Abstract
Energy balance, including diet, weight, adiposity, and physical activity is associated with carcinogenesis. Epidemiological studies indicate that obesity and sedentary and/or active behavior are risk factors for breast cancer in postmenopausal women and survival in both pre-and postmenopausal breast cancer patients. Thus, understanding energy balance modulation’s influence on changes in gene expression patterns in the normal mammary gland is important for understanding mechanisms linking energy balance and breast cancer. In a six week long study, female C57BL/6 mice (9 weeks old) were randomized into four groups: 1) food consumed ad libitum (AL); 2) AL with access to running wheels (AL+EX); 3) 30% Calorie Restricted (CR); and 4) 30% CR with access to running wheels (CR+EX). CR mice received 70% of calories but 100% of all other nutrients compared to AL mice. Diet and exercise treatments individually and combined, had significant effects on body composition and physical activity. Affymetrix oligo-microarrays were used to explore changes in gene expression patterns in total RNA samples from excised whole mammary glands. Contrasting AL versus CR resulted in 425 statistically significant expression changes whereas AL versus AL +EX resulted in 45 changes, with only 3 changes included the same genes, indicating that CR and EX differentially influence expression patterns in non-cancerous mammary tissue. Differential expression was observed in genes related to breast cancer stem cells, the epithelial -mesenchymal transition, and the growth and survival of breast cancer cells. Thus, CR and EX appear to exert their effects on mammary carcinogenesis through distinct pathways.
Keywords: Energy balance, calorie restriction, exercise, body composition, bone density, gene expression, mammary gland, mice
Introduction
A growing literature indicates that elements of energy balance, including obesity, caloric intake, and levels of physical activity, influence cancer risk at multiple sites (1,2). Breast cancer risk appears to be influenced by several of these aspects of energy balance (3–5), particularly energy intake and obesity, but an emerging literature indicates that exercise (EX) also delays postmenopausal breast cancer, as well as colon and prostate cancer (6–8). A major and unsolved question concerning these diverse results is to what extent epidemiological associations among diet, body weight, physical activity, and carcinogenesis arise through the same mechanistic pathways.
Animal models have proved useful in characterizing the mechanisms underlying the epidemiologic associations between energy balance and carcinogenesis. Calorie restriction (CR), the most commonly recommended dietary strategy in humans to prevent or reverse obesity, dramatically reduces or inhibits the incidence of spontaneous, chemically, and virally induced mammary tumorigenesis in diverse animal models (1,9–12). Experimental evidence indicates that changes in hormone/growth factor signaling and immune function mediate (at least in part) the anticancer effects of CR (1,12). Exercise (EX) is the most commonly recommended way to increase energy expenditure. However, the effects of EX on mammary carcinogenesis in rodent models have not been as consistent as results of CR interventions (2,13–22). Differences between chemically induced and spontaneous tumors, voluntary versus involuntary exercise treatments, age of initiation of physical activity, and rodent model used for the study do not appear to account for these mixed results (2,18,21–23). In mouse models of carcinogenesis, beneficial effects of CR are mediated in part by reductions in serum insulin-like growth factor (IGF)-1 levels (1). However, exercise does not appear to reduce serum IGF-1 levels in humans (24), and in some mouse studies IGF-1 is increased by exercise (2). One recent study of rats indicates that the preventive effect of EX for lung and liver cancer may be explained by increased activity of antioxidant and phase 2 enzymes (25).
A review of 22 reports of microarray analyses of the effects of CR in various organisms and tissues, most with a focus on processes related to aging, concluded that no particular genes were altered in common across all the studies (26). Nevertheless, multiple genes involved in energy metabolism, stress responses (such as heat shock and oxidative stress), and inflammation pathways consistently showed changes in these microarray analyses in response to CR. We are unaware of any past microarray studies directly comparing the effects of CR or EX, alone or in combination, using mammary tissue. One study of gene expression in mouse skin tissue reports that CR and exercise have unique effects; however, exercise with AL consumption had smaller and fewer effects on gene expression than exercise with controlled feeding (27). Importantly, there are also distinct gene expression patterns in adlib and CR fed animals in different tissues such as adipose, muscle and brain (28,29).
In order to understand more fully how energy balance influences mammary carcinogenesis, it is critical to understand the effects of energy balance on normal mammary gland growth and development. The present study was designed to determine the individual and combined effects of CR and EX on normal gene expression patterns in RNA samples from whole mammary glands of C57BL/6 mice. A combination of detailed energy balance phenotyping and measures of gene expression in normal mammary tissue were designed to explore the distinctive effects of CR and EX on gene expression in normal, non-neoplastic mammary glands. Such expression patterns represent the biological background in which carcinogenesis might occur as well as one of several potential mechanistic pathways through which tissue level preventive effects could be occurring.
Materials and Methods
Animals
Six-week-old female C57BL/6 mice (n = 96) were purchased from Charles River Laboratories (Frederick, MD). Upon arrival in the main animal facility, the mice were kept on a reverse light/dark cycle (22:00/10:00). The mice were active during the dark cycle, under red lights. During the experiment all mice were housed individually in a specific pathogen-free animal facility at the Frederick laboratories of the National Cancer Institute (NCI). The study was performed under an approved animal study protocol, and animal care was in accord with NCI Animal Care and Use Committee guidelines.
Experimental Design
Mice (at 9 weeks of age) were randomized into 4 experimental groups (n = 24 mice/group) as follows: 1) food consumed ad libitum (AL); 2) AL with free access to running wheel (AL+EX); 3) 30% CR (CR); and 4) 30% CR with free access to running wheels (CR+EX). Twelve mice/group received implanted transponders that detected body temperature and spontaneous activity levels (see below). The study was performed in 4 blocks. Blocks a and b included 12 mice each with implanted transponders, 3 in each treatment. Blocks c and d each comprised 12 mice with transponders (n = 3 in each treatment group) and 24 mice without transponders (n = 6 in each treatment group). Each block started approximately 8 weeks after the end of the previous one.
Mice on the AL diet regimen received AIN-76A diet (cat. # F05053, Bio-Serv, Frenchtown, NJ) or a CR version of the diet (cat. #F05538, Bio-Serv) delivered in 1-g dustless precision pellets and formulated such that the reduction in calories was entirely from carbohydrates. All other nutrients were increased to match AL consumption (details in (30)). CR mice were fed with AL diet until the beginning of the study and then switched to daily aliquots of the CR diet and restricted by 30% (compared to average daily intake of the AL or AL+EX groups separately).
Energy Balance Phenotyping
Mice in the EX groups were housed in cages equipped with a running wheel (MiniMitter Inc., Bend, OR). Running wheel revolutions were recorded and separate measures of spontaneous locomotor activity and body temperature were made using transponders surgically implanted (two weeks prior to study onset) in the abdominal cavity.
Body weights and food consumption (in the AL-fed animals) were recorded weekly. After six weeks on study, mice were sacrificed by continuous CO2/O2 inhalation, and serum and various tissues, including a pair of thoracic mammary glands, were collected and immediately frozen and stored at −80 °C. Body composition and bone characteristics were determined (details in (30), (31)) using dual-energy X-ray absorptiometry (DXA) (GE Lunar Piximus II, Madison, WI).
Serum IGF-1 Analysis
Serum IGF-1 concentration was measured with a rat/mouse radioimmunoassay (RIA) IGF-1 kit (Diagnostic Systems Laboratories, Inc. Webster TX). Serum IGF-1 concentrations were determined for only those mice with implanted transponders. We report average values for two replicate determinations from a single sample per animal.
RNA isolation
Total mammary gland RNA was isolated from pooled left and right thoracic mammary glands (approximately 60 mg each) for only those mice with implanted transponders using TRIzol extraction reagent (Invitrogen, Rockville, MD) and the RNeasy Midi Cleanup Kit (Qiagen, Valencia, CA). Expression data were thus obtained for 9–10 animals per group. The samples represent the entire cellular milieu of the mammary glands, including a mix of epithelial cells, adipocytes, mesenchymal cells, and immune cells. Total RNA concentration was determined by spectrophotometric evaluation of absorbance at 260 nm, and RNA integrity was confirmed by 1% agarose gel electrophoresis. RNA quality of random samples was also tested on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Microarray Analysis
Microarray analysis was performed using Affymetrix (Santa Clara, CA) GeneChip Mouse Genome 430A expression arrays (details in (32)). These arrays contain approximately 23,000 Affymetrix probe sets corresponding to about 13,000 mouse genes. Signal values and detection calls of the probe sets were determined using Affymetrix GCOS (ver. 1) software. Relative intensity variation across arrays was normalized by scaling to an average target signal level of 500 counts, excluding lowest 2% and highest 2% signals. Signal values were log2-transformed for statistical analysis.
