Abstract
Phenoxyalkanoic acid (PAA) herbicides are widely used in agriculture. Biotic degradation of such herbicides occurs in soils and is initiated by α-ketoglutarate- and Fe2+-dependent dioxygenases encoded by tfdA-like genes (i.e., tfdA and tfdAα). Novel primers and quantitative kinetic PCR (qPCR) assays were developed to analyze the diversity and abundance of tfdA-like genes in soil. Five primer sets targeting tfdA-like genes were designed and evaluated. Primer sets 3 to 5 specifically amplified tfdA-like genes from soil, and a total of 437 sequences were retrieved. Coverages of gene libraries were 62 to 100%, up to 122 genotypes were detected, and up to 389 genotypes were predicted to occur in the gene libraries as indicated by the richness estimator Chao1. Phylogenetic analysis of in silico-translated tfdA-like genes indicated that soil tfdA-like genes were related to those of group 2 and 3 Bradyrhizobium spp., Sphingomonas spp., and uncultured soil bacteria. Soil-derived tfdA-like genes were assigned to 11 clusters, 4 of which were composed of novel sequences from this study, indicating that soil harbors novel and diverse tfdA-like genes. Correlation analysis of 16S rRNA and tfdA-like gene similarity indicated that any two bacteria with D > 20% of group 2 tfdA-like gene-derived protein sequences belong to different species. Thus, data indicate that the soil analyzed harbors at least 48 novel bacterial species containing group 2 tfdA-like genes. Novel qPCR assays were established to quantify such new tfdA-like genes. Copy numbers of tfdA-like genes were 1.0 × 106 to 65 × 106 per gram (dry weight) soil in four different soils, indicating that hitherto-unknown, diverse tfdA-like genes are abundant in soils.
Phenoxyalkanoic acid (PAA) herbicides such as MCPA (4-chloro-2-methyl-phenoxyacetic acid) and 2,4-D (2,4-dichlorophenoxyacetic acid) are widely used to control broad-leaf weeds in agricultural as well as nonagricultural areas (19, 77). Degradation occurs primarily under oxic conditions in soil, and microorganisms play a key role in the degradation of such herbicides in soil (62, 64). Although relatively rapidly degraded in soil (32, 45), both MCPA and 2,4-D are potential groundwater contaminants (10, 56, 70), accentuating the importance of bacterial PAA herbicide-degrading bacteria in soils (e.g., references 3, 5, 6, 20, 41, 59, and 78).
Degradation can occur cometabolically or be associated with energy conservation (15, 54). The first step in the degradation of 2,4-D and MCPA is initiated by the product of cadAB or tfdA-like genes (29, 30, 35, 67), which constitutes an α-ketoglutarate (α-KG)- and Fe2+-dependent dioxygenase. TfdA removes the acetate side chain of 2,4-D and MCPA to produce 2,4-dichlorophenol and 4-chloro-2-methylphenol, respectively, and glyoxylate while oxidizing α-ketoglutarate to CO2 and succinate (16, 17).
Organisms capable of PAA herbicide degradation are phylogenetically diverse and belong to the Alpha-, Beta-, and Gammproteobacteria and the Bacteroidetes/Chlorobi group (e.g., references 2, 14, 29-34, 39, 60, 68, and 71). These bacteria harbor tfdA-like genes (i.e., tfdA or tfdAα) and are categorized into three groups on an evolutionary and physiological basis (34). The first group consists of beta- and gammaproteobacteria and can be further divided into three distinct classes based on their tfdA genes (30, 46). Class I tfdA genes are closely related to those of Cupriavidus necator JMP134 (formerly Ralstonia eutropha). Class II tfdA genes consist of those of Burkholderia sp. strain RASC and a few strains that are 76% identical to class I tfdA genes. Class III tfdA genes are 77% identical to class I and 80% identical to class II tfdA genes and linked to MCPA degradation in soil (3). The second group consists of alphaproteobacteria, which are closely related to Bradyrhizobium spp. with tfdAα genes having 60% identity to tfdA of group 1 (18, 29, 34). The third group also harbors the tfdAα genes and consists of Sphingomonas spp. within the alphaproteobacteria (30).
Diverse PAA herbicide degraders of all three groups were identified in soil by cultivation-dependent studies (32, 34, 41, 78). Besides CadAB, TfdA and certain TfdAα proteins catalyze the conversion of PAA herbicides (29, 30, 35). All groups of tfdA-like genes are potentially linked to the degradation of PAA herbicides, although alternative primary functions of group 2 and 3 TfdAs have been proposed (30, 35). However, recent cultivation-independent studies focused on 16S rRNA genes or solely on group 1 tfdA sequences in soil (e.g., references 3-5, 13, and 41). Whether group 2 and 3 tfdA-like genes are also quantitatively linked to the degradation of PAA herbicides in soils is unknown. Thus, tools to target a broad range of tfdA-like genes are needed to resolve such an issue. Primers used to assess the diversity of tfdA-like sequences used in previous studies were based on the alignment of approximately 50% or less of available sequences to date (3, 20, 29, 32, 39, 47, 58, 73). Primers specifically targeting all major groups of tfdA-like genes to assess and quantify a broad diversity of potential PAA degraders in soil are unavailable. Thus, the objectives of this study were (i) to develop primers specific for all three groups of tfdA-like genes, (ii) to establish quantitative kinetic PCR (qPCR) assays based on such primers for different soil samples, and (iii) to assess the diversity and abundance of tfdA-like genes in soil.
