Abstract
Here we investigated the bacterial endosymbionts of weevils of the genus Curculio. From all four species of Curculio weevils examined, a novel group of bacterial gene sequences were consistently identified. Molecular phylogenetic analyses demonstrated that the sequences formed a distinct clade in the Gammaproteobacteria, which was not related to previously known groups of weevil endosymbionts such as Nardonella spp. and Sodalis-allied symbionts. In situ hybridization revealed that the bacterium was intracellularly harbored in a bacteriome associated with larval midgut. In adult females, the bacterium was localized in the germalia at the tip of each overiole, suggesting vertical transmission via ovarial passage. Diagnostic PCR surveys detected high prevalence of the bacterial infection in natural host populations. Electron microscopy identified the reduced cell wall of the bacterial cells, and the bacterial genes exhibited AT-biased nucleotide composition and accelerated molecular evolution, which are suggestive of a long-lasting endosymbiotic association. On the basis of these results, we conclude that the novel endocellular bacteria represent the primary symbiont of Curculio weevils and proposed the designation “Candidatus Curculioniphilus buchneri.” In addition to “Ca. Curculioniphilus,” we identified Sodalis-allied gammaproteobacterial endosymbionts from the chestnut weevil, Curculio sikkimensis, which exhibited partial infection frequencies in host insect populations and neither AT-biased nucleotide composition nor accelerated molecular evolution. We suggest that such Sodalis-allied secondary symbionts in weevils might provide a potential source for symbiont replacements, as has occurred in an ancestor of Sitophilus grain weevils.
Diverse insects are obligatorily or facultatively associated with endosymbiotic bacteria, many of which provide a set of “tool kits” consisting of various genes that confer benefits to their host insects (5, 20, 30), such as supply of essential nutrients (1, 39), tolerance to high temperature (29), resistance to parasites and pathogens (33, 37), alteration of food plant range (16, 45), synthesis of toxic substances (21, 48), enhancement of energy metabolism (13), etc. On the other hand, some endosymbiotic bacteria are rather parasitic and affect their host insects negatively, either by reducing the host survival and fecundity or by manipulating the host's reproduction in a selfish manner (3, 46). The knowledge of such ubiquitous microbial associates is pivotal for understanding the ecology, adaptation, and evolution of insects.
The weevil superfamily Curculionoidea (Insecta: Coleoptera) represents the most species-rich animal group, embracing over 60,000 described species (27). Early histological studies reported that many, if not all, of them possess specialized symbiotic organs and harbor bacterial endosymbionts therein (4, 5, 26, 31, 32, 34, 38). Despite the potential diversity of weevil endosymbionts, modern microbiological characterization has been performed on a quite limited number of weevil groups: mostly weevils of the family Dryophthoridae and a few species from the subfamilies Molytinae and Cryptorhynchinae. A clade of gammaproteobacterial endosymbionts, Nardonella spp., has been identified from diverse dryophthorid weevils of the genera Cosmopolites, Matamasius, Rhynchophorus, Scyphophorus, Sphenophorus, and Yuccaborus (24); two molytine weevils of the genus Hylobius (6); and a cryptorhynchine weevil, Euscepes postfasciatus (15). The evolutionary origin of Nardonella is thought to be quite ancient: dating back to an ancestor of those weevil groups some 125 million years ago (6). Meanwhile, within several dryophthorid lineages, Nardonella has been lost and replaced by different endosymbiotic bacteria: the grain weevils of the genus Sitophilus harbor Sodalis-allied gammaproteobacterial endosymbionts (14), and the weevils of the genera Trigonotarsus and Diocarandra are associated with a distinct and unnamed gammaproteobacterial clade (24).
The acorn or seed weevils of the genus Curculio (Curculionidae: Curculioninae) are characterized by their extremely elongated snouts (see Fig. 2A), which female insects utilize for boring a hole on their host plant seed and deposit an egg into the hole. Larvae feed on the content of the seed, escape the seed upon maturity, and pupate in the soil (17, 25, 28). Some Curculio species are notorious agricultural pests, represented by the chestnut weevil, C. sikkimensis, which damages chestnut production (19). In two species of Curculio (= Balaninus Germar 1817) weevils, Buchner (4) observed that filamentous bacteria are harbored in an aggregation of tiny bacteriocytes located at the posterior convolution of the midgut in young larvae. Since the early histological description, however, the microbiological nature of the Curculio endosymbiont has been elusive to date.
FIG. 2.
(A) An adult insect of C. sikkimensis. (B to F) Localization of the primary symbiont in dissected tissues of C. sikkimensis. Red and green signals indicate 16S rRNA of the primary symbiont and nuclear DNA of the insect cells, respectively. (B) Location of the bacteriome visualized on a dissected intestine of a final instar larva; (C and D) enlarged images of the bacteriome, wherein many bacteriocytes loosely aggregate and surround the midgut tube; (E) an enlarged image of the bacteriocytes, whose cytoplasm is filled with filamentous bacterial cells; (F) the apex of an ovary dissected from an adult female, wherein the symbiont signals are seen; (G) freshly isolated symbiont cells stained with SYTOX green.