Statistical and Functional Analysis of Microarray Data
Class comparisons were first explored using two-way ANOVA considering EX and CR as independent variables (R Foundation for Statistical Computing, Vienna, Austria) after adjusting for block variation. P-values were adjusted by the Benjamini & Hochberg method for all features on the array (22,691), and 1978 features were found with a false discovery rate (FDR) of < 0.05. For the interaction between EX*CR, none were found to be significant at FDR < 0.05. This finding indicated a limited interaction between EX and CR. Thus, we examined CR and EX effects separately using Affymetrix Microarray Suite Version 5.0 (MAS5) Software. First, block-dependent scale factors and shifts of data were adjusted by transforming to Z-scores. Class comparisons were done using two-sample t-tests. T he false discovery rates and random probability of number of significant genes were estimated by multivariate permutation testing using BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html). Average gene expression changes between classes were calculated using untransformed data. Genes altered by at least 1.5 fold at p ≤ 0.005, and with the geometric mean signal value of one of the classes > 100, were selected for further examination. We note that by this pair-wise comparison method, a different list of features of potential interest resulted than the one from the two-way ANOVA approach. For example, by two-way ANOVA, CR effects indicated that 469 features were altered by ≥ 1.5-fold at FDR < 0.05 and with at least one signal value above 100. Sixty-seven percent of these features were shared between this analysis and the two sample comparison. The two-way ANOVA list can be calculated from our data, accessible through GEO Series accession number GSE14202 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14202), or is available from the authors upon request."
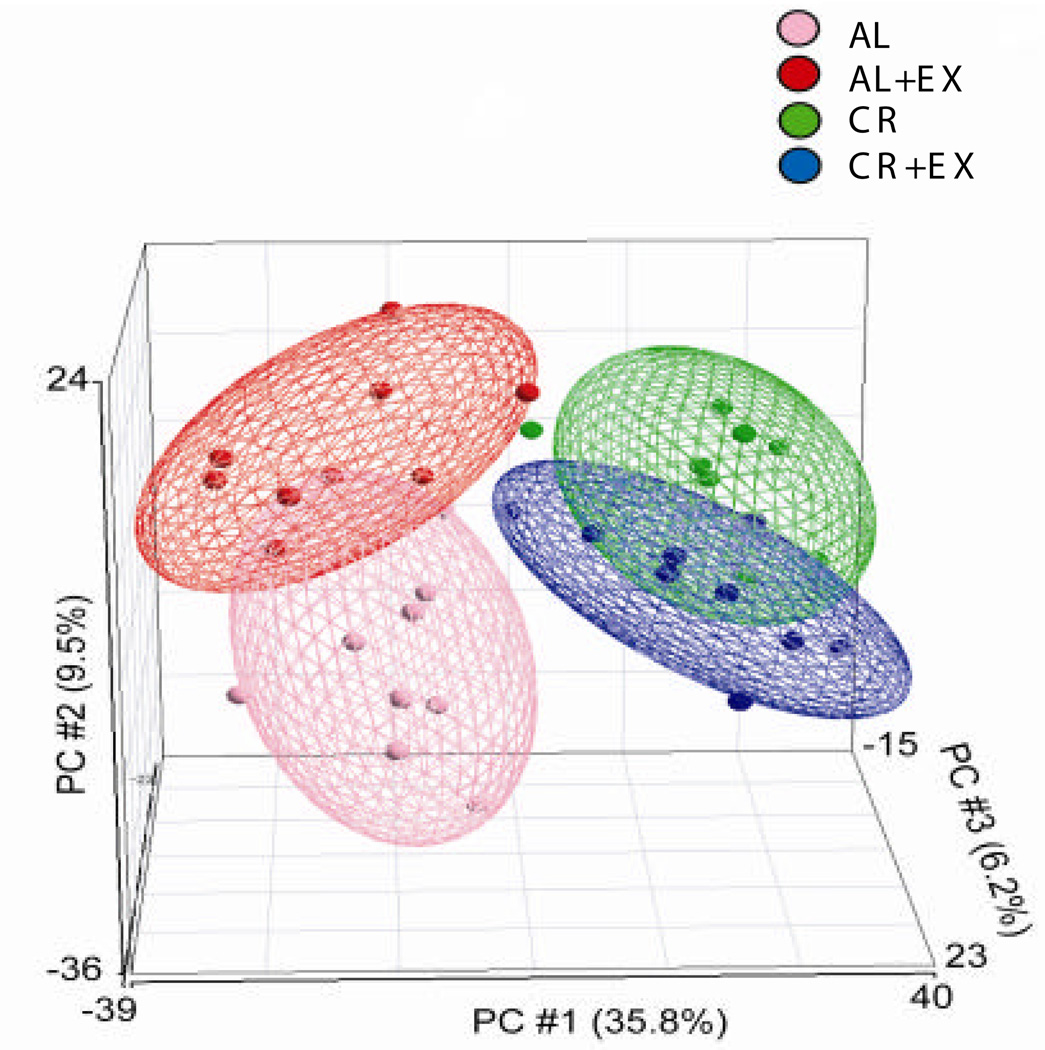
Global expression changes in each intervention group as compared to AL mice were examined by principal component analysis (PCA) using Partek Pro discovery software (Partek Inc., St. Louis, Missouri). PCA analysis included all the genes detected in at least 50% of the arrays (12814 probe sets). Further filtering was done by eliminating expression data that were associated with high standard deviations. Specifically, we eliminated genes having a standard deviation > 1 in any sample - AL, EX, CR or CR+EX. PCA of the remaining 1,649 probe sets found not to alter the patterns. Ellipsoids of 2 standard deviations (95% probability) enclosed most of the arrays (Figure 1).
Figure 1. Principal components analysis of mouse mammary gland gene expression after diet and exercise treatment.
An array representing one animal is shown as a single point in the 3-dimensional plot, with the distance between points representing a measure of dissimilarity of expression patterns between the arrays. AL animals are in pink, AL+EX in red, CR in green, and CR+EX in blue. The three largest principal components accounted for 51.5% of the total variance.
Hierarchical clustering was also used to explore changes in groups of these genes. Genes were clustered by the similarity of their expression profiles based on logarithmic values of expressions using (1-correlation) as the distance metric (33,34). To investigate functional categories of genes altered by CR and CR+EX interventions, we used 1) comprehensive literature searches and 2) gene ontology terms (http://www.geneontology.org/). Genes were grouped into functional categories using the Expression Analysis Systematic Explorer (EASE) software application for rapid biological interpretation of gene lists that result from microarray analysis (35).
Real-Time Reverse Transcriptase (RT)-PCR Quantification of mRNA
Real-time RT-PCR following manufacturers’ recommended protocols was used to confirm array results of gene expression changes (Applied Biosystems, Foster City, CA) using two µg of total RNA for the first-strand reaction and VIC-labeled β-actin (Cat # 4352933E) as the reference. mRNA expression of the following genes was quantified: leptin (Cat # Mm00434759_m1), Wee1 (Cat # Mm00494175_m1), Elov-6 (Cat # Mm00851223_s1), Igfbp4 (Cat # Mm00494922_m1), and Uble1a (Cat # Mm00502282_m1). Reactions used the TaqMan universal PCR master mix (Applied Biosystems) in a total volume of 30 µl on an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Amplifications were performed in triplicate for each sample; PCR optimal conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The threshold cycle (Ct) method was used to generate expression values, and results for individual genes were quantified relative to the β-actin control. Three replicate measurements on 5–8 mice were completed.
Statistical Analysis of Phenotypes
Effects of diet and exercise on body composition, running wheel performance, and IGF-1 serum levels were examined using one- or two-way analysis of variance to account for different experimental designs. We included body weight as a covariate in the analysis of bone mineral density (BMD) and bone mineral content (BMC). Tukey’s HSD was used to compare means of all treatment groups. All models included a block effect. Block had small but statistically significant effects on body weight and other phenotypic variables (p ~ 0.025); interaction effects between block and treatment were not statistically significant (p > 0.1). These analyses were performed using SAS JMP (SAS Institute, Inc., Cary, NC).
Results
Energy Balance Phenotypes
Table 1 summarizes effects of CR and EX on body composition. CR resulted in significantly decreased body mass (p < 0.0001), with reductions in both lean (p<0.0001) and fat mass (p = 0.0087). Access to running wheels resulted in a small decrease in body weight in both AL+EX and CR+EX mice; this difference was not statistically significant. CR significantly reduced overall bone mineral density (BMD) (p < 0.0001), tibial BMD (p < 0.0001), and vertebral BMD (p <0.0001) and also decreased BMC (p = 0.0202). In contrast, EX increased BMD in both AL+EX and CR+EX mice (p = 0.0009). The increase in BMD was greater in AL+EX mice than in CR+EX mice, even after adjustment for the effects of body size (p(interaction) = 0.04). Qualitatively similar results were obtained for mice with and without implanted with transponders.
Table 1.