MATERIALS AND METHODS
Soil samples and bacterial strain.
Samples from four different agricultural or forest soils were used (Table 1). The agricultural soil was collected at the Klostergut Scheyern experimental farm (48°30′00″N, 11°20′07″E) and was treated with MCPA in 2002. The other three soil samples were obtained from different spruce forests in Germany, which were not treated with PAA herbicides (Table 1). All soil samples were obtained from a 0- to 10-cm depth and were stored in the dark on ice during transfer. Each soil sample was homogenized, sieved (2 mm), and stored at −80°C prior to use. Cupriavidus necator JMP134 (DSM 4058; formerly Ralstonia eutropha) was cultured according to the recommendations of the DSMZ (Braunschweig, Germany) and used for testing of primer specificity (Table 2).
TABLE 1.
Soil characteristics and abundances of tfdA-like genes
| Site | pH | Clay (%) | Silt (%) | Sand (%) | C/N ratio | Reference(s) | 16S rRNA gene copy no. (× 109 g−1a) |
tfdA-like gene copy no. (× 106 g−1) |
tfdA-like gene copy no. per 16S rRNA gene (× 10−3) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 2b | Groups 2 and 3c | Groups 1 to 3d | Group 2b | Groups 2 and 3c | Groups 1 to 3d | ||||||||
| Agricultural field | |||||||||||||
| (Scheyern) | 5.8 | 22 | 36 | 42 | 10 | 48 | 2.5 ± 0.1 | 12.9 ± 2.0 | 64.9 ± 0.7 | 3.0 ± 0.7 | 4.8 ± 0.6 | 26.0 ± 0.9 | 1.2 ± 0.3 |
| Spruce forests | |||||||||||||
| Solling | 3.3 | 28 | 58 | 14 | 33 | 7 | 1.4 ± 0.1 | 2.0 ± 0.7 | 41.5 ± 2.7 | 1.3 ± 0.1 | 1.4 ± 0.4 | 30.0 ± 0.2 | 0.9 ± 0.1 |
| Steigerwald | 3.8 | 18 | 24 | 58 | 19 | 8, 42 | 3.0 ± 0.3 | 4.7 ± 0.8 | 40.8 ± 13.6 | 1.3 ± 0.6 | 1.6 ± 0.4 | 13.9 ± 5.8 | 0.5 ± 0.2 |
| Unterluess | 3.1 | 3 | 23 | 74 | 38 | 7 | 3.5 ± 0.9 | 3.3 ± 1.4 | 9.8 ± 3.4 | 1.0 ± 0.6 | 0.9 ± 0.2 | 2.7 ± 0.4 | 0.3 ± 0.1 |
TABLE 2.
PCR primers and conditions evaluated in this study
| Primer seta | Sequence (5′-3′)b | Experimental |
Target | Approx product length (bp) | PCR product of agricultural field soilc |
Reference(s) | ||
|---|---|---|---|---|---|---|---|---|
| Annealing temp (°C) | Mg2+ (mM) | Native | JMP134 spiked | |||||
| 1 | ||||||||
| TfdA21F | CCTTCATCCWCTTTTCGYCG | 63.0 | 3.0 | tfdA (group 1) | 815 | − | ++ | This work |
| TfdA836R | GCTCGGCGMAGYTCACGC | |||||||
| 2 | ||||||||
| TfdA334F | CTSTGGCATTSCGACAGYTC | 60.0 | 3.0 | tfdA (group 1) | 260 | − | ++ | This work |
| TfdA608R | CSGSGTGSGTICGVYCAG | |||||||
| 3 | ||||||||
| TfdAα52F | GGCGTCGATCTGCGCAAGCC | 70.0 (65.0)d | 4.0 (3.0)d | tfdAα (group 2) | 360 | ++ | ++ | This work |
| TfdAα408R | GTTGACGACGCGCGCCGACA | |||||||
| 4 | ||||||||
| TfdAα421F | ACSGAGTTCGSIGAYATSC | 63.6 (68.0)d | 2.5 (3.0)d | tfdAα (groups 2 and 3) | 360 | ++ | ++ | Modified from references 29 and 73 |
| TfdAα779R | CAGCGGTTGTCCCACATCAC | |||||||
| 5 | ||||||||
| TfdA421dF | ACSGARTTCKSIGACATGC | 53.0 | 3.0 | tfdA-like (groups 1 to 3) | 360 | ++ | ++ | Modified from references 29 and 73 |
| TfdA778vkR | AGCGGTTGTCCCACATCAC | |||||||
| 6 | ||||||||
| TfdA421Fv | ACSGAGTTCTGYGAYATG | 59.0 | 3.0 | tfdA-like (group 1) | 360 | +− | ++ | Modified from reference 73 |
| TfdA783R | AACGCAGCGRTTRTCCCA | |||||||
| 7 | ||||||||
| Eub341F | CCTACGGGAGGCAGCAG | 55.7 | 3.0 | 16S rRNA | 190 | ++ | ++ | 49 |
| Eub534R | ATTACCGCGGCTGCTGG | |||||||
Numbers in primer names indicate 5′ position of binding sites relative to the reference tfdA sequence of Cupriavidusnecator JMP134 (AY365053) (primer sets 1, 2, 4, 5, and 6) and of Alphaproteobacterium RD5-C2 (AB074490) (primer set 3). F, forward; R, reverse.