In this study, we examined Japanese Curculio weevils for their endosymbiotic bacteria and identified previously unknown types of weevil endosymbionts: gammaproteobacterial primary symbionts constituting a novel endosymbiont clade and Sodalis-allied gammaproteobacterial secondary symbionts.
MATERIALS AND METHODS
Insect materials.
Table 1 summarizes the insect materials used in this study. Fagaceous seeds collected in summer of 2006 to 2008 were kept in the laboratory, from which final instar larvae of C. sikkimensis, C. dentipes, and C. robustus that emerged for overwintering and pupation were harvested. About a half of the larvae of C. sikkimensis were individually introduced into plastic cases filled with soil, from which emerging adult insects were collected in next spring. Adult insects of C. dentipes and C. robustus were obtained in the same way. These adult insects were preserved in acetone for DNA analyses (9). The other half of the larvae of C. sikkimensis were immediately preserved in acetone upon collection. Some of the larvae and adults of C. sikkimensis were kept alive and subjected to in situ hybridization and electron microscopy. We also collected camellia seeds to obtain the larvae of C. camelliae (44), which were reared in the soil until eclosion.
TABLE 1.
Curculio weevils used in this study, their symbiont infections, and nucleotide sequence accession numbers for the symbiont genes
| Insect species or sample code | Localitya | Latitude and longitude | Host plant | Sex/stage of insectb | % (no. confirmed/total) identified by diagnostic PCR |
Accession no. (gene) |
|||
|---|---|---|---|---|---|---|---|---|---|
| Curculioniphilusc | Sodalis | Nardonella | Curculioniphilusc | Sodalis | |||||
| C. sikkimensis | |||||||||
| TKB | Mt. Tsukuba, Ibaraki | 36°13′N 133°12′E | Quercus serrata | Female | 100 (15/15) | 20 (3/15) | 0 (0/15) | AB514502 (16S rRNA) | AB514505 (16S rRNA) |
| Male | 100 (15/15) | 27 (4/15) | 0 (0/15) | AB514497 (groEL) | AB514501 (groEL) | ||||
| HCD | Hacchodaira, Kyoto | 35°15′N 135°50′E | Quercus crispula | Female | 87 (13/15) | 0 (0/15) | 0 (0/15) | AB514503 (16S rRNA) | |
| Male | 87 (13/15) | 0 (0/15) | 0 (0/15) | ||||||
| SHB | Shohbara, Hiroshima | 34°52′N 133°12′E | Quercus serrata | Female | 100 (15/15) | 47 (7/15) | 0 (0/15) | AB514504 (16S rRNA) | AB517595 (16S rRNA) |
| Male | 100 (15/15) | 47 (7/15) | 0 (0/15) | ||||||
| Total | 96 (86/90) | 23 (21/90) | 0 (0/90) | ||||||
| C. camelliae | Kyohgatake, Nagasaki | 32°59′N 130°05′E | Camellia japonica | Female | AB507716 (16S rRNA) | ||||
| AB514498 (groEL) | |||||||||
| C. dentipes | Shimoda, Shizuoka | 34°40′N 138°59′E | Quercus phillyraeoides | Female | AB507714 (16S rRNA) | ||||
| AB514499 (groEL) | |||||||||
| C. robustus | Nokonoshima, Fukuoka | 33°37′N 130°19′E | Quercus acutissima | Female | AB507715 (16S rRNA) | ||||
| AB514500 (groEL) | |||||||||
All localities are in Japan.
Either the adult female or adult male.
Name proposed in this study.
DNA extraction, cloning, and sequencing.
From each of the acetone-preserved larvae, gut was dissected in 70% ethanol. From each of the acetone-preserved adults, gut plus ovary/testis was dissected. These dissected tissue samples were individually subjected to DNA extraction using a QIAamp DNA minikit (Qiagen). From the DNA samples, the following bacterial genes were amplified by PCR: a 1.5-kb fragment of the bacterial 16S rRNA gene with primers 16SA1 (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16SB1 (5′-TACGGYTACCTTGTTACGACTT-3′) (11), a 0.9-kb fragment of the bacterial groEL gene with primers groEL2F (5′-ATGGGBGCTCAAATGGTKAAA-3′) and groEL2R (5′-CTCTTTCATTTCAACTTCNGTBGCA-3′), and a 1.3-kb fragment of the Sodalis-allied bacterial groEL gene with GroELSod200F (5′-GAACATGGGCGCCCAGATGGTG-3′) and GroELSod1480R (5′-CGGGTTACCTTGGTCGGATCCAG-3′). The PCR products were subjected to cloning, up to eight clones for each of the samples were genotyped by restriction fragment length polymorphism (RFLP), and multiple clones representing each of the identified genotypes were sequenced using BigDye Terminator cycle sequencing kit (Applied Biosystems) and an ABI 3130 DNA sequencer (Applied Biosystems).
Molecular phylogenetic analysis.