Effect of calorie restriction and access to running wheels on body composition of female C57BL/6 mice
| Treatment Group |
N | Body Weight (g) |
Fat Mass1 (g) | Lean Mass (g) | Percent Body Fat |
Bone Mineral Density (BMD)1 g/cm2 × 1000) |
Bone Mineral Content (BMC)1(g) |
Tibial Bone Mineral Density (g/cm2 × 1000) |
Vertebral Bone Mineral Density (g/cm2 × 1000) |
|---|---|---|---|---|---|---|---|---|---|
| AL | 24 | 23.4 ± 0.4a | 7.1 ± 0.4a | 17.0 ± 0.2a | 28.1 ± 1.3a | 48.3 ± 0.6a | 0.43 ± 0.01b | 53.4 ± 0.1a | 50.3 ± 0.01a |
| AL+EX | 23 | 22.8 ± 0.4a | 6.4 ± 0.4a | 17.2 ± 0.2a | 26.1 ± 1.3a | 50.1 ± 0.6b | 0.46 ± 0.01a | 55.6 ± 0.1a | 53.5 ± 0.01b |
| CR | 21 | 18.7 ± 04b | 3.1 ± 0.4b | 15.2 ± 0.2a,b | 21.2 ± 1.3b | 45.7 ± 0.6c | 0.41 ± 0.01b | 47.0 ± 0.1b | 45.3 ± 0.01c |
| CR+EX | 23 | 17.8 ± 0.4b | 2.5 ± 0.4b | 14.5 ± 0.2b | 21.2 ± 1.3b | 46.2 ± 0.6c | 0.42 ± 0.01b | 47.6 ± 0.1b | 45.8 ± 0.01c |
| p(Diet) | <0.0001 | 0.0087 | <0.0001 | <0.0001 | <0.0001 | 0.0202 | <0.0001 | <0.0001 | |
| p(Exercise) | 0.0461 | 0.0765 | 0.2730 | 0.4160 | 0.0009 | 0.0014 | 0.0732 | 0.0184 | |
| p(Interaction) | 0.7142 | 0.9461 | 0.0651 | 0.4287 | 0.0439 | 0.014 | 0.0752 | 0.0787 | |
Lean mass included as a covariate for fat mass; total body weight included as a covariate for bone mineral density and content
Means that do not share the same superscript are significantly different
Weekly food consumption was recorded for AL and AL+EX mice. AL+EX mice consumed significantly more (27.2 ± 0.4 g) food than AL mice (24.7 ± 0.4 g) (p < 0.0001). Mice with implanted transmitters consumed less food (25.2 ± 0.3 g) than mice without transmitters (26.6 ± 0.4 g) (p = 0.0154). There was no interaction between EX and transmitter status (p = 0.8280). CR and CR+EX mice received 70% of the AL and AL+EX food consumption level.
AL+EX mice with free access to running wheels ran 4.3 ± 0.5 km per day, whereas CR+EX mice ran 1.4 ± 0.5 miles/day (Table 2). CR significantly reduced body temperature (p = 0.0001), and there was a significant interaction between calorie intake and exercise (p = 0.0004). Running wheel access increased body temperature in AL+EX mice but reduced it in CR+EX mice (p = 0.008).
Table 2.
Physical activity and body temperature of mice implanted with transponders
| Treatment Group |
Distance (km/day) |
Activity (counts/15- minutes) |
Average Body Temperature (°C) |
|---|---|---|---|
| AL (n=12) |
NA | 133 ± 6a | 37.3 ± 0.06a |
| AL+EX (n=12) |
4.3 ± 0.5a | 167 ± 6a | 37.5 ± 0.06a |
| CR (n=10) |
NA | 143 ± 7a | 35.9 ± 0.06b |
| CR+EX (n=11) |
1.4 ± 0.5b | 132 ± 7a | 35.5 ± 0.06c |
| p(Diet) | <0.0002 | 0.0490 | 0.0001 |
| p(Exercise) | - | 0.0759 | 0.0893 |
| p(Interaction) | - | 0.0014 | 0.0001 |
Average kilometers per day tested with t-test; remaining variables tested with two-way analysis of variance
Means within a column that do not share the same superscript are significantly different
NA, not applicable
IGF-1 serum levels were 302.7 ± 30.4 ng/ml in AL mice and declined significantly in response to the diet intervention with or without exercise (CR: 181.2 ± 30.4 ng/ml; CR+EX: 176.3 ± 33.8 ng/ml) (p = 0.007). EX alone did not significantly affect IGF-1 serum levels (AL+EX: 325.3 ± 30.4 ng/ml), and there was no statistically significant interaction between CR and EX (p = 0.66).
Microarray Analysis
Principal Component Analysis
Global gene expression patterns in a subset of AL, AL+EX, CR, and CR+EX treated mice were examined by PCA. All genes detected in at least half of the arrays (12,814 probe sets) revealed that the CR interventions are more dissimilar from AL than EX from AL. Elimination of data with high standard deviations (leaving 1649 genes) did not alter these patterns. The first three principal components (PCs) accounted for 51.5% of the total variance in these 1649 genes and illustrate a strong diet effect (Figure 1). The ellipsoids of 2 standard deviations (95% probability) enclosed most of the arrays. CR-dependent changes were observed in the variance along PC#1 (35.8%), and EX-dependent changes were seen in the variance along PC#2 (9.5%).
Differential gene expression
To further explore these data, we first performed two-way ANOVA. This analysis revealed that 418 genes were altered by ≥ 1.5-fold and had at least one signal value above 100. There were 267 features at p < 0.005 for the interaction of EX*CR, all with an FDR > 0.05. This finding indicated a limited interaction between EX and CR. As described in the Methods section, we therefore chose to examine CR and EX effects separately using signals from MAS5 software in order to identify a more complete list of candidate genes influenced by CR and/or EX that could then be later followed for validation.
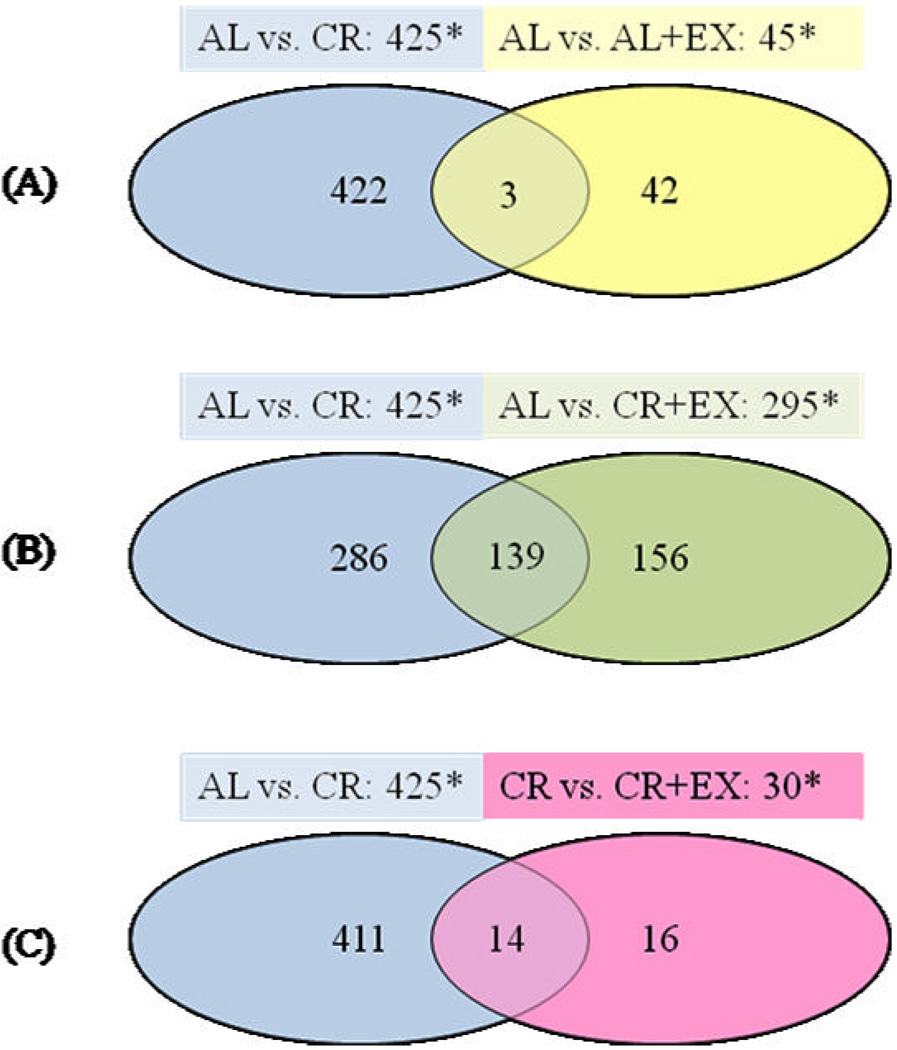
Differential gene expression compared to AL or CR treatments (expression changed by ≥ 1.5 fold (p < 0.005)) is illustrated using Venn diagrams (Figure 2A–C). Diet and exercise altered distinctly different sets of genes. More than 400 genes changed in the CR vs. AL comparison with only about 1/10th as many changes seen in the AL vs. AL+EX and CR vs. CR + EX comparisons (Figure 2A,C). The CR+EX intervention resulted in significant changes in the expression of 295 genes as compared to the AL group (134 up-regulated and 161 down-regulated), and, of these, 139 were significantly altered in the same direction by CR (Figure 2B). Of the 30 genes changed in the CR vs. CR + EX comparison, 14 were found to be common to the list of genes significantly different between AL and CR without exercise; however, the changes in expression of all 14 genes (either increased or decreased by CR) were in the opposite direction in CR+EX.
Figure 2. Venn diagrams comparing mouse mammary gene expression patterns among treatment groups.
Statistically significant gene expression changes (p ≤ 0.005; fold change ≥1.5) were identified from comparisons of the treatment groups by microarray analysis. Total numbers of changes in gene expression are indicated above each diagram by asterisks. Numbers of genes differentially expressed in different treatments are indicated inside the circles, with the number of genes common to between-group comparisons noted inside the overlapping areas of circles. A) CR and AL+EX interventions compared to AL. B) CR and CR+EX interventions compared to AL. C) CR vs. AL and CR vs. CR+EX comparisons.