S, C/G; Y, C/T; V, A/C/G; W, A/T; M, A/C; K, G/T; R, A/G.
As indicated by agarose gel electrophoresis. ++, bright band; +−, weak band; −, no band of correct size.
Numbers in parentheses refer to qPCR chemicals (see Materials and Methods).
Nucleic acid extraction.
DNA was extracted from pelleted cells of 1 ml of C. necator JMP134 culture (approximately 108 cells) or 0.5 g (wet weight) of soil according to a bead-beating protocol (22) with the following modifications: (i) 100 mM AlNH4(SO4)2 was added in soil samples during bead beating to precipitate the humic acid in soil (9); (ii) cell lysis was performed twice for 30 s each at a speed of 5.5 m/s with intermittent cooling on ice for more than 1 min to minimize heating of the samples; and (iii) nucleic acids were precipitated on ice instead of at room temperature for more than 2 h (5). DNA was usually further purified with the MinElute PCR product purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA extracts were stored at −20°C until further analyses.
Alignment and primer design.
Fifty-four gene sequences from groups 1, 2, and 3 were obtained from publicly available databases (GenBank, http://www.ncbi.nlm.nih.gov/GenBank) and aligned with the software package ARB (http://www.arb-home.de [40]), and several degenerate primers were designed according to the conserved regions identified in alignments (Table 2). The GenBank accession numbers of the sequences from the first alignment were AY540995, AF1811982, AY238495, AB025033, EF375720, BPU43196, DQ644552, DQ360372, DQ360394, DQ360397, DQ360398, DQ360399, BCU87394, AB212778, AB212779, AB212780, AB299620, AY238492, AY238493, AY238496, BSU25717, BSU43197, AFATFDA, BPU43276, AY078159, HGU22499, AY365053, AB299626, AB299627, AF182758, AY238497, AF176240, AY238494, EF600728, EF600734, EF600737, EF600748, EF600752, EF600753, DQ356907, DQ356910, DQ356912, AY554187, AY554191, AY554193, AB074490, AB074491, AB074492, AY193866, and AY554199. The GenBank accession numbers of the sequences from the second alignment containing only group 2 sequences were AB074490, AB074491, AB074492, AY193866, AY193867, AY193868, AY193869, and AY193870.
Primer evaluation and PCR conditions.
Primer specificity was evaluated in silico using GenBank's BLAST tool (1), and PCR conditions were optimized by variation of Mg2+ and annealing temperature (Table 2). 16S rRNA genes were amplified with published primers (Table 2). Each amplification reaction was performed in a total volume of 25 μl, and the reaction mixture contained 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 1.5 mM Mg2+ [5-Prime, Hamburg, Germany]) supplemented with 1.0 to 2.5 mM MgCl2, 1× TaqMaster PCR enhancer (5-Prime), 60 ng/μl bovine serum albumin (BSA), 200 μM deoxynucleoside triphosphate (dNTP) mix (5-Prime), 0.2 to 1 μM of each primer (Biomers, Ulm, Germany), 1 μl of template DNA (approximating 50 ng of DNA), and 0.5 U Taq DNA polymerase (5-Prime). BSA was included in the reaction mixture to reduce PCR inhibition from humic substances coextracted with DNA from soil (38). The PCR was performed with a T-Gradient cycler (Biometra, Göttingen, Germany) with 10 min of initial denaturation at 95°C, followed by 45 cycles, each consisting of denaturation at 95°C for 1 min, annealing at primer-dependent temperatures (Table 2) for 1 min, and elongation at 72°C for 1 min. The final elongation was at 72°C for 5 min.