Multiple alignments of the DNA sequences were generated using the program ClustalW (43). For the alignments, the best-fit substitution models were selected using the program Kakusan3 (42). Bayesian phylogenies were inferred using the program MrBayes v3.1.2 (36). Maximum likelihood phylogenies were estimated using the program PhyML v3 (12) with SPR branch-swapping and 10 neighbor-joining starting trees. The reliability of each clade was evaluated by 100 bootstrap replicates.
Diagnostic PCR.
From three localities in Japan, Mt. Tsukuba, Hacchodaira, and Shohbara, 15 adult females and 15 adult males of C. sikkimensis were collected and individually subjected to DNA extraction and diagnostic PCR (Table 1). The quality of the DNA samples was evaluated by amplification of a 0.1-kb fragment of elongation factor 1α (EF1α) gene of the host insect with primers RferEF1a546F (5′-TCCAGCCAACATCACCACTG-3′) and RferEF1a617R (5′-CCGGGTACAGCTTCTTGCA-3′). The presence of Nardonella spp. was checked by amplification of a 0.3-kb fragment of the 16S rRNA gene with primers 16SA1 and Nar16S250R (5′-CTCATCWWWKGRYRTAAGGTT-3′), wherein a DNA sample of the dryophthorid weevil Rhynchophorus ferrugineus was used as a positive control. The infection frequencies of the novel symbiont were examined by amplification of a 0.5-kb fragment of a 16S rRNA gene with primers 16SA1 and PrmSikk464R (5′-TGCATTGTTGCCTTCTTTCCTA-3′) and a 0.5-kb fragment of the groEL gene with primers GroPrmSik1000F (5′-TACTACTACTATTATTGATGGAGC-3′) and GroEL2R (5′-CTCTTTCATTTCAACTTCNGTBGCA-3′). The infection frequencies of the Sodalis-allied symbiont were examined by amplification of a 0.5-kb fragment of the 16S rRNA gene with primers Sodalis370F (5′-CGRTRGCGTTAAYAGCGC-3′) and 16SSod590R (5′-AACAGACCGCCTGCGTACG-3′) and a 0.5-kb fragment of the groEL gene with primers GroELSod200F (5′-GAACATGGGCGCCCAGATGGTG-3′) and GroELSod500R (5′-CCSGAACCCTCTTCCACGGTGATG-3′). For each of the symbionts, two genes were targeted to confirm the reproducibility of the symbiont detection. A DNA sample of C. sikkimensis, from which 16S rRNA genes of both the novel symbiont and the Sodalis-allied symbiont had been cloned and sequenced, was used as a positive control.
In situ hybridization.
Gut of larvae and ovary of adult females of C. sikkimensis were dissected in 70% ethanol and immediately preserved in Carnoy's solution (ethanol-chloroform-acetic acid at 6:3:1). After an overnight fixation, these tissues were treated with 6% hydrogen peroxide in 80% ethanol for a week to eliminate autofluorescence (23). Then the samples were thoroughly washed with 100% ethanol and subjected to whole-mount in situ hybridization. An oligonucleotide probe specific to the 16S rRNA sequence of the novel symbiont of C. sikkimensis, PrmSikk464R (5′-TGCATTGTTGCCTTCTTTCCTA-3′), with an Alexa 555 fluorescence dye labeled on the 5′ end, was used. The tissues were incubated in a hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, and 30% formamide) containing 100 nM probe and 500 nM SYTOX green (Invitrogen). After an overnight incubation at room temperature, the tissues were thoroughly washed in Dulbecco's phosphate-buffered saline (Sigma-Aldrich), mounted in Slowfade Gold antifade reagent (Invitrogen), and observed under an epifluorescent microscope (Axiophot; Carl Zeiss) and a laser-scanning confocal microscope (LSCM Pascal5; Carl Zeiss).
Electron microscopy.
The bacteriome-associated midgut sections were dissected and prefixed in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) at 4°C overnight and postfixed in 2% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4) at 4°C for 90 min. After dehydration through an ethanol series, the tissues were embedded in Spurr resin (Nisshin-EM). Ultrathin sections were made on an ultramicrotome (Ultracat-N; Leichert-Nissei), mounted on collodion-coated copper meshes, stained with uranyl acetate and lead citrate, and observed under a transmission electron microscope (H-7000; Hitachi).
RESULTS
Bacterial 16S rRNA gene sequences from Curculio weevils.
A 1.5-kb fragment of the bacterial 16S rRNA gene was amplified by PCR and cloned from DNA samples of C. sikkimensis of different geographic origins. RFLP genotyping of the 16S rRNA gene clones from each of the insects was performed, and two major genotypes, A-type and B-type, were identified from four insects of the Tsukuba population and seven insects of the Shohbara population, while only the A-type was detected in an insect of the Hacchodaira population. An insect sample from each of the species C. camelliae, C. dentipes, and C. robustus was also subjected to the analysis, and only the A-type was detected as a major clone. In addition to the A-type and B-type clones, several minor genotypes were also identified from those samples.