Hierarchical clustering
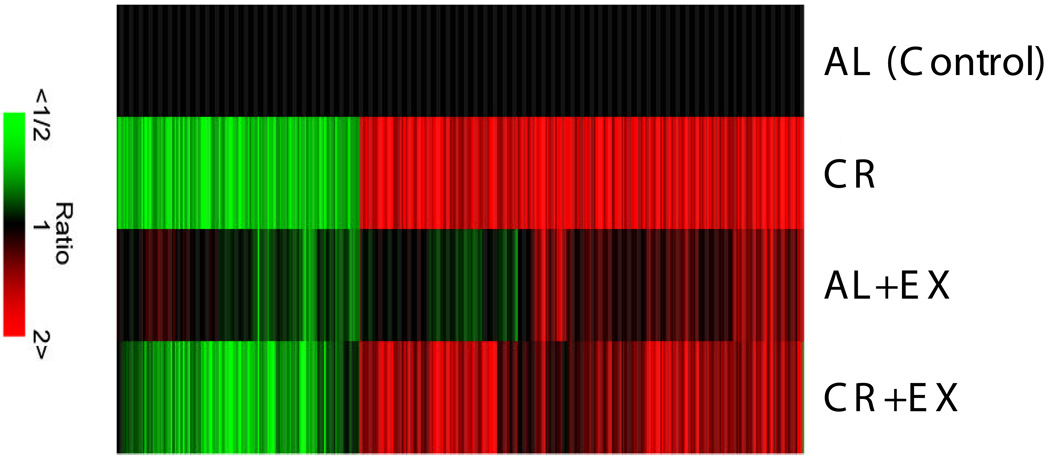
Hierarchical clustering of the 425 genes significantly altered by the CR intervention was performed in order to visualize the patterns of changes among the experimental groups. The gene expression data for the AL+EX, CR, and CR+EX groups are compared to AL and presented as a heat map (Figure 3). Only genes with statistically significant changes in the AL vs. CR comparison are shown. CR, with or without EX, had a stronger effect on expression of this group of genes than EX alone and changes were in a similar direction for both CR and CR+EX, whereas the AL+EX intervention affected a different and distinct group of genes.
Figure 3. Heat map of genes for which CR significantly altered expression vs. AL.
Cluster analysis of genes significantly changed by the CR intervention as compared to AL (p ≤ 0.005, fold change ≥1.5) was performed as described in Materials and Methods. Findings are depicted as a heat map, with red indicating up-regulated and green indicating down-regulated genes. Columns represent different treatment groups and rows represent different genes. Changes for all treatment groups are shown for the genes for which CR as compared to AL significantly changed expression, including those treatments that did not achieve statistical significance vs. AL.
Functional gene categories altered by the CR and EX interventions
Genes were categorized according to functional categories (using EASE analysis and literature review) and further sorted by their transcription fold change (Table 3). Gene expression changes occurred in diverse functional categories, including cell cycle regulation and proliferation, differentiation and morphogenesis, apoptosis, DNA repair, and lipid metabolism. Additionally, there were statistically significant changes in genes in cancer-related pathways (e.g., in the WNT, MAP kinase, and IGF-1 pathways).
Table 3.
Gene expression changes in CR vs. AL and CR+EX vs. AL
| Fold change |
|||
|---|---|---|---|
| Gene category/name |
Gene description | CR vs. AL | CR+EX vs. AL |
| Cell cycle regulation and proliferation | |||
| Wee1b | Wee1-like protein kinase | 4.84a | 3.63a |
| Ccng1b | Cyclin-G1 | 1.69a | 1.52 |
| Hspa9ab | Heat shock protein 9A | 2.51a | 1.48 |
| Mycnb | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) | 2.01 | 2.44a |
| Ccnl2c | cyclin L2 | 0.61a | 0.80 |
| Ccnd1c | cyclin D1 | 0.91 | 0.65a |
| Differentiation and morphogenesis | |||
| GHrb | Growth hormone receptor | 1.81a | 1.55a |
| stk16/Krctb | Stk16 serine/threonine kinase 16 | 1.52a | 1.27a |
| Hspa9ab | Heat shock protein 9A | 2.51a | 1.48 |
| Cd24ac | CD24a antigen | 0.58a | 0.59a |
| Cldn3c | Claudin 3 | 0.63 | 0.60a |
| Cldn4c | Claudin 4 | 0.63 | 0.52a |
| Prlrc | Prolactin receptor | 0.59 | 0.66a |
| Apoptosis | |||
| Dapk1b | Death associated protein kinase 1 | 1.82a | 1.62 |
| Casp7b | Caspase 7 | 1.61a | 1.40 |
| DNA Repair/Response to DNA damage stimulus | |||
| Rad23ab | RAD23a homolog (S. cerevisiae) | 1.95a | 1.21 |
| GADD45c | growth arrest and DNA-damage-inducible 45β | 0.48a | 0.40a |
| Lipid metabolism and biosynthesis | |||
| Cyp51b | cytochrome P450, family 51 | 1.60a | 1.80a |
| Gpamb | glycerol-3-phosphate acyltransferase, mitochondrial | 2.60a | 1.51 |
| Lipgb | lipase, endothelial | 2.05a | 2.04 |
| Fads2b | fatty acid desaturase 2 | 1.93a | 1.53 |
| Fabp5b | fatty acid binding protein 5, epidermal | 2.53a | 2.40a |
| Pcxb | pyruvate carboxylase | 3.02a | 1.82a |
| Ptdss2b | phosphatidylserine synthase 2 | 2.14a | 1.30 |
| Acaa2b | acetyl-Coenzyme A acyltransferase 2 | 1.77a | 1.32 |
| Pitpnbb | phosphatidylinositol transfer protein β | 1.87a | 1.64 |
| Hsd17b12b | hydroxysteroid (17-β) dehydrogenase 12 | 1.91a | 1.65a |
| Elovl5b | ELOVL family member 5, elongation of long chain fatty acids | 1.56a | 1.52a |
| Elovl6b | ELOVL family member 6, elongation of long chain fatty acids | 2.18a | 1.91a |
| Acot3b | acyl-CoA thioesterase 3 | 1.99a | 1.52a |
| Ptgesb | prostaglandin E synthase | 1.99a | 1.71 |
| Dgat2b | diacylglycerol O-acyltransferase 2 | 1.79a | 1.37 |
| Pmvkb | phosphomevalonate kinase | 1.71a | 1.82 |
| Aacsb | acetoacetyl-CoA synthetase | 1.92a | 1.83 |
| Aclyb | ATP citrate lyase | 3.19a | 2.49 |
| Fdpsb | farnesyl diphosphate synthetase | 2.20a | 1.81 |
| Acaa1 b | acetyl-Coenzyme A acyltransferase 1 | 1.70a | 1.38 |
| Plce1b | phospholipase C, epsilon 1 | 1.46 | 1.52a |
| Mogat2b | monoacylglycerol O-acyltransferase 2 | 1.46 | 2.16a |
| Ptgdsc | prostaglandin D2 synthase (brain) | 0.42a | 0.49a |
| WNT pathway | |||
| Wisp2b | WNT1 inducible signaling pathway protein 2 | 1.70a | 1.62a |
| Frzbb | frizzled-related protein | 2.07a | 1.87 |
| Ctnnb1b | catenin (cadherin associated protein) β1 | 1.61a | 1.12 |
| Apcdd1c | adenomatosis polyposis coli down-regulated 1 | 0.59a | 0.55a |
| Nlkc | nemo like kinase | 0.47a | 0.44a |
| Wnt4c | frizzled-related protein | 0.69 | 0.60a |
| MAP kinase pathway | |||
| Mapk6b | Mitogen - activated protein kinase 6 | 2.70a | 1.95 |
| Mapk8ip1b | Mitogen - activated protein kinase 8 interacting protein 1 | 2.52a | 1.70 |
| Mapk14b | Mitogen - activated protein kinase 14 | 2.16a | 1.15 |
| IGF-1 pathway | |||
| Igfbp4b | insulin-like growth factor binding protein 4 | 2.77a | 1.75 |
| Igfalsb | insulin-like growth factor binding protein, acid labile subunit | 3.23a | 3.07a |
| Other pathways | |||
| Camk2db | calcium/calmodulin-dependent protein kinase II, delta (Camk2d), transcript variant 3 | 1.62a | 1.43 |
| Lepc | leptin | 0.23a | 0.26 |
genes altered by ≥ 1.5 fold (p ≤ 0.005)
genes up-regulated
genes down-regulated
Differences in expression of certain genes for the CR and CR+EX interventions compared to AL were not always both statistically significant, but the direction of change was always consistent (up- or down-regulated vs. AL), as illustrated in the heat map presentation (Figure 3). Notable examples include the cell cycle regulation and cell proliferation genes Wee1 kinase, Cyclin-G1 (Ccng1), Cyclin D1 (Ccnd1), and Mycn (Table 3). Furthermore, death-associated protein kinase 1 (Dapk1), caspase 7 (Casp7), and Rad23a were all significantly up-regulated by CR vs. AL, while GADD45 was significantly down-regulated in mammary glands of both CR and CR+EX mice compared to AL mice.
CR and/or CR+EX compared to AL influenced the expression of genes involved in fatty acid synthesis and elongation (Table 3). These included elongation of long-chain fatty acids (ELOVL5 and ELOVL6); fatty acid binding protein 5, epidermal (fabp5); endothelial lipase (lipg); and fatty acid desaturase 2 (fads2). Several genes involved in mammary gland development showed changes in expression. Growth hormone receptor (GHr) was significantly over-expressed compared to AL, and CD24a antigen (CD24a), which plays an important role in cell differentiation, was significantly down-regulated. Stk 16 serine/threonine kinase 16 (stk16/Krct) and heat shock protein 9A (hspa9a) were up-regulated in mammary glands of both CR and CR+EX mice, and claudins 3 and 4 (cldn3 and cldn4) and prolactin receptor (prlr) were all down-regulated (although not all comparisons were significant for both treatments).