Quantification of genes in soil.
qPCRs were performed with an iQ5 Real-Time qPCR cycler (Bio-Rad, Munich, Germany). All qPCRs were set up in duplicate. Negative controls with sterilized water instead of DNA template were included in every PCR setup. Standard curves were set up by serially diluting M13uni/rev PCR products of a pGEM-T vector with the appropriate insert from 108 to 101 target gene copies μl−1 for every primer set. qPCRs were performed in 20-μl reaction mixtures that were composed of iQ SYBR green Supermix (Bio-Rad), 60 ng/μl BSA, 0.2 to 1.6 pM of each primer (Biomers, Ulm, Germany), 5 μl of template DNA, and sterilized deionized water. Initial denaturation was at 95°C for 8 min, followed by 45 cycles of denaturation at 95°C for 40 s, annealing at temperatures appropriate for each primer set (Table 2) for 30 s, and elongation at 72°C for 15 s when fluorescence signal was recorded (i.e., a 3-step protocol was utilized). The final PCR elongation step was at 72°C for 5 min. qPCR with primer set 4 (Table 2) yielded a specific product with a melting temperature (Tm) of approximately 91°C and an unspecific product with a Tm of approximately 79°C (see Fig. S1A in the supplemental material). Agarose gel electrophoresis indicated that the latter product was primer dimers. The lower limit of quantification was 103 gene copy numbers μl−1 of DNA extract (see Fig. S1C). Thus, two more steps at 85°C (i.e., 6 s for preheating and 15 s for signal recording) were added after elongation to melt up primer dimers prior to signal recording (i.e., a 5-step protocol was utilized). After application of the 5-step protocol, primer dimers were not detected during melting curve analysis (see Fig. S1B). Melting curve analyses were performed from 70 to 95°C with increments of 0.2°C per cycle. 16S rRNA gene copy numbers were determined concomitantly for all environmental samples (Table 2) in order to quantify tfdA-like genes in soil relative to total bacterial 16S rRNA genes.
Inhibition of qPCR by environmental DNA extracts.
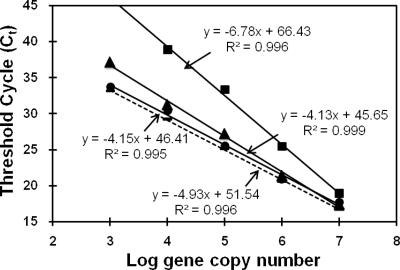
Humic acids and other organic compounds coextracted from soil with nucleic acids can inhibit PCR significantly (53, 72, 75, 76). Dilution of DNA extracts and correction of threshold cycle (CT) values are two strategies for overcoming such inhibition. For testing the inhibitory potential of environmental DNA extracts, such extracts were treated with DNase I (Fermentas, St. Leon-Rot, Germany) at 37°C for 4 h according to the manufacturer's protocol to obtain DNA-free soil extracts. Equal amounts of the DNase-treated soil DNA extracts (nondiluted and 10- and 50-fold diluted) were mixed with standards at 103 to 107 gene copy numbers μl−1 prior to qPCR with primer set 4 (Fig. 1). Standard curves obtained in the presence of nondiluted and 10-fold-diluted DNase-treated soil DNA extracts were shifted toward higher CT values than were those obtained with pure standard DNA (controls), indicating strong PCR inhibition. The standard curve obtained in the presence of 50-fold-diluted DNase-treated soil DNA extracts was similar to that for controls, suggesting that inhibition was small. Thus, 50-fold dilutions of soil DNA were used routinely in this study for qPCR. However, inhibition still occurred at 50-fold dilution of DNA extracts, and inhibitory effects may vary with DNA extracts. Gene copy numbers in environmental samples were calculated according to the standard curve and were corrected for inhibition. Soil DNA was spiked with pure standard DNA to increase target gene copy numbers per reaction by more than 10-fold. Gene copy numbers obtained by qPCR of spiked and nonspiked DNA extracts were termed S and NS, respectively. C was the gene copy number measured for the standard DNA. The inhibition factor (IF) was calculated as IF = (S − NS)/C and theoretically ranges from 0 (complete inhibition) to 1 (no inhibition). NS was divided by IF to approximate real gene copy numbers in DNA extracts. IF varied from 0.1 to 0.91 for DNA extracts obtained from different soil samples. Representative PCR products were analyzed by agarose gel electrophoresis, and products were cloned and sequenced to check for specific amplification of target genes.
FIG. 1.
Effect of inhibitors present in soil DNA extracts on standard curves generated during qPCR with primer set 4. Solid lines indicate standards spiked with various concentrations of DNA-free soil extract (▪, nondiluted; ▴, 10-fold diluted; •, 50-fold diluted); the dashed line indicates the nonspiked standard curve. Equations of linear regressions and R2 values of every curve are given.
The recovery of target DNA from soil was estimated by spiking soil with defined amounts of a linearized, tfdA-like gene harboring pGEM-T. pGEM-T was linearized by digestion with SacI (New England Biolabs) overnight at 37°C, quantified by recording the A260 (ND1000; Peqlab), and spiked to soil prior to DNA extraction and qPCR.
Cloning, screening, and sequencing.