We sequenced the A-type clones from insects of three C. sikkimensis populations. The 1,463-bp sequences were almost identical to each other (99.7% [1,459/1,463] to 99.8% [1,460/1,463] identities). We also sequenced the A-type clones from C. camelliae, C. dentipes, and C. robustus. The 1,466- to 1,471-bp sequences were similar to each other (94.9% [1,399/1,474] to 97.8% [1,441/1,473] identities) and also similar to the sequences from C. sikkimensis (95.4% [1,400/1,467] to 98.0% [1,441/1,471] identities). BLAST searches with the A-type sequence from C. sikkimensis as the query retrieved gammaproteobacterial sequences. The top hit sequence was Sodalis glossinidius, an endosymbiont of the tsetse fly Glossina morsitans, with moderate sequence similarity (93.4% [1,370/1,467] identity; accession no. AP008232).
We also sequenced the B-type clones from two C. sikkimensis populations. The 1,454-bp sequences were almost identical to each other (99.4% [1446/1454] identity). BLAST searches with the B-type sequence from C. sikkimensis as the query retrieved gammaproteobacterial sequences. The top hit sequence was an endosymbiont of the dryophthorid grain weevil Sitophilus oryzae with high sequence similarity (99.0% [1,439/1,455] identity; accession no. AF548137).
Phylogenetic placement of the bacterial 16S rRNA gene sequences from Curculio weevils.
The A-type and B-type 16S rRNA gene sequences were subjected to molecular phylogenetic analysis with 16S rRNA gene sequences of gammaproteobacterial representatives (Fig. 1). The A-type sequences obtained from Curculio spp. constituted a monophyletic group with high statistical supports. Within the group, the sequences from different populations of C. sikkimensis formed a robust clade. The bacterial clade was distinct from any of the weevil endosymbiont groups previously described: Nardonella spp. universally detected from dryophthorid, molytine, and cryptorhynchine weevils (6, 15, 24); Sodalis-allied endosymbionts associated with dryophthorid weevils of the genus Sitophilus (14); and so-called “D-clade” endosymbionts of dryophthorid weevils of the genera Trigonotarsus and Diocarandra (24). Meanwhile, the B-type sequences from C. sikkimensis populations clustered with the endosymbiont of the tsetse fly, Sodalis glossinidius (7), endosymbionts of Sitophilus grain weevils (14), and the endosymbiont of the Columbicola pigeon louse (10).
FIG. 1.
Molecular phylogenetic analysis of the endosymbionts of Curculio weevils on the basis of 16S rRNA gene sequences. A Bayesian tree inferred from 1,180 unambiguously aligned nucleotide sites is shown, whereas a maximum likelihood analysis gave substantially the same topology (data not shown). Posterior probabilities for the Bayesian analysis and bootstrap values (>50%) for the maximum likelihood analysis are shown above and below the nodes, respectively. Sequence accession numbers and AT contents of the nucleotide sequences are in brackets and parentheses, respectively. As for insect endosymbionts, name of the host insect is also indicated in parentheses. P-symbiont, primary symbiont; S-symbiont, secondary symbiont.
Analysis of bacterial groEL gene sequences from Curculio weevils.
A 0.9-kb segment of the bacterial groEL gene was amplified by PCR and cloned from DNA samples of Curculio weevils. The obtained 923-bp sequences were similar to each other (85.9% [793/923] to 94.0% [868/923] identities) between host species. BLAST searches with the C. sikkimensis sequence as the query retrieved the top hit sequence as the groEL gene sequence of Blochmannia laevigatus, an endosymbiont of the carpenter ant, Camponotus laevigatus, with relatively low sequence similarity (80.5% [743/923] identity; accession no. AY334434).
From DNA samples of C. sikkimensis wherein the A-type and B-type 16S rRNA gene sequences were identified, a 1.3-kb groEL gene segment was amplified by PCR with Sodalis-targeted primers, cloned, and sequenced. The obtained 1,289-bp sequence retrieved the BLAST top hit as an endosymbiont of the dryophthorid grain weevil Sitophilus oryzae with high sequence similarity (98.9% [1,274/1,289] identity; accession no. AF005236). The two types of bacterial groEL gene sequences from C. sikkimensis showed relatively low sequence similarity (77.2% [713/923] identity).
These sequences were subjected to molecular phylogenetic analysis with groEL gene sequences of gammaproteobacterial representatives (see Fig. S1 in the supplemental material). The former sequences formed a monophyletic group with high statistical supports. The bacterial clade was allied to neither Nardonella nor Sodalis. Meanwhile, the latter sequences clustered with Sodalis glossinidius and the Sitophilus endosymbiont.
Prevalence of the symbionts in C. sikkimensis populations.
