angles (e), the smallest d.r.m.s. (f) and the smallest atomic r.m.s. displacement (g) relative to the mature dimer. The Cα atom of Gly 51 at the tip of the flap is shown as a sphere to guide the eye. The isotopically labelled and spin-labelled subunits are shown in red and grey, respectively; the blue subunit corresponds to the orientation relative to the red subunit seen in the mature dimer. h, Histogram of the d.r.m.s. metric for the Ne = 4 structures (total of 20 × 4 = 80 conformers) at a population of 5% heterodimer.
The distribution of the spin-labelled monomer relative to the isotopically labelled monomer in the computed ensemble of SFNFPR(D25N) encounter complexes is shown in Fig. 2c. The predominant interactions between the two monomers involve the same residues that comprise the dimer interface in the mature dimer, and one subunit of the mature dimer is embedded within the ensemble distribution of the spin-labelled subunit. The orientation of the subunits in the encounter complex ensembles can be described by spherical angles describing the orientation of the vector joining the centre of masses of the two subunits to the coordinate axis frame. Many members within the calculated ensemble are clustered around the values corresponding to the mature dimer (Fig. 2d). This is reflected in the distribution of the distance root mean square (d.r.m.s.; see Methods) deviation metric where over one-half of the ensemble members have d.r.m.s. values less than 6 Å (Fig. 2h). However, the structures with spherical angles close to the mature dimer (indicated by arrows in Fig. 2d) and low d.r.m.s. values have a widespread range of relative self-rotations, as illustrated by three examples comprising the ensemble members with the closest spherical angles to the mature dimer (Fig. 2e), the smallest d.r.m.s. (Fig. 2f) and the smallest Cα atomic r.m.s. displacement (Fig. 2g). The difference from the mature dimer in rotation angle about the axis joining the centre of masses of the two subunits ranges from 13° (Fig. 2g) to 135° (Fig. 2e), with an intermediate rotation angle of 70° for the structure in Fig. 2f (see Supplementary Fig. 3 for definitions). One can therefore conclude that the actual occupancy of a structure within the encounter complex ensemble corresponding to the mature dimer is very small.
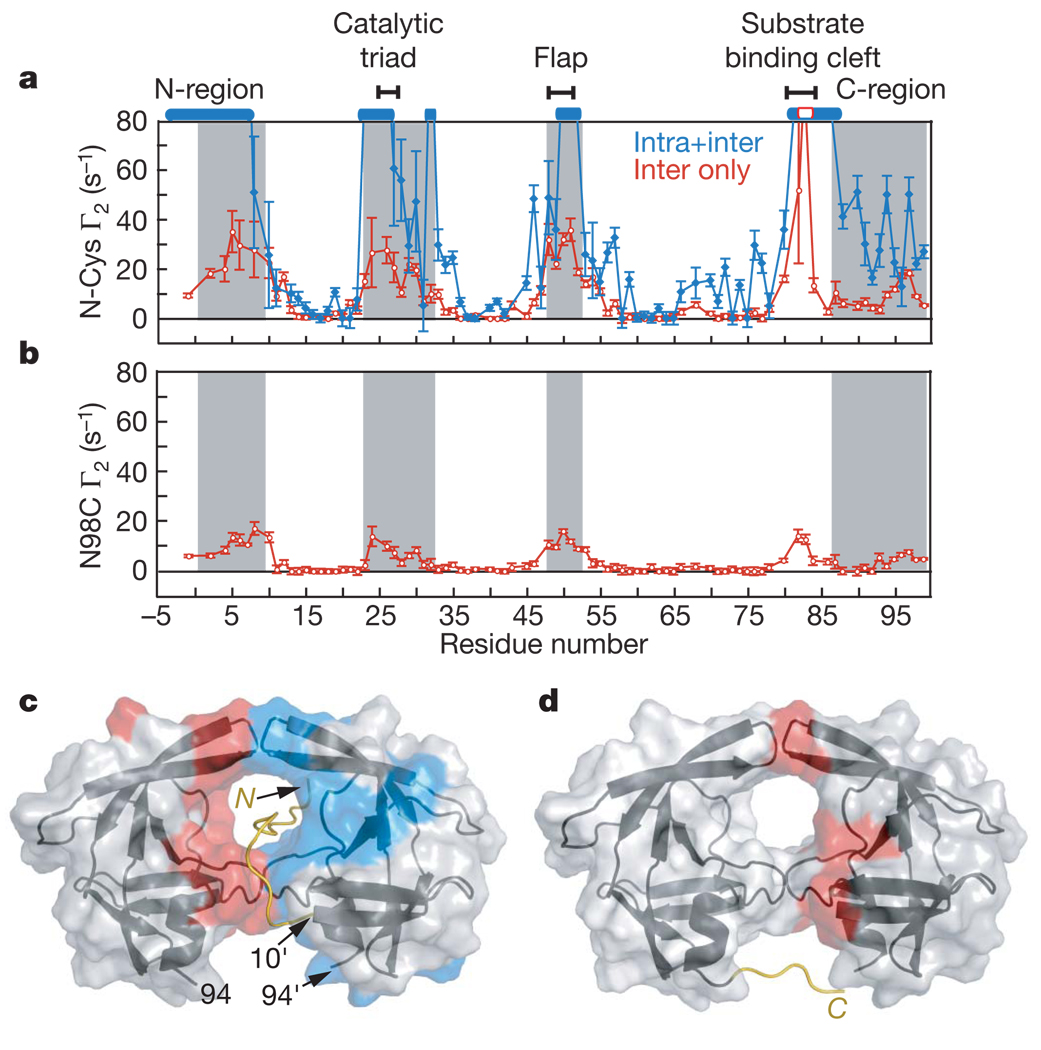
To probe the conformational space sample by the disordered N-terminal flanking sequence of the SFNFPR(D25N) precursor we introduced a spin label on a Cys residue inserted immediately after the N-terminal serine (S(C)FNFPR(D25N)). PRE measurements were carried out on a 1:1 mixture of 0.2 mM U-[2H/13C/15N]–SFNFPRD25N precursor and 0.2 mM spin-labelled, natural isotopic abundance S(C)FNFPR(D25N) to detect intermolecular PREs, and on a sample of 0.2 mM spin-labelled, U-[2H/13C/15N]-labelled S(C)FNFPR(D25N) to observe both