(q)PCR products used for cloning were analyzed by gel electrophoresis on 1% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer (AppliChem, Darmstadt, Germany). The gels contained ethidium bromide and were visualized under UV light. PCR products were routinely purified by cutting out the band of the expected size (Table 2) to remove primers and primer dimers with subsequent extractions using the MiniElute gel extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. PCR products of all target samples were ligated into pGEM-T (Promega, Mannheim, Germany), which was used for transformation of Escherichia coli JM109 competent cells (Promega, Madison, WI) according to the manufacturer's protocol. For screening of the clone libraries from (q)PCR products, the fragments were directly amplified from 1 μl of resuspended clones and electrophoresed on agarose gels (1%). The gene libraries were screened by restriction fragment length polymorphism (RFLP). PCR products of clones generated with primer set 3 (Table 2) were digested with 1 U of AluI and BsuRI (New England BioLabs, Ipswich, MA) in separate reactions, and those generated with primer sets 4 and 5 (Table 2) were likewise digested with 1 U of MspI and TaqAI (New England BioLabs) in separate reactions for 3 h at 37°C (TaqAI at 65°C). The fragments were separated on agarose gels (3%). One to 10 representative clones of each RFLP pattern per library were selected for sequencing. Representative PCR products were purified (PCR cleanup plates; Millipore, Schwalbach, Germany) according to the manufacturer's protocol and commercially sequenced (Macrogen, South Korea). Sequence comparisons and database searches were carried out by using the BLAST service at the NCBI website (http://blast.ncbi.nlm.nih.gov/).
Phylogenetic analyses.
Phylogenetic analyses were performed with the ARB software package and Mega4 software (http://www.megasoftware.net/) (69). tfdA-like genes were retrieved from NCBI, translated in silico, and prealigned with the ClustalW algorithm, and the alignment was refined manually. The DNA was aligned according to the aligned proteins. Regions of primer binding sites were excluded from further sequence analysis. Phylogenetic trees were calculated based on amino acid sequences using the neighbor-joining algorithm (55). Tree topologies of neighbor-joining trees were verified by bootstrap analysis with 1,000 replicates.
Prealigned 16S rRNA gene sequences were retrieved from SILVA (http://www.arb-silva.de/) (52). Distance matrices for the correlation of pairwise similarities of tfdA-like and 16S rRNA gene sequences were generated with the ARB software package (40).
Analysis of tfdA-like genotype diversity.
Sequences that had the same RFLP patterns as those of sequenced tfdA-like gene fragments were represented by the fully sequenced fragments. Distance matrices were generated from aligned amino acid sequences. DOTUR (http://schloss.micro.umass.edu/software/index.html) (57) was applied to define genotypes based on the amino acid sequence dissimilarity of proteins derived from translated tfdA-like sequences by the furthest-neighbor method. Sequences with maximal 3% or 20% dissimilarity (i.e., distance D ≤ 3 or 20%) were thereby assigned to one genotype. Additional information on DOTUR is available at the publisher's website (http://schloss.micro.umass.edu/software/index.html). Coverage (C) is the number of the detected genotypes relative to their expected total number in a gene library and was calculated as C = (1 − n × N−1) × 100, where n is the number of genotypes that occurred only once and N is the number of clones screened (21). The diversity of genotypes represented in gene libraries of tfdA-like genes was analyzed by rarefaction analysis (28), Shannon diversity index, and the genotype richness estimator Chao1 (24). All calculations were done with DOTUR (57). Rarefaction curves were produced using the Analytic Rarefaction software (version 1.2; http://www.uga.edu/strata/software/index.html).
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in the EMBL nucleotide sequences database (http://www.ebi.ac.uk) under accession numbers FN376574 to FN376845.
RESULTS
Novel primers for amplification of tfdA-like genes.
Eight conserved regions of tfdA-like genes were used to design consensus primers (data not shown). Five different degenerated primer sets were designed (1 to 5, Table 2) to amplify (i) almost full-length fragments of the tfdA gene group 1, (ii) a partial gene fragment of tfdA group 1, (iii) a partial gene fragment of tfdAα group 2, (iv) a partial gene fragment of tfdAα covering group 2 genes, and (v) a partial gene fragment of tfdA-like genes targeting group 1 and 2 genes. Nevertheless, primer sets 4 and 5 amplified group 3 tfdA-like genes (Table 2; Fig. 2). In silico analyses indicated that primers of sets 1 and 2 had 1 to 4 mismatches to group 2 tfdA-like genes. Primer set 3 targeted a subset of group 2 tfdA-like genes and had more than 4 mismatches to group 1 tfdA. Primer set 4 covered all known group 2 tfdA-like genes and had one mismatch to group 1 tfdA. Primer set 5 had no mismatches to any known tfdA-like gene. In silico analyses of primer sets 4 and 5 were not performed with group 3 tfdAα fragments since the fragments publicly available to date were truncated. PCR products generated with primer set 6 from Scheyern soil contained only low concentrations of targets with the expected size (Table 2). However, all primer sets that were tested readily amplified fragments of the expected sizes (Table 2) from Scheyern soil that was amended with 104, 106, and 108 cells of C. necator JMP134 (harboring group 1 tfdA) per gram of soil, indicating that all primer sets detected target genes in soil (data not shown). The proportion of group 1 tfdA sequences per gene library (25 to 29 sequences per library) generated from Scheyern soil with primer set 5 approximated 15, 76, and 100% for soil amended with 104, 106, and 108 cells of C. necator JMP134 per gram of soil, respectively, indicating that the detection limit of group 1 tfdA in Scheyern soil with primer set 5 is below 104 copies per gram of soil (assuming one copy of group 1 tfdA per cell). Gene libraries generated from nonspiked Scheyern soil with primer sets 4 and 5 contained group 2- and 3-related tfdA-like genes. Primer set 3 yielded likewise specific products which were most closely related to group 2 tfdA-like genes (see Fig. S2 in the supplemental material). The results showed that the designed primers allowed a specific targeting of tfdA-like genes from soil.
