Abstract
The t(8;21) translocation between two genes known as AML1 and ETO is seen in approximately 12–15% of all acute myeloid leukemia (AML) and is the second-most-frequently observed nonrandom genetic alteration associated with AML. AML1 up-regulates a number of target genes critical to normal hematopoiesis, whereas the AML1/ETO fusion interferes with this trans-activation. We discovered that the fusion partner ETO binds to the human homolog of the murine nuclear receptor corepressor (N-CoR). The interaction is mediated by two unusual zinc finger motifs present at the carboxyl terminus of ETO. Human N-CoR (HuN-CoR), which we cloned and sequenced in its entirety, encodes a 2,440-amino acid polypeptide and has a central domain that binds ETO. N-CoR, mammalian Sin3 (mSin3A and B), and histone deacetylase 1 (HDAC1) form a complex that alters chromatin structure and mediates transcriptional repression by nuclear receptors and by a number of oncoregulatory proteins. We found that ETO, through its interaction with the N-CoR/mSin3/HDAC1 complex, is also a potent repressor of transcription. This observation provides a mechanism for how the AML1/ETO fusion may inhibit expression of AML1-responsive target genes and disturb normal hematopoiesis.
Acute myeloid leukemia (AML) is a hematologic malignancy characterized by the proliferation of a transformed clone of myeloid progenitor cells. The t(8;21)(q22;q22) translocation is one of the most frequently observed nonrandom genetic alterations and is associated with AML with maturation (M2 morphology) (1). Juxtaposition of the AML1 gene on chromosome 21 to the ETO gene on chromosome 8 fuses the amino-terminal portion of AML1 with near-full-length ETO, creating the AML1/ETO chimeric fusion (2–4). The portion of AML1 contained in the fusion includes a central 118-amino acid domain homologous to the Drosophila segmentation gene runt (3), which serves to bind the enhancer core DNA sequence TGT/cGGT (5). AML1 is able to form a heterodimer with core-binding factor β (CBFβ). The AML1-CBFβ transcription factor is an important regulator of a number of target genes involved in hematopoiesis, many of which are homeobox-containing HOX genes (1, 6). Murine embryos with targeted mutations in AML1 lacked fetal liver hematopoiesis, reinforcing the idea that AML1 is critical to normal blood cell development (7).
The AML1/ETO fusion retains the ability to interact with the enhancer core DNA sequence via the runt homology domain (RHD) and interferes with the expression of AML1-responsive target genes (8, 9). In mice heterozygous for a “knocked-in” AML1/ETO allele, hematopoiesis was profoundly impaired (10) as in the AML1 knock-out mice (7), suggesting that the chimeric fusion blocks wild-type AML1 function in a trans-dominant manner. The AML1/ETO fusion contains nearly full-length ETO, missing only a small region with no DNA-binding or transcription regulation motifs. ETO is a phosphoprotein that is normally expressed in brain tissue (4) and in CD34+ hematopoietic cells (11). Ectopic expression of ETO in NIH 3T3 cells, however, leads to transformation (12). With two zinc finger motifs and proline-rich or proline/serine/threonine-rich regions, ETO structurally resembles a transcription factor (4, 13), although DNA-binding properties have not yet been confirmed. Mutation analysis has identified ETO sequences within the chimeric fusion as being required for the dominant repression of transcription of AML1 target genes (14).
Recently, other oncoregulatory proteins involved in transcriptional repression have been found to interact with corepressor factors that subserve important functions in modifying chromatin structure by histone deacetylation (15, 16). Mad and Mxi1 proteins are antagonists of the Myc family of transcription factors. Mxi1-mediated inhibition of Myc requires interaction with mammalian Sin3 (mSin3A or mSin3B) proteins (16). The nuclear receptor corepressor (N-CoR) and histone deacetylase 1 (HDAC1) are two other members of a resultant complex that represses transcription by enzymatic deacetylation of histones and creation of a repressive chromatin structure (15, 16).