We examined the prevalence of the novel bacterial gene sequences and the Sodalis-allied gene sequences in three populations of C. sikkimensis by diagnostic PCR. The novel bacteria exhibited consistently high infection frequencies: 100% (30/30) in Tsukuba, 87% (26/30) in Hacchodaira, and 100% (30/30) in Shohbara. On the other hand, the Sodalis-allied bacteria were of low infection frequencies: 23% (7/30) in Tsukuba, 0% (0/30) in Hacchodaira, and 47% (14/30) in Shohbara. In the diagnostic PCR experiments described above, the results with the 16S rRNA gene were perfectly concordant with those with the groEL gene. Notably, the 16S rRNA gene sequence of Nardonella was detected from none of the insects (Table 1).
Localization and morphology of the novel symbiont in C. sikkimensis.
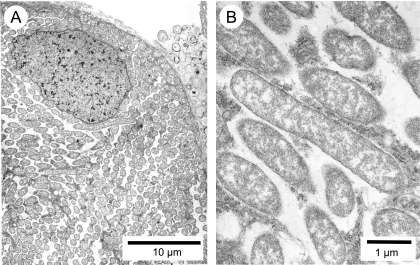
Dissected tissues of larvae and adults of C. sikkimensis were subjected to whole-mount in situ hybridization targeting 16S rRNA of the novel symbiont. In larvae, the symbiont signals were localized in a bacteriome located at the posterior end of a voluminous stomach-like midgut region (Fig. 2B). The bacteriome was a loose assemblage of a number of bacteriocytes surrounding the outside of a tubular section of the midgut (Fig. 2C and D). Numerous filamentous bacteria were harbored in the cytoplasm of the bacteriocytes (Fig. 2E). Freshly smeared bacteriocyte specimens showed the dimensions of the bacterial cells as 14.1 ± 2.8 μm in length and 0.6 ± 0.1 μm in width (mean ± standard deviation [SD]; n = 16) (Fig. 2G). Electron microscopy revealed that the symbiont cells had a reduced cell wall and were present in the cytoplasm without being encased in a host-derived membrane (Fig. 3A and B). In adult females, the symbiont signals were detected in the germaria located at the tip of the ovarioles (Fig. 2F).
FIG. 3.
Transmission electron microscopy of the bacteriocyte of C. sikkimensis. (A) A bacteriocyte, whose cytoplasm is full of slender bacterial cells; (B) an enlarged image of the symbiont cells.
Accelerated molecular evolution in the novel symbiont from Curculio weevils.
The evolutionary rates of the 16S rRNA and groEL gene sequences in the lineage of the novel symbionts of Curculio weevils were significantly higher than those in the lineages of free-living gammaproteobacteria, respectively. Meanwhile, the evolutionary rates in the Sodalis-allied symbionts of Curculio weevils were not significantly different from those of free-living gammaproteobacteria (Table 2).
TABLE 2.
A relative-rate test for comparing the molecular evolutionary rates of 16S rRNA and groEL gene sequences between the lineages of the primary and secondary symbionts of Curculio weevils and their free-living relatives
| Gene | Lineage 1 | Lineage 2 | Outgroup | K1a | K2b | Difference of distance between K1 and K2 | K1/K2 ratio | P valuec |
|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene of the primary symbionts (“Ca. Curculioniphilus”) | P-symbionts of Curculio spp.d | Escherichia coli and Salmonella enterica serovar Typhie | V. choleraef | 0.059 | 0.041 | 0.018 | 1.4 | 0.049 |
| groEL gene of the primary symbionts (“Ca. Curculioniphilus”) | P-symbionts of Curculio spp.g | E. coli and S. Typhih | V. choleraei | 0.080 | 0.019 | 0.061 | 4.2 | 3.4 × 10−6 |
| 16S rRNA gene of the secondary symbionts (Sodalis-allied) | S-symbionts of Curculio spp.j | E. coli and S. Typhie | V. choleraef | 0.029 | 0.031 | −0.002 | 0.94 | 0.82 |
| groEL gene of the secondary symbionts (Sodalis-allied) | S-symbionts ofCurculio spp.k | E. coli and S. Typhih | V. choleraei | 0.030 | 0.014 | 0.016 | 2.1 | 0.066 |
Estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
Estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
P values were generated using the program RRTree (35).
P-symbiont of C. sikkimensis (AB514502), C. camelliae (AB507716), C. dentipes (AB507714), and C. robustus (AB507715).
V. cholerae (X74694).
V. cholerae (CP000627).
S-symbiont of C. sikkimensis (AB514501).
AT-biased nucleotide composition in genes of the novel symbiont from Curculio weevils.
The 16S rRNA gene sequences constituting the novel symbiont clade exhibited AT contents ranging from 49.5% to 49.9%, which were remarkably higher than the AT contents of free-living gammaproteobacteria of around 45%. On the other hand, the 16S rRNA gene sequences of the Sodalis-allied symbiont from C. sikkimensis were 44.5% AT, which was similar to the values in free-living gammaproteobacteria (Fig. 1).
The groEL gene sequences representing the novel symbiont clade exhibited AT contents ranging from 59.8% to 61.4%, which were drastically higher than the AT contents of free-living gammaproteobacteria, ranging from 43% to 47%. Meanwhile, the groEL gene sequence of the Sodalis-allied symbiont from C. sikkimensis was 44.0% AT, which was comparable to the values in free-living gammaproteobacteria (see Fig. S1 in the supplemental material).