inter- and intramolecular PRE effects. Although the overall PRE profiles for the two samples are similar (although differences in detail are apparent), the magnitude of the PREs for the second sample is much larger than for the first, reflecting the contribution from intramolecular PREs (Fig. 3a). The N-terminal residues −4 to 9, and residues comprising the active site, flap and substrate-binding cleft, display large inter- and intramolecular PREs (Fig. 3a, c). The intermolecular PREs involving residues 82–84 are fully consistent with the large intermolecular PREs observed on the N-terminal residues from spin-labelled V82C (Fig. 1d). These data indicate that the N-terminal tail can insert itself into the active site and make transient contact with both subunits in the encounter complex ensemble. The spin label is located four residues proximal to the scissile peptide bond, and the observation that large PREs are observed for both sides of the active site (see Fig. 3c) suggests that the tail shuttles back and forth within the substrate binding cleft formed by the two subunits in the context of a dimer. Such translational movement is a functional requirement, as the protease precursor cuts the N-terminal transframe region in two major locations before cleaving its C terminus (Fig. 1a)9,10. This is confirmed by the observation of a very similar intermolecular PRE profile from full-length TFR–PR(D25N) spin-labelled at position −44, four residues downstream from the TFP–p6pol cleavage site at residues −48/−49, to U-[2H/13C/15N]–SFNFPR(D25N) (Supplementary Fig. 2b).
Figure 3. PRE profiles with spin labels attached at the N- and C termini of the SFNFPR(D25N) mini-precursor.
a, Intermolecular PREs (red) observed for a 1:1 mixture (0.2 mM each) of N-terminal spin-labelled S(C)FNFPR(D25N) at natural isotopic abundance and U-[2H/13C/15N]–SFNFPR(D25N), and the sum of the inter- and intramolecular PREs (blue) observed for 0.2 mM N-terminal spin-labelled U-[2H/13C/15N]–S(C)FNFPR(D25N). Residues broadened beyond detection are denoted by open bars. b, Intermolecular PREs observed for a 1:1 mixture (0.2 mM each) of U-[2H/13C/15N]–SFNFPR(D25N) and C-terminal spin-labelled (at N98C) SFNFPR(D25N) at natural isotopic abundance. Grey shaded areas in a and b delineate residues that are buried at the dimer interface in the mature protease. Error bars in a and b represent 1 s.d. c, d, Inter- and intramolecular PREs with Γ2 rates >10 s−1 colour-coded in red and blue, respectively, onto the molecular surface of the mature protease dimer originating from the N-terminal (c) and the C-terminal (d) spin labels. The intramolecular PRE rates are given by the difference in PRE rates between the blue and red profiles in a. Cartoons of modelled N-terminal (residues −4 to 9) and C-terminal (residues 95–99) regions bearing the spin labels are included in c and d, respectively.