In our experiments, we sought to better understand the transcriptional regulation properties of ETO by examining its interaction with other proteins. Our findings uncovered a previously unrecognized link between the ETO oncoprotein and the N-CoR/mSin3/HDAC1 transcription repression pathway.
MATERIALS AND METHODS
Two-Hybrid Methodology.
The entire cDNA coding region of human ETO (MTG8a) was generated by polymerase chain amplification (PCR) using pCRII/ETO as a template (12). The amplified fragment was inserted into the pGBT9 plasmid (CLONTECH). DNA sequencing was performed to confirm the in-frame fusion between ETO and the GAL4 DNA-binding domain (DBD).
A human fetal brain cDNA library (CLONTECH) inserted into the pGAD10 plasmid containing the GAL4 activation domain was screened by using the pGBT9-ETO cDNA as bait. HF7c yeast cells were transformed with pGBT9-ETO and the library plasmid DNA and grown on tryptophan−, leucine−, and histidine− selective medium plates. The colonies were transferred onto filter paper and frozen in liquid nitrogen to lyse the yeast cells. β-Galactosidase assays (performed multiple times to exclude false positives) were performed to identify potential positive colonies. Plasmids were extracted from yeast and used to transform Escherichia coli HB101 cells. Plasmids extracted from E. coli were then analyzed by DNA sequencing.
In Vitro Protein Interaction Analysis: Glutathione S-Transferase (GST) Pull-Down Assay.
The B2 insert was recovered and cloned into pGEX-5X-1 (Pharmacia), creating an in-frame fusion to GST. GST and GST-B2 fusion proteins were expressed in BL21 E. coli cells, and equal amounts of each were immobilized onto glutathione-Sepharose beads. The beads were incubated for 12 hr with 35S-labeled full-length ETO protein produced by in vitro translation (Promega). The beads were washed with washing buffer (10 mM Tris⋅HCl/150 mM NaCl/1 mM EDTA/1% Nonidet P-40/1% deoxycholic acid containing 10 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride) three times. Proteins, eluted by glutathione elution buffer, were subjected to SDS/polyacrylamide gel electrophoresis (PAGE) and autoradiography.
Cloning of the Full-Length HuN-CoR cDNA.
Using the B2 insert fragment as a probe, we screened human fetal brain cDNA libraries (D. Tagle, National Institutes of Health). A total of six overlapping fragments were obtained and analyzed by automated DNA sequencing (Perkin–Elmer).
Immunoprecipitation and Western Blot Experiments.
293 cells were transfected by calcium phosphate coprecipitation with mammalian expression plasmids expressing ETO or AML1/ETO (S. Hiebert, Vanderbilt University, Nashville, TN) alone or with Flag epitope-tagged HDAC1 (S. Schreiber, Harvard University, Cambridge, MA) or Flag-tagged N-CoR (M. G. Rosenfeld, University of California, San Diego). The cells were cultured for 2 days after transfection, collected, resuspended in cell lysis buffer (PBS containing 0.1% Nonidet P-40, 1 mM EDTA, 1 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride), and sonicated. Cell lysates were obtained after centrifugation at 10,000 rpm in an Eppendorf microcentrifuge for 2 min. The M2 monoclonal antibody against the Flag epitope (Sigma), rabbit polyclonal antibody against mSin3A (Santa Cruz Biotechnology), or rabbit polyclonal antibody against ETO (Calbiochem) and 20 μl of protein A/G agarose were added to cell lysates. Immunoprecipitation was performed at 4°C for 12 hr. After centrifugation, washing with cell lysis buffer, and denaturation, immunoprecipitated proteins were applied to SDS/PAGE gels for electrophoresis. Proteins were transferred onto nitrocellulose membranes, and blocking was performed in TBST buffer (10 mM Tris⋅HCl/150 mM NaCl/0.05% Tween-20) containing 5% nonfat milk. Western blotting was done with rabbit polyclonal antibodies against either AML1/RHD (Calbiochem) or ETO. Proteins were visualized by anti-rabbit IgG conjugated with alkaline phosphatase (Promega).