DISCUSSION
We found herein a novel group of bacterial endosymbionts from the weevil genus Curculio, which represents the primary symbiont of Curculio weevils. A well-defined gammaproteobacterial clade, Nardonella spp., comprises the ancient endosymbiont lineage associated with diverse weevils representing the Dryophthoridae, the Molytinae, and the Cryptorhynchinae, whose evolutionary origin is estimated to be some 125 million years ago (6, 15, 24). However, the Nardonella-weevil association has not been perfectly coherent. Symbiont replacements have occurred in several dryophthorid lineages, such as the clade of grain weevils, Sitophilus spp., and the clade of Trigonatarsus-Diocarandra (24). In a recent extensive molecular phylogenetic analysis of the Curculionoidea (27), molytine weevils and curculionine weevils were placed in the same clade, suggesting that the common ancestor of Curculio weevils was also associated with Nardonella. Hence, it seems likely that the symbiont replacement from Nardonella to the primary symbiont occurred in an ancestor of the Curculio weevils.
The ancestral weevil endosymbionts of the Nardonella spp. are harbored in a U- or ring-shaped large bacteriome at larval stages. The bacteriome is a well-defined organ consisting of many bacteriocytes surrounding the alimentary tract at the foregut-midgut junction (5, 6, 31). In Sitophilus, Trigonotarsus, and Diocalandra species in which Nardonella has been replaced by different bacteria, the bacteriome remains in the same location and exhibits a similar histological configuration (5, 31). Hence, upon these symbiont replacement events, the symbiotic bacteria have been taken over while the symbiotic organ has been conserved. In the Curculio weevils, on the other hand, no bacteriome was present at the foregut-midgut junction, but, strikingly, a bacteriome of quite different histological configuration was found at the posterior end of a stomach-like midgut region (Fig. 2). Interestingly, the symbiont replacement event in an ancestor of Curculio weevils entailed development of a different symbiotic organ, whose evolutionary origin comprises an enigma.
As for the primary symbiont of Curculio weevils, its high prevalence in host populations (Table 1), high AT contents (Fig. 1 and see Fig. S1 in the supplemental material) and accelerated molecular evolution (Table 2) suggest an intimate host-symbiont association over evolutionary time and also an important biological role of the symbiont for the host. Curculio weevils feed on acorns, seeds, or galls of their specific host plants at their larval stages (17). The symbiont may provide some nutritional components that are deficient in those plant parts, as many insect symbionts do for their hosts (8). Alternatively, the symbiont might be involved in detoxification of defense substances accumulated in those plant parts. Fagaceous acorns are known to contain high concentrations of tannins (40, 47). Some insect-induced plant galls also contain high levels of tannins, which were traditionally utilized for industrial production of gallotannin (2). Many plant seeds accumulate various toxic substances as antiherbivore agents (18, 49). In some wild mice that mainly feed on fagaceous acorns, their resistance to tannins was shown to be attributed to tannase-producing gut bacterial symbionts (41). Whether the primary symbiont of Curculio weevils plays a similar role deserves future studies.
When adult weevils collected from three natural populations of C. sikkimensis were examined by diagnostic PCR for the primary symbiont infection, the Tsukuba population and Shohbara population exhibited 100% infection frequencies, whereas the Hacchodaira population contained some uninfected insects, attaining 87% infection frequency (Table 1). This result suggests that the primary symbiont may not always be essential for C. sikkimensis. In the grain weevils, Sitophilus spp., aposymbiotic insects are able to grow and reproduce while suffering several defects, including retarded growth and lower fecundity than symbiotic insects (14). In the alydid bug Riptortus pedestris (= R. clavatus), symbiont-free insects become adults and lay viable eggs, but their body size is smaller than that of symbiont-infected insects (22). Similarly, the primary symbiont of Curculio weevils might be such a “beneficial but not essential” endosymbiont. Considering that the diagnostic PCR analysis was performed on adult insects only, the primary symbiont might be essential for larvae but not for adults of Curculio weevils. Alternatively, the primary symbiont might be conditionally needed for Curculio weevils, depending on ecological, physiological, and/or environmental factors the insects have experienced. To address which of these hypotheses is the most appropriate, experimental studies on symbiotic and aposymbiotic Curculio weevils are needed.
On account of the distinct genetic, phylogenetic, and microbiological traits described above, we propose the designation “Candidatus Curculioniphilus buchneri” for the primary symbiotic bacteria identified from the weevils of the genus Curculio. The generic name refers to the affinity to Curculio weevils. The specific name honors Paul Buchner, who first described the endosymbiotic bacteria of Curculio weevils (4).