The C-terminal region of the SFNFPR(D25N) precursor was spin-labelled at N98C. The resulting intermolecular PREs are much smaller than those with the spin label at the N terminus, but the PRE profiles are similar (Fig. 3b, d). Thus, the C-terminal flexible region can also make intermolecular contacts with the active site and substrate-binding cleft in the context of the precursor encounter complex ensemble. Because the N- and C termini are highly mobile, intermolecular PREs between the N- and C termini will be significantly attenuated. Nevertheless, intermolecular PREs are observed on residues 95–97 from the spin label at the N terminus (Fig. 3a), and on residues 5–8 (Fig. 3b) from the spin label at the C terminus (Fig. 3b). Small intermolecular PREs are also observed from the N98C spin label to the C-terminal region (residues 95–99). These observations might suggest the existence of transient, loose interactions between the N- and C termini that may partially approximate a portion of the intersubunit β-sheet in the mature dimer.
The PRE data presented here demonstrate that although the HIV-1 protease precursor is predominantly monomeric, transient encounter complex dimers are formed using the same interface as that of the mature dimer but with a wide range of relative subunit orientations. Only a very small fraction of the encounter complexes adopt the same subunit orientation as in the mature protease, accounting for the very low enzymatic activity of the precursor. This small subset, which may be partially stabilized by transient, loose interactions involving the N- and C-terminal regions, can accommodate transient insertion of the N-terminal region including the N-terminal cleavage site in the substrate binding cleft, thereby providing a structural model for autoprocessing at the N terminus of the protease leading to the formation of a stable dimer with mature catalytic activity.
METHODS SUMMARY
Sample preparation and NMR spectroscopy
Protein expression, mutagenesis, purification and conjugation of engineered surface cysteine residues to 3-iodomethyl-(1-oxy-2,2,5,5-tetramethylpyrroline) are described in the Methods. Samples for NMR were in 20 mM sodium phosphate buffer, pH 5.8. NMR experiments were collected at 20 °C at a 1H spectrometer frequency of 600 MHz. 1HN PRE data were acquired using a two-dimensional 1H–15N correlation-based pulse scheme with an interleaved two time-point measurement21.
Simulated annealing calculations
Conjoined rigid-body/torsion angle dynamics simulated annealing calculations on the basis of the PRE data were carried out using Xplor-NIH22 as described14.
Supplementary Material
Acknowledgements
We thank R. Ishima for providing initial backbone assignments for the SFNFPR(D25N) protease construct; C. Schwieters for many discussions; Y. Sheng for help with the CS-Rosetta calculations; Y. Kim for providing the code for structure clustering and d.r.m.s. calculations; and J. Sayer for MALDI measurements. This work was supported by funds from the Intramural Program of the NIH, NIDDK and the AIDS Targeted Antiviral program of the Office of the Director of the NIH (to G.M.C.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 2.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: structure, dynamics and inhibition. Adv. Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 3.Wlodawer A, Erikson J. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 4.Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 5.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J. Biol. Chem. 2003;278:43311–43319. doi: 10.1074/jbc.M307549200. [DOI] [PubMed] [Google Scholar]
- 6.Miller M, et al. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;246:1149–1152. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 7.Louis JM, Nashed NT, Parris KD, Kimmel AR, Jerina DM. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc. Natl Acad. Sci. USA. 1994;91:7970–7974. doi: 10.1073/pnas.91.17.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Co E, et al. Proteolytic processing mechanisms of a miniprecursor of the aspartic protease of human immunodeficiency virus type 1. Biochemistry. 1994;33:1248–1254. doi: 10.1021/bi00171a027. [DOI] [PubMed] [Google Scholar]
- 9.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 10.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nature Struct. Biol. 1999;6:868–874. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 11.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 2004;78:8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J. Biol. Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 13.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 14.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 15.Volkov AN, Worall JA, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc. Natl Acad. Sci. USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 17.Tessmer U, Kräusslich H-G. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 1998;72:3459–3463. doi: 10.1128/jvi.72.4.3459-3463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig C, Leiherer A, Wagner G. Importance of protease cleavage sites within and flanking human immunodeficiency virus type 1 transframe protein p6* for spatiotemporal regulation of protease activation. J. Virol. 2008;82:4573–4584. doi: 10.1128/JVI.02353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wondrak EM, Nashed NT, Haber MT, Jerina DM, Louis JM. A transient precursor of the HIV-1 protease: isolation, characterization and kinetics of maturation. J. Biol. Chem. 1996;271:4477–4481. doi: 10.1074/jbc.271.8.4477. [DOI] [PubMed] [Google Scholar]
- 20.Cherry E, et al. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease-reverse transcriptase fusion proteins. J. Mol. Biol. 1998;284:43–56. doi: 10.1006/jmbi.1998.1968. [DOI] [PubMed] [Google Scholar]
- 21.Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwieters CD, Kuszewski J, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 23.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 24.Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochimie. 1991;73:1391–1396. doi: 10.1016/0300-9084(91)90169-2. [DOI] [PubMed] [Google Scholar]
- 25.Schwieters CD, Clore GM. Reweighted atomic densities to represent ensembles of NMR structures. J. Biomol. NMR. 2002;23:221–225. doi: 10.1023/a:1019875223132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.