Construction of ETO Truncation Expression Plasmids.
Carboxyl-terminal deletions of pGBT9/ETO were constructed (Erase-a-Base System, Promega). PCR was applied with pCRII/ETO (12) as a template, the unique antisense primer 5′-CGC GGA TCC CAG TTC TGA GTT CAC GTC-3′, and the following sense primers: 5′-CG GAA TTC TCA AGC GAG AGT TGC TGG-3′ (to amplify ETO amino acids 484–578) and 5′-CG GAA TTC AAC ACA GCC CGA TAC TGT-3′ (to amplify ETO amino acids 503–578). The amplified fragments were ligated into the pGBT9 vector (fused in-frame with the GAL4 DBD).
Mammalian Expression Vector Construction.
To construct an expression vector for ETO, PCR was performed using pCRII/ETO as template with the following primers: 5′-GCTCTAGAACCTGATCGTACTGAG-3′ and 5′-CGGGGTACCTCGCGTTGGTTGTGTT-3′. PCR fragments were inserted into the pFA-CMV plasmid (Stratagene) to create GAL4 DBD/ETO.
Mammalian Cell Transfection.
Transfections of CV-1 (ATCC) cells were performed with various amounts (indicated in Fig. 5) of GAL4 DBD/ETO, 2.0 μg of the luciferase reporter plasmid, 1.0 μg of the CMX-β-galactosidase plasmid (R. Evans and D. Chen, Salk Institute, La Jolla, CA) as an internal control, and various amounts of the pUC19 plasmid to maintain equal amounts of transfected DNA among the different experimental groups. The reporter plasmid contains four copies of the GAL4 binding site upstream of the TK (thymidine kinase) promoter (B. O’Malley, Baylor Medical School, Houston, TX). The RARα LBD plasmid was from R. Evans and D. Chen. Luciferase activity was normalized to β-galactosidase determinations.
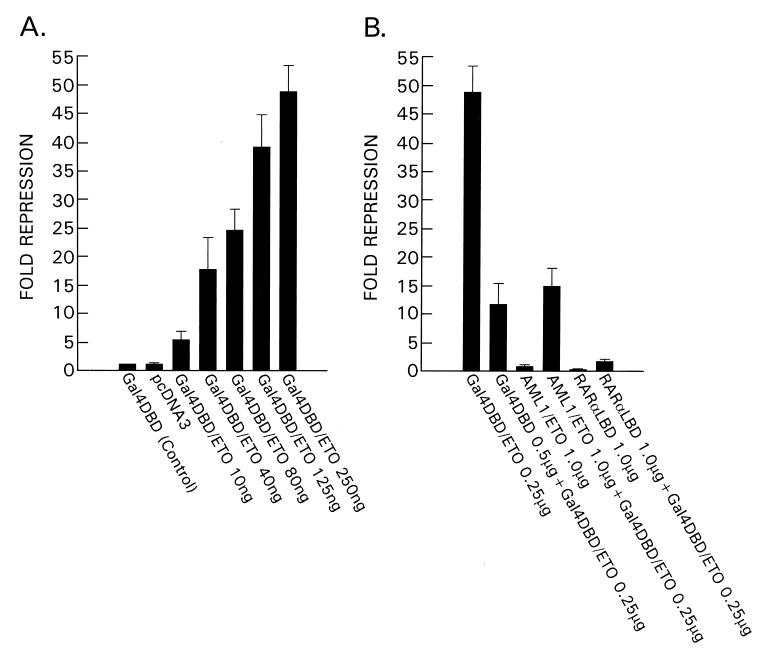
Figure 5.