Finally, several genes involved in pathways (e.g., WNT, MAP kinase, and IGF-1) shown to play an important role in carcinogenesis were also significantly altered by CR as compared to AL. As mentioned above, these genes were altered in the same direction by CR+EX vs. AL, and several also reached statistical significance. In addition, the leptin (lep) gene was significantly down-regulated in mammary tissue of CR mice compared to the AL group.
A similar analysis with PubMed searches and EASE was also performed on the genes altered in the comparisons AL vs. AL+EX (45 genes) and CR vs. CR+EX (30 genes). The complete list of the 75 genes altered, the direction of change, and gene functions are reported in Table 4; some of the genes identified were related to motile and catabolic processes, but other categories of gene function were not obvious in this small sample of genes.
Table 4.
Gene expression changes in EX vs AL and CR+EX vs CR: selected genes altered by 1.5 fold or more in the analysis (p ≤ 0.005)
| Gene category/name | Gene description | Function * | Fold change | |||
|---|---|---|---|---|---|---|
| EX/AL | CR/AL | CR+EX/AL | CR+EX/CR | |||
| 2900097C17Rik | RIKEN cDNA 2900097C17 gene | 3.53 | 3.91 | 1.73 | 0.44 | |
| Acy1 | aminoacylase 1 | Catalytic activity | 2.25 | 1.63 | 1.29 | 0.79 |
| Hspb1 (or HSP25) | heat shock protein 1 | The 25-kDa heat-shock protein (Hsp25) is a member of the small heat-shock protein family but its function remains largely unknown. | 2.23 | 0.60 | 0.66 | 1.09 |
| Tmem38a | transmembrane protein 38a | 2.22 | 1.98 | 1.29 | 0.65 | |
| Pitx2 | paired-like homeodomain transcription factor 2 | Regulates both morphogenesis and gene expression in developing extraocular muscles | 2.05 | 0.97 | 1.28 | 1.31 |
| Clpb | ClpB caseinolytic peptidase B homolog (E. coli) | Transcription factor activity | 1.98 | 1.83 | 1.23 | 0.67 |
| Pik3c2a | phosphatidylinositol 3-kinase, C2 domain containing, α polypeptide | Catalytic activity | 1.89 | 1.50 | 1.37 | 0.92 |
| Casq2 | calsequestrin 2 | High-capacity, moderate affinity, calcium-binding protein and thus acts as an internal calcium store in muscle. The release of calcium bound to calsequestrin through a calcium release channel triggers muscle contraction. | 1.88 | 1.79 | 1.72 | 0.96 |
| Sgcg | Sarcoglycan, gamma (dystrophin-associated glycoprotein) (Sgcg), mRNA | Component of the sarcoglycan complex | 1.87 | 0.80 | 0.97 | 1.21 |
| Ptgs1 | prostaglandin-endoperoxide synthase 1 | May play an important role in regulating or promoting cell proliferation in some normal and neoplastically transformed cells | 1.86 | 1.17 | 1.02 | 0.87 |
| Bdh | 3-hydroxybutyrate dehydrogenase (heart, mitochondrial) | Catalytic activity | 1.74 | 0.84 | 0.39 | 0.46 |
| Wdr77 | WD repeat domain 77 | 1.73 | 1.09 | 1.49 | 1.37 | |
| Dusp18 | dual specificity phosphatase 18 | Catalytic activity | 1.73 | 0.87 | 0.86 | 0.98 |
| Ptk9l | protein tyrosine kinase 9-like (A6-related protein) | Actin-binding protein involved in motile and morphological processes - Catalytic activity | 1.70 | 0.69 | 0.61 | 0.89 |
| Golga4 | golgi autoantigen, golgin subfamily a, 4 | 1.69 | 0.74 | 0.60 | 0.82 | |
| St3gal5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | Catalytic activity; Transferase activity - Glycosylation. | 1.68 | 0.98 | 0.83 | 0.85 |
| Mpp5 | membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | Catalytic activity | 1.68 | 1.50 | 1.34 | 0.90 |
| Luc7l2 | LUC7-like 2 (S. cerevisiae) | 1.66 | 0.56 | 0.60 | 1.08 | |
| 5430435G22Rik | RIKEN cDNA 5430435G22 gene | Catalytic activity | 1.65 | 1.28 | 1.27 | 0.99 |
| Cnot7 | CCR4-NOT transcription complex, subunit 7 | Ubiquitous transcription factor | 1.63 | 1.44 | 1.38 | 0.96 |
| BC027174 | cDNA sequence BC027174 | 1.59 | 1.10 | 1.02 | 0.92 | |
| Rsnl2 | restin-like 2 | 1.59 | 1.04 | 0.89 | 0.85 | |
| Ddx49 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 | 1.59 | 1.48 | 1.30 | 0.88 | |
| 6530403A03Rik | RIKEN cDNA 6530403A03 gene | 1.56 | 0.79 | 1.16 | 1.47 | |
| Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | Catalytic activity; sterol metabolizing enzyme | 1.55 | 1.41 | 0.96 | 0.68 |
| Agl | amylo-1,6-glucosidase, 4-alpha-glucanotransferase | Catalytic activity; glycogen metabolism | 1.55 | 1.09 | 1.12 | 1.03 |
| Pstpip2 | proline-serine-threonine phosphatase-interacting protein 2 | 1.53 | 1.27 | 1.01 | 0.80 | |
| Art1 | ADP-ribosyltransferase 1 | Catalytic activity; Transferase activity | 1.52 | 1.13 | 0.96 | 0.85 |
| Tceb1 | transcription elongation factor B (SIII), polypeptide 1 | 1.52 | 1.53 | 1.50 | 0.98 | |
| Tmepai | transmembrane, prostate androgen induced RNA | 0.66 | 0.61 | 0.61 | 1.00 | |
| Blm | Bloom syndrome homolog (human) | Catalytic activity | 0.65 | 0.77 | 0.89 | 1.15 |
| Hoxc8 | homeo box C8 | 0.65 | 0.86 | 1.12 | 1.30 | |
| Klk21 | kallikrein 21 | Catalytic activity | 0.65 | 0.62 | 0.64 | 1.04 |
| Mgll | monoglyceride lipase | Catalytic activity; catalyzes the last step in the hydrolysis of stored triglycerides in the adipocyte. | 0.64 | 1.13 | 1.03 | 0.91 |
| Mtap | methylthioadenosine phosphorylase | Catalytic activity; Transferase activity | 0.63 | 1.00 | 0.91 | 0.92 |
| Rabepk | Rab9 effector protein with kelch motifs | 0.61 | 0.72 | 0.89 | 1.23 | |
| 3230401M21Rik | RIKEN cDNA 3230401M21 gene | 0.61 | 0.86 | 0.75 | 0.87 | |
| 2310007F12Rik | RIKEN cDNA 2310007F12 gene | 0.60 | 0.79 | 0.79 | 1.00 | |
| 9130211I03Rik | RIKEN cDNA 9130211I03 gene | 0.59 | 0.81 | 0.64 | 0.78 | |
| Rassf2 | Ras association (RalGDS/AF-6) domain family 2 | Potential tumor suppressor. acts as a kras-specific effector protein. May promote apoptosis and cell cycle arrest. | 0.59 | 1.07 | 0.85 | 0.79 |
| 6720460F02Rik | RIKEN cDNA 6720460F02 gene | 0.59 | 0.68 | 0.73 | 1.08 | |
| C630004H02Rik | RIKEN cDNA C630004H02 gene | 0.57 | 0.65 | 0.57 | 0.87 | |
| Fkbp10 | FK506 binding protein 10 | Catalytic activity | 0.56 | 0.82 | 0.94 | 1.15 |
| Slc22a8 | solute carrier family 22 (organic anion transporter), member 8 | Ion transporter activity | 0.56 | 0.88 | 0.70 | 0.80 |
| Pkd2l2 | polycystic kidney disease 2-like 2 | Ion and cation channel activity | 0.43 | 0.60 | 0.70 | 1.17 |
| EX/AL | CR/AL | CR+EX/AL | CR+EX/CR | |||
| Uble1a | ubiquitin-like 1 (sentrin) activating enzyme E1A | Catalytic activity - Ligase activity - Ubiquitin activating enzyme activity | 1.05 | 0.51 | 0.99 | 1.95 |
| Cyp2b10 | cytochrome P450, family 2, subfamily b, polypeptide 10 | Monooxygenases | 1.01 | 0.91 | 1.43 | 1.57 |
| Zfp160 | zinc finger protein 160 | 1.00 | 0.60 | 0.92 | 1.54 | |
| Asph | aspartate-beta-hydroxylase | Cell motility - muscle contraction | 0.95 | 0.76 | 1.16 | 1.53 |
| Ak3 | adenylate kinase 3 | Adenylate kinase and transferase activity | 1.23 | 2.15 | 1.41 | 0.66 |
| 2410026K10Rik | RIKEN cDNA 2410026K10 gene | 1.07 | 1.43 | 0.93 | 0.65 | |
| Immt | inner membrane protein, mitochondrial | 1.18 | 1.85 | 1.18 | 0.64 | |
| Akp5 | alkaline phosphatase 5 | Metabolism | 1.07 | 1.24 | 0.79 | 0.64 |
| Sf3b5 | splicing factor 3b, subunit 5 | 1.08 | 1.68 | 1.06 | 0.63 | |
| Stom | stomatin (probe set ID: 1419098_at) | Thought to regulate cation conductance | 1.29 | 2.59 | 1.62 | 0.62 |
| Ahcyl1 | S-adenosylhomocysteine hydrolase-like 1 | Hydrolase activity - Involved in the one-carbon compound metabolism | 1.09 | 1.77 | 1.10 | 0.62 |
| Sorbs1 | sorbin and SH3 domain containing 1 | Protein binding involved in transport processes | 1.00 | 1.73 | 1.07 | 0.62 |
| Evpl | envoplakin | Component of the cornified envelope of keratinocytes | 0.72 | 0.88 | 0.54 | 0.61 |
| 2410026K10Rik | RIKEN cDNA 2410026K10 gene | 1.21 | 1.75 | 1.06 | 0.60 | |
| Msn | moesin | Cell motility | 0.90 | 1.68 | 1.00 | 0.60 |
| Dgcr2 | DiGeorge syndrome critical region gene 2 | 1.47 | 1.78 | 1.01 | 0.56 | |
| Gja1 | gap junction membrane channel protein alpha 1 | Gap junction - Cell motility - muscle contraction | 1.14 | 2.80 | 1.52 | 0.54 |
| Sema6c | sema domain, transmemb. domain (TM), and cytopl. domain, (semaphorin) 6C | Involved in the maintenance and remodeling of neuronal connections | 1.27 | 1.83 | 0.98 | 0.53 |
| Nubp1 | nucleotide binding protein 1 | ATP binding | 1.28 | 1.32 | 0.69 | 0.53 |
| 4833439L19Rik | RIKEN cDNA 4833439L19 gene | 1.64 | 2.73 | 1.39 | 0.51 | |
| Zfp574 | zinc finger protein 574 | 1.21 | 1.63 | 0.80 | 0.49 | |