In addition to the primary symbiont “Ca. Curculioniphilus,” we identified Sodalis-allied gammaproteobacterial endosymbionts from C. sikkimensis (Fig. 1 and see Fig. S1 in the supplemental material). The diagnostic PCR survey revealed relatively low infection frequencies of the Sodalis-allied bacteria: 23% in the Tsukuba population, 0% in the Hacchodaira population, and 47% in the Shohbara population (Table 1). Meanwhile, the bacterial genes were not detected from the other Curculio species, although the samples examined in this study are limited in number (Table 1). The bacterial gene sequences exhibited neither AT-biased nucleotide composition (Fig. 1 and Fig. S1) nor accelerated molecular evolution (Table 2). On the basis of these results, we conclude that the Sodalis-allied bacteria represent the secondary symbiont of C. sikkimensis.
As mentioned above, in the grain weevils of the genus Sitophilus, the ancient endosymbiont Nardonella has been lost and replaced by a Sodalis-allied endosymbiont (24), which exhibits bacteriome-specific localization and benefits the host's growth and reproduction (14). It appears plausible that the bacteriome-associated Sodalis-allied primary symbiont of Sitophilus spp. might have evolved from a Sodalis-allied secondary symbiont of weevils. In this context, it is of great interest how commonly Sodalis-allied facultative symbionts are found in the diversity of extant weevils. Comparative studies on the Sodalis-allied primary symbiont of Sitophilus spp. and the Sodalis-allied secondary symbiont of C. sikkimensis would provide insights into how a less-specialized endosymbiont has established the bacteriome-specific localization and the mutualistic association.
Supplementary Material
Acknowledgments
We thank M. Higaki and the laboratory of pear/chestnut horticulture, National Institute of Fruit Tree Science, for technical advice on rearing of C. sikkimensis; Y. Higashiura and Y. Sato for information on sampling localities; K. Fuse, I. Hanya, M. Hanya, and S. Yokota for weevil samples; S. Hanada for bacterial nomenclature; and H. Anbutsu, M. Kutsukake, Y. Matsuura, K. Tanaka, and T. Tsuchida for advice on laboratory experiments.
This study was funded by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and also by a grant-in-aid (no. 204352) from the Ministry of Education, Science, Sports and Culture, Japan. H.T. was supported by the Japan Society for the Promotion of Science (JSPS) postdoctoral fellowship for young scientists.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Bernays, E. A., G. C. Driver, and M. Bilgener. 1989. Herbivores and plant tannins. Adv. Ecol. Res. 19:263-302. [Google Scholar]
- 3.Bourtzis, K., H. Braig, and T. Karr. 2003. Cytoplasmic incompatibility, p. 217-246. In K. Bourtzis and T. Miller (ed.), Insect symbiosis. CRC, Boca Raton, FL.
- 4.Buchner, P. 1933. Studien an intracellularen symbionten. Zoomorphology 26:709-777. [Google Scholar]
- 5.Buchner, P. 1965. Endosymbionts of animals with plant microorganisms. Interscience, New York, NY.
- 6.Conord, C., L. Despres, A. Vallier, S. Balmand, C. Miquel, S. Zundel, G. Lemperiere, and A. Heddi. 2008. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol. Biol. Evol. 25:859. [DOI] [PubMed] [Google Scholar]
- 7.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov., and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49:267-275. [DOI] [PubMed] [Google Scholar]
- 8.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 9.Fukatsu, T. 1999. Acetone preservation: a practical technique for molecular analysis. Mol. Ecol. 8:1935-1945. [DOI] [PubMed] [Google Scholar]
- 10.Fukatsu, T., R. Koga, W. A. Smith, K. Tanaka, N. Nikoh, K. Sasaki-Fukatsu, K. Yoshizawa, C. Dale, and D. H. Clayton. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heddi, A., F. Lefebvre, and P. Nardon. 1993. Effect of endocytobiotic bacteria on mitochondrial enzymatic activities in the weevil Sitophilus oryzae (Coleoptera, Curculionidae). Insect Biochem. Mol. Biol. 23:403-411. [Google Scholar]
- 14.Heddi, A., and P. Nardon. 2005. Sitophilus oryzae L.: a model for intracellular symbiosis in the Dryophthoridae weevils (Coleoptera). Symbiosis 39:1-11. [Google Scholar]
- 15.Hosokawa, T., and T. Fukatsu. Nardonella endosymbiont in the West Indian sweet potato weevil Euscepes postfasciatus (Coleoptera: Curculionidae). Appl. Entomol. Zool., in press.
- 16.Hosokawa, T., Y. Kikuchi, M. Shimada, and T. Fukatsu. 2007. Obligate symbiont involved in pest status of host insect. Proc. R. Soc. B 274:1979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, J., and A. P. Vogler. 2004. Ecomorphological adaptation of acorn weevils to their oviposition site. Evolution 58:1971-1983. [DOI] [PubMed] [Google Scholar]
- 18.Hulme, P., and C. W. Benkman. 2002. Granivory, p. 132-154. In C. M. Herrera and O. Pellmyr (ed.), Plant-animal interactions: an evolutionary approach. Blackwell Publishing, Oxford, United Kingdom.