In the mammalian expression plasmid GAL4 DBD/ETO, ETO is fused with the GAL4 DBD (amino acids 1–147). The firefly luciferase reporter gene is driven by the TK promoter with four copies of the GAL4 DNA-binding site upstream. (A) ETO exhibited potent, dose-dependent, transcriptional repression. (B) GAL4 DBD alone partially relieved repression by GAL4 DBD/ETO, due to competition for GAL4 binding sites. When AML1/ETO or RARα LBD, both driven by the cytomegalovirus promoter, were cotransfected with GAL4 DBD/ETO, the repressive effects of ETO were almost abrogated. The AML1/ETO fusion and RARα LBD may compete with ETO for HuN-CoR binding, leading to relief of repression.
RESULTS
To better understand transcriptional regulation by ETO, we applied the yeast two-hybrid method to identify potential ETO-binding proteins. The ETO gene fused to the GAL4 DBD was used as bait to screen a human fetal brain cDNA library fused to the GAL4 activation domain. Putative ETO-binding proteins should recruit the GAL4 activation domain to the promoter of the reporter gene, resulting in expression of the reporter. From the cDNA library, we isolated a clone that strongly interacted with ETO. The sequence of this 2.4-kb insert cDNA, clone B2, had greater than 90% homology with that of the murine N-CoR (17). The ETO-interacting protein was therefore named the human nuclear receptor corepressor or HuN-CoR. Historically, the murine N-CoR (17, 18) and a related corepressor known as SMRT (silencing mediator for retinoid and thyroid-hormone receptors) (19) were identified as molecules interacting with DNA-bound nuclear receptors for thyroid hormone (T3R) and retinoic acid (RAR). These receptors can heterodimerize with the retinoid-X receptor (RXR). The ligand-binding domain (LBD) of T3R and RAR interacts with the murine N-CoR to repress basal (ligand-independent) transcription of target genes (17, 18). Histone deacetylation has been proposed as a major mechanism underlying this transcriptional repression, as a result of recruitment of a repressor complex including N-CoR (or SMRT), mSin3A, and HDAC1 (15, 20).
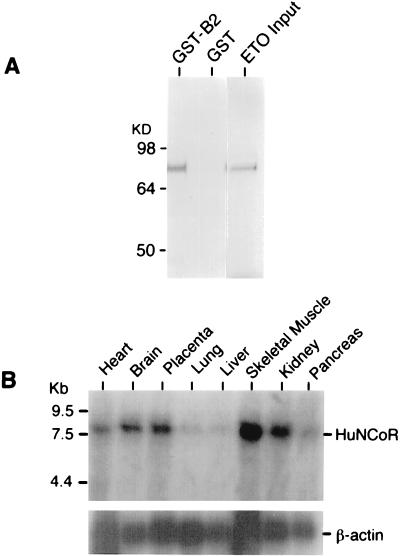
To confirm that the ETO and the HuN-CoR proteins interact in vitro, we performed GST coprecipitation assays (Fig. 1A). The B2 DNA fragment was inserted into pGEX-5X-1 to fuse the fragment with GST. GST and GST-B2 fusion proteins were expressed in E. coli and purified. Equal amounts of protein were immobilized on glutathione beads and incubated with in vitro-translated ETO labeled with [35S]methionine. After extensive washing, the eluted proteins were subjected to electrophoresis and autoradiography. ETO was specifically precipitated by GST-B2 but not by GST alone (Fig. 1A). This result confirmed the physical interaction between ETO and the B2 HuN-CoR fragment.
Figure 1.
(A) The B2-HuN-CoR fragment was fused with GST. 35S-labeled ETO protein generated by in vitro translation was specifically coprecipitated by GST-B2 but not by GST alone. (B) mRNA from various human tissues (CLONTECH) was subjected to Northern blot analysis using the B2 probe. The analysis revealed that the hybridizing mRNA (labeled HuNCoR) was approximately 8,000 bases in size, indicating that the B2 fragment was a partial cDNA. β-Actin mRNA controls are shown below.