| 0610025L06Rik | RIKEN cDNA 0610025L06 gene | 1.07 | 0.88 | 0.42 | 0.48 | |
| Gprk6 | G protein-coupled receptor kinase 6 | Specifically phosphorylates the activated forms of g protein-coupled receptors | 1.03 | 1.35 | 0.63 | 0.47 |
| Pfkp | phosphofructokinase, platelet | Carbohydrate degradation | 1.30 | 1.99 | 0.92 | 0.46 |
| Stom | stomatin (probe set ID: 1419099_x_at) | Thought to regulate cation conductance | 1.85 | 3.59 | 1.66 | 0.46 |
| Banp | Btg3 associated nuclear protein | 0.76 | 1.45 | 0.66 | 0.45 | |
| Ctse | cathepsin E | Hydrolase activity | 1.60 | 1.91 | 0.79 | 0.41 |
| Gna12 | guanine nucleotide binding protein, alpha 12 | Involved as modulators or transducers in various transmembrane signaling systems | 1.70 | 2.51 | 1.03 | 0.41 |
| BC010304 | cDNA sequence BC010304 | 1.02 | 2.57 | 1.04 | 0.40 | |
| Ppp2r5c | Protein phosphatase 2, reg. subunit B (B56), γ isoform (Ppp2r5c), mRNA | Chaperone. Isoform 2 may function as an endogenous inhibitory regulator of hsc70 by competing the cochaperones | 1.65 | 2.47 | 0.66 | 0.27 |
On bold: Statistically significant gene change (p≤0.005)
Real-Time RT-PCR analysis
Real-time RT-PCR analysis was used to validate array results. The genes were selected to include targets related to energy balance that increased, decreased, or did not change in the microarray analysis (Elov6, Leptin, Igfbp4, Uble1A, and Wee1). Gene expression levels of the selected genes were qualitatively similar in RT-PCR and microarray analyses. For example, contrasts between CR and AL fed animals were 3.52 (0.72), 0.17 (0.07), 1.11 (0.19), 0.92, and 1.41 (0.21) based on replicate RT-PCR measurements from multiple animals (Standard errors in parentheses for the five genes listed above and 2.18, 0.23, 2.77, 0.97 (N.S.), and 4.84 from microarray analysis N.S. = not significantly different from unity. CR + EX vs. Al contrasts gave also indicated that RT-PCR and microarray estimated expression changes were in the same direction.
Discussion
This study shows, for the first time, that CR and EX have distinct effects on normal mammary gland gene expression. In addition, the study clearly demonstrated that CR in combination with or without EX has profound effects on animal phenotype, with the response to EX contingent on calorie intake. The results of this study highlight the complex effects of calorie intake compared to exercise on gene expression in mammary gland and contribute to efforts aimed at understanding molecular and epidemiological associations between energy balance and breast cancer.
Exercise alone (AL+EX) altered an almost completely distinct set of genes in our mouse mammary glands compared to CR alone (Figure 2A). Furthermore, the effects on gene expression in CR+EX mice differed substantially from the effects of CR (Figure 2B); no genes were common to both the CR+EX or AL+EX vs. AL comparisons. These results and our observation that there were statistically significant interactive effects of diet and running wheel access on major aspects of metabolism, including body temperature, spontaneous locomotor activity, and bone characteristics, all support the argument that metabolic effects of physical activity depend on energy balance. Lu, et al., used a very similar experimental design to examine the interactive effects of CR and EX on gene expression in skin tissues from SENCAR mice (27). Though Lu, et al., used treadmill exercise instead of running wheels, a different target tissue, 20% CR, and 10 weeks of exercise, these authors also found that CR significantly altered many more genes than EX and that there was little overlap in the specific genes altered in the different treatment groups.
The relatively large number of genes unique to AL vs. CR (286) as compared to AL vs. CR+EX (156), and the limited effects of EX in CR mice, are most parsimoniously explained as the consequences of a moderate EX intervention. However, CR+EX did reverse some of the gene expression changes caused by CR alone or AL+EX alone. Further work, possibly in a model with more intense exercise treatment, is needed to determine if these represent interactive as well as distinct effects of CR vs EX. A more intense exercise regime might reveal more complete changes of gene expression pathways in a few major areas. The results of this study provide a strong case for such work. An alternative exercise regime such as swimming, or the use of mice bred to be more active may be required to elicit such exercise patterns more consistently than the present voluntary regimen.
The moderate-intensity exercise performed by mice in this study had significant effects on normal mammary gland gene expression. Most of the transcripts identified as responsive to EX alone are involved in metabolism (catalytic activity, ion transporter activity, transferase activity, etc.) and in particular in metabolism of lipids (Table 3). The effect of exercise on lipid metabolism may be more general; a recent study in male C57BL/6J mice reports that exercise reverses many of the effects of a high-fat diet on hepatic gene expression, particularly for a number of genes related to fatty acid metabolism (36).
In contrast to the modest effects seen for EX alone in terms of overall number of genes changed relative to AL, CR alone significantly altered the expression of a large number of genes (425 total). This finding is consistent with the notion that CR affects cancer risk in both humans and animal models by changing the expression of genes in many pathways and in many tissues, including the mammary gland. Analysis of specific gene changes revealed that there were 139 gene expression changes common to both CR and CR+EX vs. AL mice. These genes represent 32.7% of the overall gene changes in the first comparison and 47.1% in the second. It is also noteworthy that all of them were altered in the same direction in both comparisons and generally fall into categories/pathways relevant to mammary gland development and carcinogenesis (Figure 3 and Table 32). The categories/pathways into which they grouped included differentiation and morphogenesis; regulation of cell cycle and proliferation; apoptosis; DNA repair/response to DNA damage stimulus; lipid metabolism and biosynthesis; the WNT, MAP kinase and IGF-1 pathways; and the leptin pathway. We discuss some aspects of these results below.
Genes that are differentially expressed in the mammary glands of CR and CR+EX mice are known to be involved in mammary gland growth, differentiation, and morphogenesis. The CD24a antigen gene was down-regulated in both CR vs. AL and CR+EX vs. AL. This protein plays a pivotal role in cell differentiation and was originally identified as down-regulated in primitive ectoderm, mesoderm, and ventral endoderm when organogenesis is completed (37). In addition, the Krct and heat shock protein 9 genes were up-regulated in mammary glands of both CR and CR+EX mice. Krct (or Stk16/Krct) is a member of the family of serine/threonine protein kinases that is involved in end bud morphogenesis in murine mammary gland development and plays an important role in regulating stromal-epithelial interactions during ductal morphogenesis (38). In a transgenic mouse model, a modest over-expression of Krct in the mammary gland during puberty results in duplication of the terminal end bud axis (38). Future studies examining the effects of CR and/or EX on the growth and morphologic development of the mammary gland and whether there are downstream effects on cancer risk would contribute to our mechanistic understanding of the influence of these interventions.
The claudin 3 and 4 genes were both down-regulated by CR and CR+EX, though the results reached statistical significance only in the CR+EX mice. The claudins are members of a family of transmembrane proteins that are critical in the maintenance of epithelial and endothelial tight junctions and may play a role in the cytoskeleton in cell signaling (39). Claudins 3 and 4 are differentially expressed during normal mammary gland development in the mouse and mammary gland differentiation processes of pregnancy, lactation, and involution, suggesting a role for these proteins at different stages of mammary gland function (40). Specific claudins are over-expressed in a wide variety of cancer types, although their functional role in cancer progression remains unclear (39).