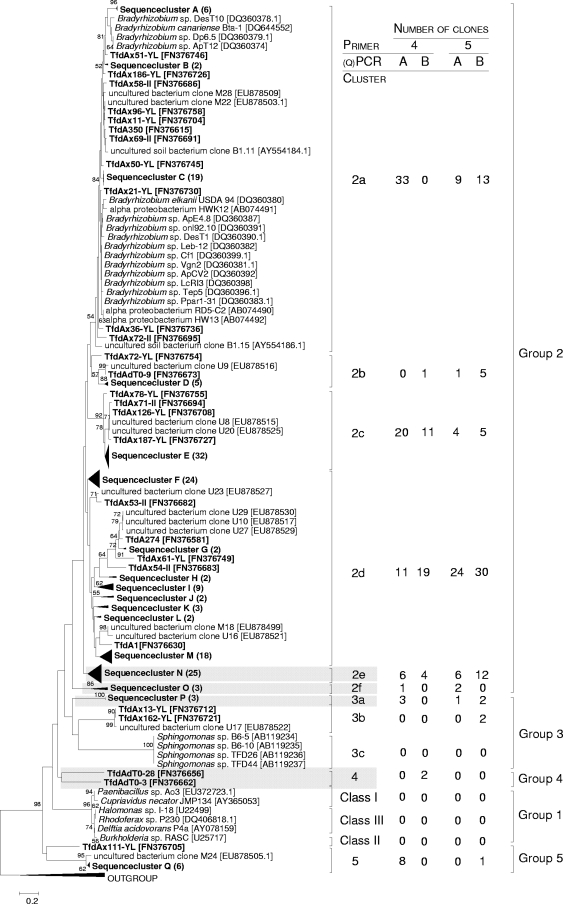
FIG. 2.
Phylogenetic neighbor-joining tree of representative tfdA-like genes generated with primer sets 4 and 5 (approximately 110 amino acids; indicated in boldface). Accession numbers are shown in brackets, and bootstrap values above 50% are shown at nodes. The abundances of sequences in the gene library are given in parentheses. Clusters of tfdA-like genes containing novel sequences only are highlighted with gray boxes. The following TauD (α-ketoglutarate-dependent taurine dioxygenase) sequences were used as the outgroup: CP000699, CP000854.1, CP000124.1, AP009048, and AL590842. In the associated table, sequences generated with PCR and qPCR are indicated by A and B, respectively. The scale bar represents an estimated sequence dissimilarity of 20%.
Detected diversity of tfdA-like genes.
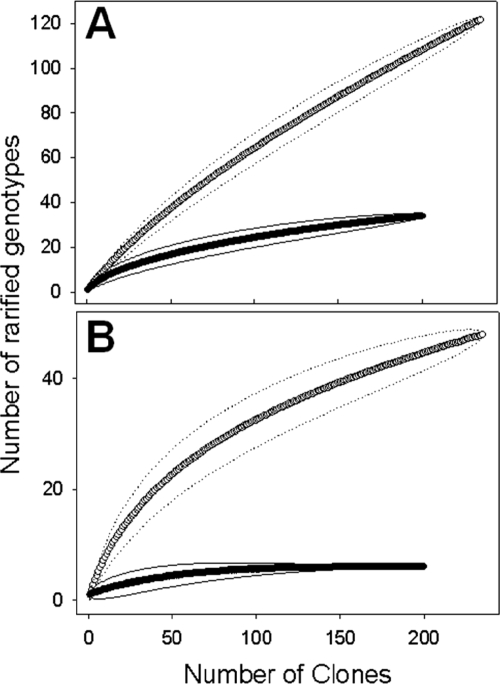
A total number of 437 tfdA-positive clones (201, 119, and 117 clones for primer sets 3, 4, and 5, respectively) were screened by RFLP, and 210 representative clones were sequenced. Coverages approximated 93 and 100% at D ≤ 3% and D ≤ 20%, respectively, for the gene library generated with primer set 3 targeting a fraction of group 2 tfdA-like genes (Table 3; see also Fig. S2 and S3 in the supplemental material). Coverages approximated 62 and 91% at D ≤ 3% and D ≤ 20%, respectively, for the combined gene libraries generated with primers 4 and 5 targeting tfdA-like genes of groups 1 to 3 (Table 3; Fig. 2 and 3), indicating that the gene libraries were sufficiently sampled.
TABLE 3.