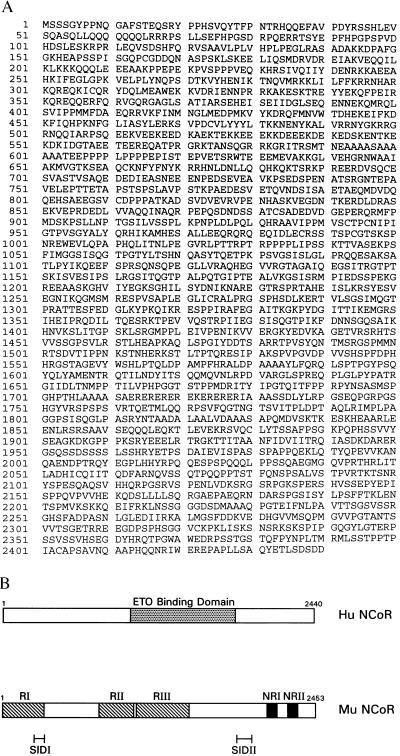
The B2 HuN-CoR fragment identified from the yeast two-hybrid assays was 2.4 kb in size. Using this fragment as a probe, Northern blot hybridization analysis revealed an approximately 8-kb transcript (Fig. 1B). This result indicated that the HuN-CoR cDNA fragment isolated by our yeast two-hybrid screen was only a partial cDNA sequence. To clone full-length HuN-CoR, a human fetal brain cDNA library was screened. Six overlapping fragments were obtained that constituted the full-length HuN-CoR, encoding a 2,440-amino acid polypeptide with 96% similarity and 92% identity to the murine N-CoR (Fig. 2A). In functional assays, the HuN-CoR acted similarly to the murine N-CoR in suppressing RAR/RXR-induced transcriptional activation (R.L.R. and J.W., unpublished data). The structure of the murine N-CoR can be divided into distinct functional domains that mediate repression (RI, RII, RIII) (17), interact with the nuclear receptor (NRI, NRII) (17), or interact with the Sin3 corepressor complex (SIDI, SIDII) (15) (Fig. 2B). Our yeast two-hybrid analyses demonstrated that the region of the HuN-CoR that binds ETO includes residues 988-1816. By comparison with the functional domains of the murine N-CoR, the ETO-binding domain therefore lies between SIDI and SIDII and roughly corresponds to RIII (Fig. 2B).
Figure 2.
(A) Using B2 as a probe, we screened a human fetal brain cDNA library to obtain the full-length sequence of the ETO-binding protein, HuN-CoR. Shown is the complete amino acid sequence (GenBank accession no. AF044209). (B) The structure of the murine N-CoR (Mu N-CoR) can be divided into domains that mediate repression (RI, RII, RIII) (17), interact with the nuclear receptor (NRI, NRII) (17), or interact with the Sin3 corepressor complex (SIDI, SIDII) (15). By comparison with the functional domains of murine N-CoR, the ETO-binding domain of HuN-CoR lies between SIDI and SIDII and roughly corresponds to RIII.