- 19.Ihara, F., M. Toyama, M. Higaki, K. Mishiro, and K. Yaginuma. 2009. Comparison of pathogenicities of Beauveria bassiana and Metarhizium anisopliae to chestnut pests. Appl. Entomol. Zool. 44:127-132. [Google Scholar]
- 20.Janson, E. M., J. O. Stireman III, M. S. Singer, and P. Abbot. 2008. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution 62:997-1012. [DOI] [PubMed] [Google Scholar]
- 21.Kellner, R. L. L. 2002. Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae). Insect Biochem. Mol. Biol. 32:389-395. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi, Y., T. Hosokawa, and T. Fukatsu. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koga, R., T. Tsuchida, and T. Fukatsu. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44:281-291. [Google Scholar]
- 24.Lefevre, C., H. Charles, A. Vallier, B. Delobel, B. Farrell, and A. Heddi. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21:965-973. [DOI] [PubMed] [Google Scholar]
- 25.Maeto, K., and K. Ozaki. 2003. Prolonged diapause of specialist seed-feeders makes predator satiation unstable in masting of Quercus crispula. Oecologia 137:392-398. [DOI] [PubMed] [Google Scholar]
- 26.Mansour, K. 1930. Preliminary studies on the bacterial cell mass (accessory cell mass) of Calandra oryzae: the rice weevil. Q. J. Microsc. Sci. 73:421-436. [Google Scholar]
- 27.McKenna, D. D., A. S. Sequeira, A. E. Marvaldi, and B. D. Farrell. 2009. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl. Acad. Sci. U. S. A. 106:7083-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menu, F. 1993. Strategies of emergence in the chestnut weevil Curculio elephas (Coleoptera, Curculionidae). Oecologia 96:383-390. [DOI] [PubMed] [Google Scholar]
- 29.Montllor, C. B., A. Maxmen, and A. H. Purcell. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189-195. [Google Scholar]
- 30.Moran, N. A. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. U. S. A. 104:8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardon, P., C. Lefevre, B. Delobel, H. Charles, and A. Heddi. 2002. Occurrence of endosymbiosis in Dryophthoridae weevils: cytological insights into bacterial symbiotic structures. Symbiosis 33:227-241. [Google Scholar]
- 32.Nardon, P., C. Louis, G. Nicolas, and A. Kermarrec. 1985. Mise en évidence et éude des bactéries symbiotiques chez deux charançons parasites du bananier: Cosmopolites sordidus (GERMAR) et Metamasius hemipterus (L.) (Col. Curculionidae). Ann. Soc. Entomol. Fr. 21:245-258. [Google Scholar]
- 33.Oliver, K. M., J. A. Russell, N. A. Moran, and M. S. Hunter. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 100:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierantoni, U. 1927. L'organo simbiotico nello sviluppo di Calandra oryzae. Rend. Reale Acad. Sci. Fis. Mat. Napoli 35:244-250. [Google Scholar]
- 35.Robinson-Rechavi, M., and D. Huchon. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296-297. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 37.Scarborough, C. L., J. Ferrari, and H. C. J. Godfray. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. [DOI] [PubMed] [Google Scholar]
- 38.Scheinert, W. 1933. Symbiose und Embryonalentwicklung bei Rüsselkäfern. Z. Morphol. Oekol. Tiere 27:76-128. [Google Scholar]
- 39.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Shimada, T. 2001. Nutrient compositions of acorns and horse chestnuts in relation to seed-hoarding. Ecol. Res. 16:803-808. [Google Scholar]
- 41.Shimada, T., T. Saitoh, E. Sasaki, Y. Nishitani, and R. Osawa. 2006. Role of tannin-binding salivary proteins and tannase-producing bacteria in the acclimation of the Japanese wood mouse to acorn tannins. J. Chem. Ecol. 32:1165-1180. [DOI] [PubMed] [Google Scholar]
- 42.Tanabe, A. S. 2007. kakusan: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol. Ecol. Notes 7:962-964. [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toju, H., and T. Sota. 2006. Adaptive divergence of scaling relationships mediates the arms race between a weevil and its host plant. Biol. Lett. 2:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. [DOI] [PubMed] [Google Scholar]
- 46.Werren, J. H., L. Baldo, and M. E. Clark. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741-751. [DOI] [PubMed] [Google Scholar]
- 47.Xiao, Z., M. K. Harris, and Z. Zhang. 2007. Acorn defenses to herbivory from insects: implications for the joint evolution of resistance, tolerance and escape. For. Ecol. Manag. 238:302-308. [Google Scholar]
- 48.Yoshida, N., K. Oeda, E. Watanabe, T. Mikami, Y. Fukita, K. Nishimura, K. Komai, and K. Matsuda. 2001. Protein function: chaperonin turned insect toxin. Nature 411:44. [DOI] [PubMed] [Google Scholar]
- 49.Zangerl, A. R., and M. R. Berenbaum. 2005. Increase in toxicity of an invasive weed after reassociation with its coevolved herbivore. Proc. Natl. Acad. Sci. U. S. A. 102:15529-15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.