Summary of data from amino acid analyses of in silico-translated tfdA-like gene sequence fragments retrieved from Scheyern agricultural soil
| Primer set(s)a | Target | nb | Dc (%) | Sd | Ce (%) | Chao1f | H′g |
|---|---|---|---|---|---|---|---|
| 3 | tfdAα (group 2) | 201 | 3 | 34 | 92.5 | 47 | 2.5 |
| 20 | 6 | 100.0 | 6 | 0.5 | |||
| 4 and 5 | tfdA-like | 236 | 3 | 122 | 61.9 | 389 | 4.3 |
| (groups 1 to 3) | 20 | 48 | 91.1 | 78 | 3.2 |
See Table 2.
n, number of clones screened.
D, maximal amino acid sequence dissimilarity used for the assignment of sequences to genotypes (see Materials and Methods).
S, number of genotypes.
C, coverage; see Materials and Methods for details.
Richness estimator; indicative of estimated number of genotypes per gene library.
H′, Shannon index.
FIG. 3.
Rarefaction curves of cloned tfdA-like gene fragments for an evolutionary distance of 3% (A) or 20% (B). tfdA-like sequences were retrieved from Scheyern soil with primer sets 4 and 5 (○) and primer set 3 (•). Dotted and solid lines indicate 95% confidence intervals.
The gene library targeting group 2 tfdA-like genes (primer set 3) contained 34 and 6 genotypes at D ≤ 3% and D ≤ 20%, respectively (Table 3). The estimated numbers of genotypes present in the gene library were 47 and 6, respectively; Shannon's diversity indices were 2.5 and 0.5, respectively (Table 3). All sequences were related to tfdA-like genes of cultured Alphaproteobacteria (Bradyrhizobium spp.) or tfdA-like genes retrieved from other soils (see Fig. S2 in the supplemental material). Group 2 tfdA-like genes retrieved from Scheyern agricultural soil were assigned to 2 phylogenetic clusters (see Fig. S2). Cluster 2 contained sequences from Scheyern soil only, indicating that phylogenetically novel tfdA-like genes occur in Scheyern agricultural soil. The maximal phylogenetic distance of cluster 1 sequences was 23.3%, and the cluster contained Bradyrhizobium-related sequences. Cluster 1 sequences generated with primer set 3 (see Fig. S2) were represented by cluster 2a in gene libraries generated with primers 4 and 5 (maximal phylogenetic distance, 23.7% [Fig. 2]).
Primer sets 4 and 5 amplified similar fragments and detected similar groups of tfdA-like genes (Fig. 2). Thus, the gene libraries generated with such primers were combined for further analysis and contained 122 and 48 genotypes at D ≤ 3% and D ≤ 20%, respectively (Table 3). The estimated numbers of genotypes present in the gene library were 389 and 78, respectively; Shannon's diversity indices were 4.3 and 3.2, respectively (Table 3). All sequences of the gene library generated with primer sets 4 and 5 were related to tfdA-like genes from the Alphaproteobacteria Bradyhizobium spp. and Sphingomonas spp. or uncultured bacteria of other soils and were assigned to 11 clusters. Most of the clusters belonged to group 2 (90% of sequences) of tfdA-like genes (Fig. 2). Approximately 5% of the sequences were distantly related to any known group of tfdA-like genes and assigned to two novel groups, 4 and 5 (Fig. 2). Four out of the 11 clusters contained sequences from Scheyern soil only, reinforcing the conclusion that Scheyern soil harbors novel and diverse tfdA-like genes. The numbers of genotypes, the genotype richness estimators, and the phylogenetic diversities of tfdA-like genes were higher within gene libraries of primer sets 4 and 5 than within those of primer set 3 (Table 3; Fig. 2; see also Fig. S2 in the supplemental material). Thus, the detectability of tfdA-like gene diversity in soil was considerably higher with primer sets 4 and 5 than with primer set 3, indicating that those primers are useful tools to detect novel tfdA-like sequences in the environment.
Comparison of tfdA-like and 16S rRNA gene similarity.
