Abstract
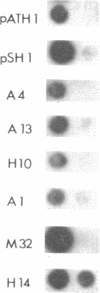
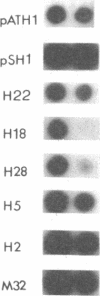
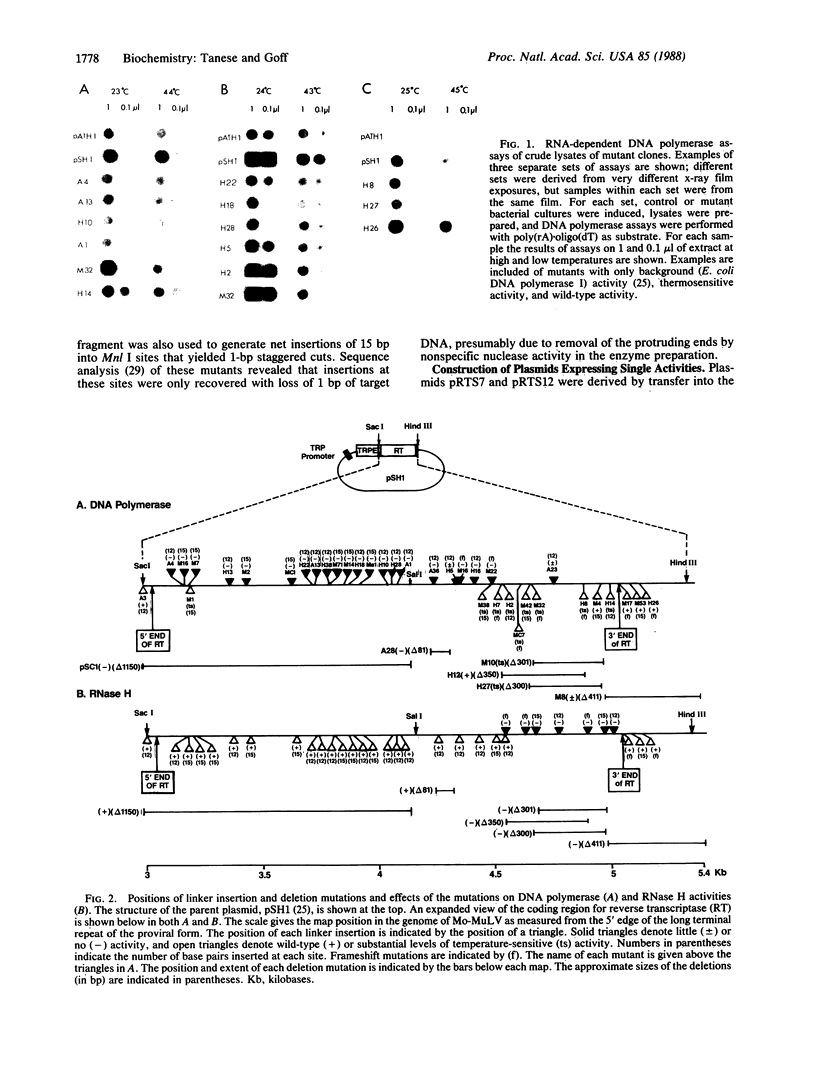
The reverse transcriptase of Moloney murine leukemia virus, like that of all retroviruses, exhibits a DNA polymerase activity capable of synthesis on RNA or DNA templates and an RNase H activity with specificity for RNA in the form of an RNA.DNA hybrid. We have generated a library of linker insertion mutants of the Moloney murine leukemia virus enzyme expressed in bacteria and assayed these mutants for both enzymatic activities. Those mutations affecting the DNA polymerase activity were clustered in the 5'-proximal two-thirds of the gene, and those affecting RNase H were in the remaining 3' one-third. Based on these maps, plasmids were made that expressed each one of the domains separately; assays of the proteins encoded by these plasmids showed that each domain exhibited only the expected activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Gilboa E., Baltimore D. Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J Virol. 1984 Mar;49(3):686–691. doi: 10.1128/jvi.49.3.686-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Dierks P., Parsons J. T., Faras A. J. RNase H hydrolysis of the 5' terminus of the avian sarcoma virus genome during reverse transcription. Nature. 1978 Mar 9;272(5649):181–184. doi: 10.1038/272181a0. [DOI] [PubMed] [Google Scholar]
- Freeman-Wittig M. J., Vinocour M., Lewis R. A. Differential effects of captan on DNA polymerase and ribonuclease H activities of avian myeloblastosis virus reverse transcriptase. Biochemistry. 1986 May 20;25(10):3050–3055. doi: 10.1021/bi00358a047. [DOI] [PubMed] [Google Scholar]
- Gerard G. F., Grandgenett D. P. Purification and characterization of the DNA polymerase and RNase H activities in Moloney murine sarcoma-leukemia virus. J Virol. 1975 Apr;15(4):785–797. doi: 10.1128/jvi.15.4.785-797.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki M., Panet A. Discrimination of DNA polymerase and RNase H activities in reverse transcriptase of avian myeloblastosis virus. Biochemistry. 1978 Jun 13;17(12):2438–2442. doi: 10.1021/bi00605a030. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Gerard G. F., Green M. A single subunit from avian myeloblastosis virus with both RNA-directed DNA polymerase and ribonuclease H activity. Proc Natl Acad Sci U S A. 1973 Jan;70(1):230–234. doi: 10.1073/pnas.70.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D., Quinn T., Hippenmeyer P. J., Oroszlan S. Structural characterization of the avian retrovirus reverse transcriptase and endonuclease domains. J Biol Chem. 1985 Jul 15;260(14):8243–8249. [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchick J. J., Jamjoom G. A., Watson K. F., Arlinghaus R. B. Biosynthesis of reverse transcriptase from Rauscher murine leukemia virus by synthesis and cleavage of a gag-pol read-through viral precursor polyprotein. Proc Natl Acad Sci U S A. 1978 Apr;75(4):2016–2020. doi: 10.1073/pnas.75.4.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai M. H., Verma I. M. Reverse transcriptase of RNA tumor viruses. V. In vitro proteolysis of reverse transcriptase from avian myeloblastosis virus and isolation of a polypeptide manifesting only RNase H activity. J Virol. 1978 Feb;25(2):652–663. doi: 10.1128/jvi.25.2.652-663.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G., Hu S. C., Rein A., Messer L. I., Gerwin B. I. Murine leukemia virus mutant with a frameshift in the reverse transcriptase coding region: implications for pol gene structure. J Virol. 