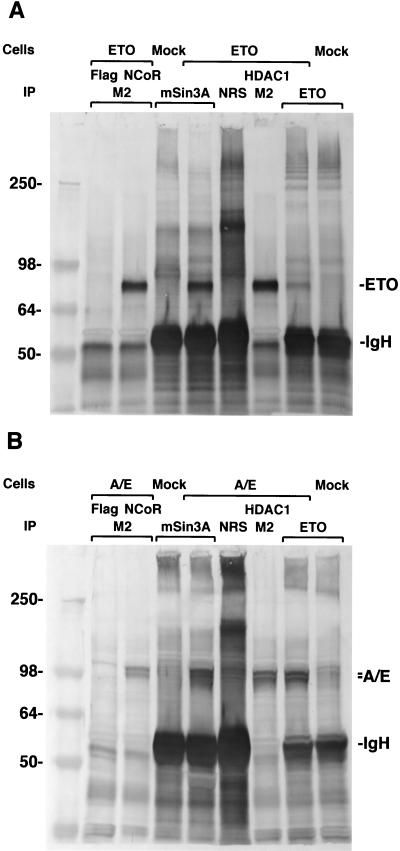
Having determined that ETO interacts with a central domain of the HuN-CoR, we then asked whether ETO is able to associate with other members of the corepressor complex. We transfected 293 cells with a construct expressing ETO alone or with a plasmid expressing the Flag epitope, N-CoR/Flag, or HDAC1/Flag. Immunoprecipitation was performed with either the M2 anti-FLAG antibody (against Flag, N-CoR/Flag, or HDAC1/Flag) or antibody against mSin3A (Fig. 3A). After immunoprecipitation, ETO specifically associated with the N-CoR/mSin3A/HDAC1 corepressor complex as demonstrated by Western blotting with an anti-ETO antibody. These results suggested that the ETO protein forms a complex with N-CoR, mSin3A, and HDAC1 in vivo. The AML1/ETO chimeric protein contains near-full-length ETO protein, implying that the AML1/ETO protein would also be associated with the corepressor complex through ETO residues. To confirm this hypothesis, 293 cells were transfected with AML1/ETO, alone or with Flag, N-CoR/Flag, or HDAC1/Flag (Fig. 3B). Cell lysates were prepared from transfected and mock-transfected 293 cells. Either the M2 antibody or an anti-mSin3A antibody was used for immunoprecipitation, after which Western blotting was done with an anti-AML1/RHD antibody. The AML1/ETO protein coprecipitated with N-CoR, mSin3A, and HDAC1, suggesting binding to the complex in vivo.
Figure 3.
(A) ETO associates with the N-CoR/mSin3A/HDAC1 complex in vivo. Cells were transfected with ETO alone or with Flag, N-CoR/Flag, or HDAC1/Flag. Immunoprecipitation (IP) was performed with either the M2 anti-FLAG antibody (against Flag, N-CoR/Flag, or HDAC1/Flag) or antibody against mSin3A. Normal rabbit serum (NRS) was used as negative control for the IP antibody. After IP, ETO specifically associated with the N-CoR/mSin3A/HDAC1 complex as demonstrated by Western blotting with an anti-ETO antibody. Cell lysates from ETO-transfected and untransfected (mock) cells, precipitated by anti-ETO antibody, were used as positive and negative controls, respectively (1/4 amount of lysate used as for the other experiments). Molecular mass markers are shown in kilodaltons. IgH, immunoglobulin heavy chain. (B) AML1/ETO (abbreviated A/E) associates with the N-CoR/mSin3A/HDAC1 complex in vivo. Cells were transfected with A/E, alone or with Flag, N-CoR/Flag, or HDAC1/Flag. After IP, proteins were subjected to Western blotting using anti-AML1/RHD antibody. Proteins from lysates of A/E-transfected or mock-transfected cells precipitated by ETO antibody and blotted by AML1/RHD antibody were used as positive and negative controls for the AML1/ETO protein (1/4 amount of lysate used as for the other experiments). The doublet band (denoted by the double tick) seen in the figure may be due to translation at different ATG start codons.
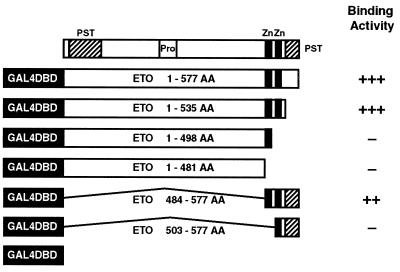
To define the region of ETO that interacted with the HuN-CoR, a series of ETO truncation mutants were constructed and inserted into the pGBT9 vector (Fig. 4). By yeast two-hybrid assays, we found that deletion of either zinc finger, located at the carboxyl terminus of the ETO protein, abrogated binding to the HuN-CoR. These results demonstrated that the HuN-CoR binding domain localized to the zinc finger motifs. The cysteine-histidine sequences within ETO’s zinc fingers are unusual and do not match those of previously defined DNA-binding zinc fingers (21). This Cys-His region is highly conserved, however, between ETO and its Drosophila homolog, the homeotic target gene nervy (22), another Drosophila protein called DEAF-1 (deformed epidermal autoregulatory factor 1) (23), and a gene involved in apoptosis known as RP-8 (24).