Prolactin receptor gene expression was also decreased by CR and CR+EX compared to AL mice, although the effect was not statistically significant in the former. This gene is an important member of the set of prolactin-regulated genes that mediate prolactin-driven mammary development (41,42). Decreased prolactin levels are associated with decreased risk of breast cancer in women (43), but this association is not found consistently in epidemiological studies (44). Ongoing work may shed additional light on this association.
Several genes involved in cell cycle regulation and cell proliferation, including Wee1 kinase, Cyclin G1, Cyclin D1 and Mycn were also all significantly over-expressed in mammary glands from CR and/or CR+EX mice (Table 3). These genes are negative regulators of different checkpoints in the cell cycle, during the G1/S transition as well as the G2 checkpoint (45–48). Over-expression of Dapk1 and Casp7 (significant only for CR) and the inhibition of GADD45 (significant both in CR and CR+EX mice) also demonstrate a possible role for CR and CR+EX in promoting the apoptotic process (49). Dunn, et al., reported that CR increases the rate of apoptosis in mouse bladder cancers (50), supporting a role for CR in increasing apoptosis. As for EX, the GADD45 transcript was also found to be down-regulated by exercise training in hepatic transcriptional profiles of mice submitted to a high-fat diet (36).
Our analyses also identified changes in many genes common to the CR and CR+EX groups that are involved in lipid metabolism, in particular up-regulation of genes related to the elongation of fatty acids chains (Table 3,4). A higher level of fatty acid serum concentration, usually due to an increased release from adipose tissue in obesity, is a possible early effect of obesity related to the development of insulin resistance and compensatory hyperinsulinaemia (3), an effect opposite of what might be happening in our model.
The gene encoding leptin was down-regulated in the CR (p < 0.05) and CR+EX (NS) mice compared to the AL mice. Circulating leptin concentrations are proportional to body fat content; thus this finding supports earlier findings of decreased leptin in the serum of CR mice (1). Although most studies have failed to relate leptin serum levels to breast cancer risk (51,52), in vitro studies show that leptin stimulates the proliferation of benign and malignant epithelial breast cells and tumor cell invasion in breast cancer cell lines (53–55). Leptin is produced by breast cancer cells, and thus local leptin production by malignant cells as well as adipocytes rather than serum leptin may play a critical role for breast cancer development (56).
Finally, we investigated the effects of EX in the context of CR as compared to CR alone (CR+EX vs. CR) and identified 30 genes that were significantly altered (4 up and 26 down). The genes differentially expressed are primarily involved in motile and catabolic processes (Table 4). No genes were found in common between CR vs. CR+EX and AL vs. AL+EX comparisons. Again, the EX effect on gene expression appears to be dependent on caloric intake level. However, we cannot determine in this experimental design if this dependence is mediated through the amount of exercise (CR animals ran less than AL animals) or via some other moderating effect of nutritional status on exercise effects.
There are some limitations to the present study. Notably, we analyzed gene expression changes in whole, undissected samples of mammary tissue that included epithelial cells, adipocytes and other cell types. Adipocytes alone, as well as other cell types are known to exhibit distinct changes in gene expression in response to diet (28,29). The present study does not allow us to determine which specific cell types are most responsive to the effects of CR versus EX However, the tissue analyzed in this study does represent the combined cellular milieu in which mammary tumors arise. The hypothesis being tested by these analyses is that CR and EX differentially effect pathways in the mammary gland related to breast cancer susceptibility. Recent evidence suggests that multiple cellular compartments within the mammary gland, in addition to the epithelium, contribute to the development and progression of breast cancer. We therefore chose to characterize global gene expression patterns in the entire mammary gland to begin to understand the common and unique effects of CR and EX on breast cancer susceptibility.
Second, past studies of exercise in rodents have had mixed results (57), and it is currently not clear what factors underlie the heterogeneity of EX effects in mouse models. The effects of CR in rodents have been much more consistent and robust, consistent with the notion that CR and EX exert there effects though different mechanisms. Thus it is not clear to what extent rodents and mice in particular, represent models for diet and PA effects on carcinogenesis. Nonetheless, mice have been used as models for the influence of CR on mammary carcinogenesis (1) and the mice in this study showed significant phenotypic responses to both diet and exercise treatments. Thus, they represent at least a model for influences of preventive interventions on the biological environment found in the mammary gland and a source of ideas and hypotheses concerning targets for further understanding mechanisms for breast cancer prevention related to energy balance.
In summary, this study demonstrates that calorie restriction has specific and significant effects on the expression of genes in the normal mouse mammary gland, including many genes implicated in carcinogenesis. Furthermore, we observed distinctive effects of calorie intake and exercise on gene expression in mouse mammary glands. Studies such as this one complement epidemiological evidence indicating that physical activity reduces risk of breast cancer in post-menopausal women independent of obesity and provide clues for future work aimed at identifying the mechanist bases of such associations and designing PA related interventions for breast cancer prevention.
Acknowledgments
M. Padovani would like to thank Dr. Stuart H. Yuspa and the members of the Laboratory of Cancer Biology and Genetics (LCBG) for housing and mentoring her during her final year at the NCI and for generously offering their valuable advice. Lisa Riffle and Dan Logsdon ably assisted with animal care and surgery and Heather L Hill performed the IGF-1 assays.
References
- 1.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 2.Colbert LH, Mai V, Tooze JA, Perkins SN, Berrigan D, Hursting SD. Negative energy balance induced by voluntary wheel running inhibits polyp development in APCMin mice. Carcinogenesis. 2006;27:2103–2107. doi: 10.1093/carcin/bgl056. [DOI] [PubMed] [Google Scholar]
- 3.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 4.Malin A, Matthews CE, Shu XO, et al. Energy balance and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:1496–1501. doi: 10.1158/1055-9965.EPI-04-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvie M, Howell A. Energy balance adiposity and breast cancer - energy restriction strategies for breast cancer prevention. Obes Rev. 2006;7:33–47. doi: 10.1111/j.1467-789X.2006.00207.x. [DOI] [PubMed] [Google Scholar]
- 6.Thune I, Furberg AS. Physical activity and cancer risk: dose-response and cancer, all sites and site-specific. Med Sci Sports Exerc. 2001;33:S530–S550. doi: 10.1097/00005768-200106001-00025. [DOI] [PubMed] [Google Scholar]
- 7.Friedenreich CM, Orenstein MR. Physical activity and cancer prevention: etiologic evidence and biological mechanisms. J Nutr. 2002;132:3456S–3464S. doi: 10.1093/jn/132.11.3456S. [DOI] [PubMed] [Google Scholar]
- 8.Slattery ML, Samowitz W, Curtin K, et al. Associations among IRS1, IRS2, IGF1, and IGFBP3 genetic polymorphisms and colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1206–1214. [PubMed] [Google Scholar]
- 9.Kritchevsky D, Weber MM, Klurfeld DM. Dietary fat versus caloric content in initiation and promotion of 7,12-dimethylbenz(a)anthracene-induced mammary tumorigenesis in rats. Cancer Res. 1984;44:3174–3177. [PubMed] [Google Scholar]
- 10.Welsch CW. Dietary fat, calories, and mammary gland tumorigenesis. Adv Exp Med Biol. 1992;322:203–222. doi: 10.1007/978-1-4684-7953-9_16. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z, Haegele AD, Thompson HJ. Effect of caloric restriction on pre-malignant and malignant stages of mammary carcinogenesis. Carcinogenesis. 1997;18:1007–1012. doi: 10.1093/carcin/18.5.1007. [DOI] [PubMed] [Google Scholar]
- 12.Weindruch R, Walford RL. The retardation of aging and disease by dietary restriction. Springfield, IL: Charles C. Thomas; 1988. p. 436. [Google Scholar]
- 13.Thompson HJ, Ronan AM, Ritacco KA, Tagliaferro AR, Meeker LD. Effect of exercise on the induction of mammary carcinogenesis. Cancer Res. 1988;48:2720–2723. [PubMed] [Google Scholar]
- 14.Thompson HJ, Ronan AM, Ritacco KA, Tagliaferro AR. Effect of type and amount of dietary fat on the enhancement of rat mammary tumorigenesis by exercise. Cancer Res. 1989;49:1904–1908. [PubMed] [Google Scholar]