1984 Aug;51(2):470–478. doi: 10.1128/jvi.51.2.470-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L. I., Goff S. P. Construction of mutants of Moloney murine leukemia virus by suppressor-linker insertional mutagenesis: positions of viable insertion mutations. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4149–4153. doi: 10.1073/pnas.81.13.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S. L., Smith S. W., Modak M. J. Reverse transcriptase-associated RNase H activity. II. Inhibition by natural and synthetic RNA. J Virol. 1978 Sep;27(3):576–581. doi: 10.1128/jvi.27.3.576-581.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalith M., Falk H., Panet A. Differential inhibition of DNA polymerase and RNase H activities of the reverse transcriptase by phosphonoformate. Mol Cell Biochem. 1982 Mar 19;43(2):97–103. doi: 10.1007/BF00423097. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moelling K. Further characterization of the Friend murine leukemia virus reverse transcriptase-RNase H complex. J Virol. 1976 May;18(2):418–425. doi: 10.1128/jvi.18.2.418-425.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Faras A. J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982 Oct;30(3):797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- Resnick R., Omer C. A., Faras A. J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984 Sep;51(3):813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Tanese N., Goff S. P. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J Biol Chem. 1985 Aug 5;260(16):9326–9335. [PubMed] [Google Scholar]
- Rucheton M., Lelay M. N., Jeanteur P. Evidence from direct visualization after denaturing gel electrophoresis that RNase H is associated with MSV-MuLV reverse transcripase. Virology. 1979 Aug;97(1):221–223. doi: 10.1016/0042-6822(79)90392-1. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Modak M. J. Reverse transcriptase-associated RNase H. Part IV. Pyrophosphate does not inhibit RNase H activity of AMV DNA polymerase. Biochem Biophys Res Commun. 1979 Dec 14;91(3):892–899. doi: 10.1016/0006-291x(79)91963-6. [DOI] [PubMed] [Google Scholar]
- Tanese N., Roth M. J., Goff S. P. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol. 1986 Aug;59(2):328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Roth M., Goff S. P. Expression of enzymatically active reverse transcriptase in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4944–4948. doi: 10.1073/pnas.82.15.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970 Jun 27;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- Verma I. M., Mason W. S., Drost S. D., Baltimore D. DNA polymerase activity from two temperature-sensitive mutants of Rous sarcoma virus is thermolabile. Nature. 1974 Sep 6;251(5470):27–31. doi: 10.1038/251027a0. [DOI] [PubMed] [Google Scholar]
- Verma I. M. Studies on reverse transcriptase of RNA tumor viruses III. Properties of purified Moloney murine leukemia virus DNA polymerase and associated RNase H. J Virol. 1975 Apr;15(4):843–854. doi: 10.1128/jvi.15.4.843-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. Studies on reverse transcriptase of RNA tumor viruses. I. Localization of thermolabile DNA polymerase and RNase H activities on one polypeptide. J Virol. 1975 Jan;15(1):121–126. doi: 10.1128/jvi.15.1.121-126.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. The reverse transcriptase. Biochim Biophys Acta. 1977 Mar 21;473(1):1–38. doi: 10.1016/0304-419x(77)90005-1. [DOI] [PubMed] [Google Scholar]