Figure 4.
To determine the HuN-CoR binding domain of ETO, a series of ETO truncation mutants were constructed and inserted into the pGBT9 vector. Yeast cells were cotransformed with the B2 plasmid and the ETO truncation mutants. β-Galactosidase assays were used to test for binding activity in vivo (scored from − to +++). Shown in the schematic is the structure of the ETO protein, with proline/serine/threonine (PST)- and proline (Pro)-rich domains as well as the two zinc finger motifs (Zn). Deletion of either one of the two zinc finger motifs in the ETO protein abrogated binding to the HuN-CoR, thus localizing the binding domain to these motifs.
To determine the effects of ETO on regulation of transcription, we placed the GAL4 DBD/ETO fusion into a mammalian expression vector. This vector was cotransfected with a luciferase reporter gene plasmid in which four copies of the GAL4 DNA-binding site have been placed upstream of the reporter gene’s TK promoter. Through the GAL4 DBD, the ETO protein should be recruited to the regulatory region of the reporter gene. In this assay system, ETO exhibited potent, dose-dependent transcriptional repression (Fig. 5A). Cotransfection of a vector expressing the AML1/ETO fusion abrogated transcriptional repression by the GAL4 DBD/ETO fusion, presumably by competing for binding to the endogenous HuN-CoR corepressor complex (Fig. 5B).
The RARα LBD is known to interact with the murine N-CoR by means of the NRI and NRII domains (17, 18). We reasoned that the RARα LBD might also compete with the GAL4 DBD/ETO fusion for binding to the endogenous HuN-CoR complex. Consistent with this hypothesis, repression by the GAL4 DBD/ETO fusion was almost completely blocked by cotransfection and addition of the RARα LBD (Fig. 5B). Although the ETO-binding site of the HuN-CoR does not overlap NRI and NRII, the RARα LBD/HuN-CoR complex may deplete HuN-CoR molecules, making them unavailable for binding to ETO.
DISCUSSION
The AML1/ETO chimeric fusion blocks trans-activation of AML1-responsive hematopoietic target genes. Downstream targets of AML1 include such important genes as those encoding myeloperoxidase, neutrophil elastase, interleukin 3, and granulocyte–macrophage colony-stimulating factor (8, 9). The fusion also can block trans-activation induced by other members of the AML1 family of transcription factors that all bind the core enhancer sequence by means of the RHD (25). Recently, AML1 has been found to interact with the multifunctional transcriptional coactivator p300 (26). Originally identified as a cellular protein that could bind to the adenovirus-E1a oncoprotein, p300 interacts with a histone acetyltransferase, P/CAF (27) and itself has acetyltransferase activity (28), serving to regulate transcription through chromatin remodeling and recruitment of basal transcription factors. Disruption of AML1 function by the AML1/ETO fusion thus affects both transcription activation and cellular differentiation. It had previously been hypothesized that this functional block resulted from direct competition for AML1 binding sites. However, our experiments implicate the HuN-CoR corepressor complex as a mediator of transcriptional repression by ETO and suggest the following model. The ETO portion of the AML1/ETO fusion interacts with the corepressor complex. The AML1/ETO fusion contains only the RHD of AML1 and lacks the carboxyl-terminal region of AML1 that interacts with the coactivator p300 (26). In place of this interaction, the runt DNA-binding domain instead recruits the N-CoR/mSin3/HDAC1 complex to the promoter of AML1-responsive target genes, resulting in histone deacetylation and transcriptional repression.