- 15.Thompson HJ, Westerlind KC, Snedden J, Briggs S, Singh M. Exercise intensity dependent inhibition of 1-methyl-1-nitrosourea induced mammary carcinogenesis in female F-344 rats. Carcinogenesis. 1995;16:1783–1786. doi: 10.1093/carcin/16.8.1783. [DOI] [PubMed] [Google Scholar]
- 16.Thompson HJ. Effects of physical activity and exercise on experimentally-induced mammary carcinogenesis. Breast Cancer Res Treat. 1997;46:135–141. doi: 10.1023/a:1005912527064. [DOI] [PubMed] [Google Scholar]
- 17.Cohen LA, Choi K, Backlund JY, Harris R, Wang CX. Modulation of N-nitrosomethylurea induced mammary tumorigenesis by dietary fat and voluntary exercise. In Vivo. 1991;5:333–344. [PubMed] [Google Scholar]
- 18.Cohen LA, Boylan E, Epstein M, Zang E. Voluntary exercise and experimental mammary cancer. Adv Exp Med Biol. 1992;322:41–59. doi: 10.1007/978-1-4684-7953-9_5. [DOI] [PubMed] [Google Scholar]
- 19.Shephard RJ. Physical activity and cancer. Int J Sports Med. 1990;11:413–420. doi: 10.1055/s-2007-1024830. [DOI] [PubMed] [Google Scholar]
- 20.Basterfield L, Reul JM, Mathers JC. Impact of physical activity on intestinal cancer development in mice. J Nutr. 2005;135:3002S–3008S. doi: 10.1093/jn/135.12.3002S. [DOI] [PubMed] [Google Scholar]
- 21.Colbert LH, Davis JM, Essig DA, Ghaffar A, Mayer EP. Exercise and tumor development in a mouse predisposed to multiple intestinal adenomas. Med Sci Sports Exerc. 2000;32:1704–1708. doi: 10.1097/00005768-200010000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Colbert LH, Mai V, Perkins SN, et al. Exercise and intestinal polyp development in APCMin mice. Med Sci Sports Exerc. 2003;35:1662–1669. doi: 10.1249/01.MSS.0000089349.54813.41. [DOI] [PubMed] [Google Scholar]
- 23.Cohen LA, Kendall ME, Meschter C, Epstein MA, Reinhardt J, Zang E. Inhibition of rat mammary tumorigenesis by voluntary exercise. In Vivo. 1993;7:151–158. [PubMed] [Google Scholar]
- 24.McTiernan A, Sorensen B, Yasui Y, et al. No effect of exercise on insulin-like growth factor 1 and insulin-like growth factor binding protein 3 in postmenopausal women: a 12-month randomized clinical trial. Cancer Epidemiol Biomarkers Prev. 2005;14:1020–1021. doi: 10.1158/1055-9965.EPI-04-0834. [DOI] [PubMed] [Google Scholar]
- 25.Duncan K, Harris S, Ardies CM. Running exercise may reduce risk for lung and liver cancer by inducing activity of antioxidant and phase II enzymes. Cancer Lett. 1997;116:151–158. doi: 10.1016/s0304-3835(97)00189-4. [DOI] [PubMed] [Google Scholar]
- 26.Han ES, Hickey M. Microarray evaluation of dietary restriction. J Nutr. 2005;135:1343–1346. doi: 10.1093/jn/135.6.1343. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Xie L, Sylvester J, et al. Different gene expression of skin tissues between mice with weight controlled by either calorie restriction or physical exercise. Exp Biol Med (Maywood ) 2007;232:473–480. [PubMed] [Google Scholar]
- 28.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, et al. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144:4773–4782. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 29.Miller RS, Becker KG, Prabhu V, Cooke DW. Adipocyte gene expression is altered in formerly obese mice and as a function of diet composition. J Nutr. 2008;138:1033–1038. doi: 10.1093/jn/138.6.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berrigan D, Lavigne JA, Perkins SN, Nagy TR, Barrett JC, Hursting SD. Phenotypic effects of calorie restriction and insulin-like growth factor-1 treatment on body composition and bone mineral density of C57BL/6 mice: implications for cancer prevention. In Vivo. 2005;19:667–674. [PubMed] [Google Scholar]
- 31.Nagy TR, Clair AL. Precision and accuracy of dual-energy X-ray absorptiometry for determining in vivo body composition of mice. Obes Res. 2000;8:392–398. doi: 10.1038/oby.2000.47. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi Y, Lavigne JA, Hursting SD, et al. Using DNA microarray analyses to elucidate the effects of genistein in androgen-responsive prostate cancer cells: identification of novel targets. Mol Carcinog. 2004;41:108–119. doi: 10.1002/mc.20045. [DOI] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alon U, Barkai N, Notterman DA, et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KY, Kim SJ, Cha YS, et al. Effect of exercise on hepatic gene expression in an obese mouse model using cDNA microarrays. Obesity (Silver Spring) 2006;14:1294–1302. doi: 10.1038/oby.2006.147. [DOI] [PubMed] [Google Scholar]
- 37.Nieoullon V, Belvindrah R, Rougon G, Chazal G. mCD24 regulates proliferation of neuronal committed precursors in the subventricular zone. Mol Cell Neurosci. 2005;28:462–474. doi: 10.1016/j.mcn.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Stairs DB, Notarfrancesco KL, Chodosh LA. The serine/threonine kinase, Krct, affects endbud morphogenesis during murine mammary gland development. Transgenic Res. 2005;14:919–940. doi: 10.1007/s11248-005-1806-6. [DOI] [PubMed] [Google Scholar]
- 39.Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med. 2006;8:1–11. doi: 10.1017/S1462399406000056. [DOI] [PubMed] [Google Scholar]
- 40.Blanchard AA, Watson PH, Shiu RP, et al. Differential expression of claudin 1, 3, and 4 during normal mammary gland development in the mouse. DNA Cell Biol. 2006;25:79–86. doi: 10.1089/dna.2006.25.79. [DOI] [PubMed] [Google Scholar]
- 41.Ormandy CJ, Naylor M, Harris J, et al. Investigation of the transcriptional changes underlying functional defects in the mammary glands of prolactin receptor knockout mice. Recent Prog Horm Res. 2003;58:297–323. doi: 10.1210/rp.58.1.297. [DOI] [PubMed] [Google Scholar]
- 42.Harris J, Stanford PM, Sutherland K, et al. Socs2 and elf5 mediate prolactin-induced mammary gland development. Mol Endocrinol. 2006;20:1177–1187. doi: 10.1210/me.2005-0473. [DOI] [PubMed] [Google Scholar]
- 43.Hankinson SE, Willett WC, Michaud DS, et al. Plasma prolactin levels and subsequent risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 1999;91:629–634. doi: 10.1093/jnci/91.7.629. [DOI] [PubMed] [Google Scholar]
- 44.Goodman G, Bercovich D. Prolactin does not cause breast cancer and may prevent it or be therapeutic in some conditions. Med Hypotheses. 2007 Jul 18; doi: 10.1016/j.mehy.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 45.Park CE, Kim YH, Jeon EH, Cha KY, Lee SH, Lee KA. Expression of wee1 and its related cell cycle components in mouse early stage follicles. Cells Tissues Organs. 2004;177:221–228. doi: 10.1159/000080135. [DOI] [PubMed] [Google Scholar]
- 46.Sherr CJ. Mammalian G1 cyclins and cell cycle progression. Proc Assoc Am Physicians. 1995;107:181–186. [PubMed] [Google Scholar]
- 47.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 48.Bell E, Lunec J, Tweddle DA. Cell cycle regulation targets of MYCN identified by gene expression microarrays. Cell Cycle. 2007;6:1249–1256. doi: 10.4161/cc.6.10.4222. [DOI] [PubMed] [Google Scholar]
- 49.Sheikh MS, Hollander MC, Fornance AJ., Jr Role of Gadd45 in apoptosis. Biochem Pharmacol. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- 50.Dunn SE, Kari FW, French J, et al. Dietary restriction reduces insulin-like growth factor I levels, which modulates apoptosis, cell proliferation, and tumor progression in p53-deficient mice. Cancer Res. 1997;57:4667–4672. [PubMed] [Google Scholar]
- 51.Mantzoros CS, Bolhke K, Moschos S, Cramer DW. Leptin in relation to carcinoma in situ of the breast: a study of pre-menopausal cases and controls. Int J Cancer. 1999;80:523–526. doi: 10.1002/(sici)1097-0215(19990209)80:4<523::aid-ijc7>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 52.Stattin P, Soderberg S, Biessy C, et al. Plasma leptin and breast cancer risk: a prospective study in northern Sweden. Breast Cancer Res Treat. 2004;86:191–196. doi: 10.1023/B:BREA.0000036782.11945.d7. [DOI] [PubMed] [Google Scholar]
- 53.Hu X, Juneja SC, Maihle NJ, Cleary MP. Leptin--a growth factor in normal and malignant breast cells and for normal mammary gland development. J Natl Cancer Inst. 2002;94:1704–1711. doi: 10.1093/jnci/94.22.1704. [DOI] [PubMed] [Google Scholar]
- 54.Okumura M, Yamamoto M, Sakuma H, et al. Leptin and high glucose stimulate cell proliferation in MCF-7 human breast cancer cells: reciprocal involvement of PKC-alpha and PPAR expression. Biochim Biophys Acta. 2002;1592:107–116. doi: 10.1016/s0167-4889(02)00276-8. [DOI] [PubMed] [Google Scholar]
- 55.Dieudonne MN, Machinal-Quelin F, Serazin-Leroy V, Leneveu MC, Pecquery R, Giudicelli Y. Leptin mediates a proliferative response in human MCF7 breast cancer cells. Biochem Biophys Res Commun. 2002;293:622–628. doi: 10.1016/S0006-291X(02)00205-X. [DOI] [PubMed] [Google Scholar]
- 56.O'Brien SN, Welter BH, Price TM. Presence of leptin in breast cell lines and breast tumors. Biochem Biophys Res Commun. 1999;259:695–698. doi: 10.1006/bbrc.1999.0843. [DOI] [PubMed] [Google Scholar]
- 57.Colbert LH. Mechanisms associating physical activity with cancer incidence: animal models. In: McTiernan A, editor. Cancer prevention and management through exercise and weight control. Boca Raton: CRC Press; 2005. pp. 325–339. [Google Scholar]