The model that we propose whereby a sequence-specific DNA-binding protein has its function altered by fusion with a protein capable of recruiting the N-CoR/mSin3/histone deacetylase complex is similar to the model suggested for the PLZF-RARα variant of acute promyelocytic leukemia (29). PLZF has also been found to interact autonomously with SMRT (as well as N-CoR, mSin3, and HDAC1), and both ETO and PLZF appear to function as transcriptional repressors in a ligand-independent manner. PLZF interacts with SMRT by means of the so-called POZ (pox viruses and zinc fingers) domain (29). We have mapped the N-CoR binding region of ETO to its zinc finger motifs, a region that does not resemble a POZ domain.
Our experiments have focused on ETO’s role in the context of the AML1/ETO fusion. The physiologic or developmental function of wild-type ETO in brain and hematopoietic tissues (where ETO is expressed) is unknown. ETO’s Drosophila homolog, nervy, is expressed in segregating neuroblasts during embryogenesis, suggesting a regulatory role in early development (22). ETO’s structure is typical for a transcription factor, but ETO has not yet been shown to bind DNA. In addition to their other homologies, both ETO and nervy also contain an area of similarity to the Drosophila coactivator TAF110 (TATA-binding protein-associated factor 110) (13). This suggests that in an appropriate context, ETO may also have gene activation properties.
Recent observations have suggested a link between chromatin remodeling and cancer. An AML-associated chromosomal translocation has been described that fuses histone acetyltransferase CREB-binding protein (CBP) to the zinc-finger domain of MOZ (monocytic leukemia zinc finger) (30). Both the Myc antagonists Mad and Mxi1 (15, 16, 31) and the retinoblastoma protein (32, 33) induce transcriptional repression through the recruitment of corepressor factors. The fusion proteins of RARα associated with acute promyelocytic leukemia have also been found to interact with the histone deacetylase complex (29, 34, 35). Our experiments indicate that the ETO oncoprotein as well as the AML1/ETO fusion protein suppresses transcription by recruitment of a multimolecular complex capable of remodeling chromatin into a repressive conformation. This pathway presents itself as a potential target for novel anticancer therapies.
Acknowledgments
We thank N. Young for support and encouragement.
ABBREVIATIONS
- AML
acute myeloid leukemia
- RHD
runt homology domain
- N-CoR
nuclear receptor corepressor
- HuN-CoR
human N-CoR
- HDAC1
histone deacetylase 1
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- GST
glutathione S-transferase
- SMRT
silencing mediator for retinoid and thyroid-hormone receptors
- RAR
retinoic acid receptor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF044209).
References
- 1.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 2.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 4.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, et al. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 7.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 8.Meyers S, Lenny N, Hiebert S W. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer SD. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 10.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 11.Erickson P F, Dessev G, Lasher R S, Philips G, Robinson M, Drabkin H A. Blood. 1996;88:1813–1823. [PubMed] [Google Scholar]
- 12.Wang J, Wang M, Liu J M. Cancer Res. 1997;57:2951–2955. [PubMed] [Google Scholar]
- 13.Erickson P F, Robinson M, Owens G, Drabkin H A. Cancer Res. 1994;54:1782–1786. [PubMed] [Google Scholar]
- 14.Lenny N, Meyers S, Hiebert S W. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 15.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 16.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 17.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Nature (London) 1995;377:397–403. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 18.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Nature (London) 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 20.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 21.Evans R M, Hollenberg S M. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 22.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross C T, McGinnis W. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 24.Owens G P, Hahn W E, Cohen J J. Mol Cell Biol. 1991;11:4177–4188. doi: 10.1128/mcb.11.8.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers S, Lenny N, Sun W, Hiebert S W. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 26.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V V, Schiltz R L, Rusanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Hong S-H, David G, Wong C-W, Dejean A, Privalsky M L. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, et al. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 31.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 32.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 33.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 34.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 35.Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]