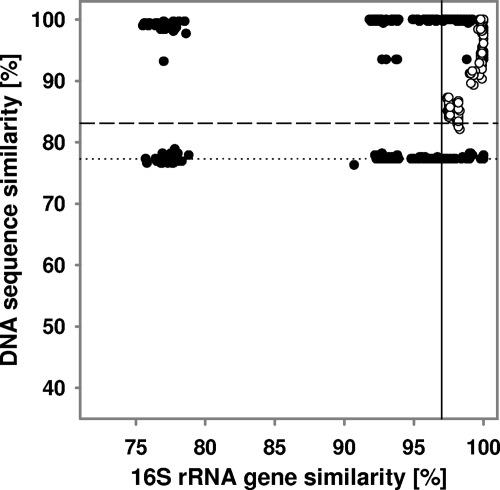
The comparative phylogeny of tfdA-like and 16S rRNA genes of bacteria harboring tfdA-like genes was assessed to determine if tfdA-like genes could be used for the identification of such organisms. Pairwise similarities of tfdA-like and 16S rRNA gene sequences ranged from 76.3 to 100% and 75.5 to 100% for group 1, respectively. For group 2 sequences, pairwise similarities of tfdA-like and 16S rRNA gene sequences ranged from 80.4 to 100% and 97.4 to 100%, respectively. Such data indicate that 16S rRNA genes are more conserved than tfdA-like genes (Fig. 4). Group 1 organisms that shared <77% tfdA-like gene similarity (or 80% protein similarity of translated tfdA-like genes) always shared <97% 16S rRNA gene similarity (a conservative threshold similarity for assigning two organisms to different species [65, 66]) (Fig. 4). The scattering of data points for group 1 organisms (Fig. 4) indicated horizontal gene transfer of group 1 tfdA, making it difficult to predict 16S rRNA gene similarities and consequently new species based on group 1 tfdA-like gene similarities. However, there was a correlation between 16S rRNA and group 2 tfdA-like gene similarity, indicating little or no horizontal gene transfer. Approximately 90% of any group 2 organisms that shared >97% 16S rRNA sequence similarity shared >82% tfdA-like gene similarity (equivalent to 84% protein similarity of translated tfdA-like genes). Thus, group 2 organisms that share ≤82% tfdA-like gene similarity (equivalent to ≤84% protein similarity of translated tfdA-like genes) have a very high probability of belonging to different species. Group 2 sequences that share ≤77% tfdA-like gene similarity (equivalent to ≤80% protein similarity of translated tfdA-like genes) have consequently an even higher probability of belonging to different species.
FIG. 4.
Correlation of 16S rRNA gene similarity with tfdA-like sequence similarity of pure cultures. Group 1 (•) and 2 (○) sequences are indicated. The solid line indicates 16S rRNA gene threshold similarity values for species delineation, and the dotted line indicates threshold similarity values below which sequences are indicative of novel species with high probability. The dashed line represents the 90% quantile of pairwise sequence comparisons with a 16S rRNA similarity of >97%.
Quantitation and recovery of tfdA-like genes.
Primer sets 3 to 5 were used for quantification of tfdA-like genes in soil by qPCR. Quantification of group 2 tfdA-like gene fragments with primer set 3 was reliable from 102 to 109 gene copy numbers μl of DNA extract−1 as indicated by the standard curve (see Fig. S3 in the supplemental material). Such qPCR products and those from environmental DNA had a Tm of approximately 90°C, and no nonspecific products were detected by agarose gel electrophoresis. Similar results were obtained with primer set 5 (see Fig. S3). Quantification of group 2 tfdA-like gene fragments with primer set 4 was reliable from 101 to 108 gene copy numbers μl of DNA extract−1 as indicated by the standard curve (see Fig. S1D).
Soils contained approximately 106 tfdA-like gene copies per gram (Table 1). Linearized pGEM-T with tfdA-like gene insert was added to triplicate Scheyern soil samples at final concentrations of 7.6 × 108 tfdA-like genes per gram of soil. tfdA-like gene copies per gram of soil measured by qPCR after DNA extraction were (1.58 ± 0.25) × 108. Thus, recovery of tfdA-like genes from soil approximated 20%, indicating that the DNA extraction protocol contributed to an underestimation of gene copy numbers in soil.
Quantification of tfdA-like genes relative to 16S rRNA genes in four different soils.
To overcome the effect of various degrees of qPCR inhibition by different DNA extracts, inhibition factors were recorded per DNA extract, and the gene copy numbers obtained by qPCR were corrected accordingly (see Materials and Methods). tfdA-like genes were abundant in all 4 different soils. Copy numbers ranged from 1 × 106 to 65 × 106 per gram of soil, depending on the primers and the soil (Tables 1 and 2). Primer set 4 consistently detected higher tfdA-like gene copy numbers than did primer set 3, which is in agreement with in silico analyses (Table 2). However, primer set 5, which theoretically targeted the broadest diversity (group 1 to 3 tfdA-like genes), in this study yielded copy numbers that were lower than those obtained with primer set 4 (groups 2 and 3), indicating that the broader specificity of primer set 5 compromised its sensitivity toward group 2 and 3 tfdA-like genes. Thus, primer set 4 is recommended for the quantification of group 2 and 3 tfdA-like genes in soils. Nevertheless, qPCR assays utilizing any of the primer sets used were successfully used for quantification of tfdA-like genes in soils. 16S rRNA gene copies ranged from 1 × 109 to 3 × 109 per gram of soil (Table 1). tfdA-like genes were most abundant in the agricultural soil of Scheyern compared to forest soils. The proportion of tfdA-like genes relative to 16S rRNA genes was likewise highest in Scheyern agricultural soil, indicating that a substantial number of bacteria harbored tfdA-like genes.
DISCUSSION
Novel and diverse tfdA-like genes were detected in soil by primers developed in this study to match a broad diversity of tfdA-like genes (Fig. 2; Table 3; see also Fig. S2 in the supplemental material). All tfdA-like genes were phylogenetically distinct from tauD (Fig. 2; see also Fig. S2), which encodes an α-KG-dependent taurine dioxygenase with approximately 30% sequence identity to tfdA-like genes (74), indicating that the novel primers discriminate well against tauD. tfdA-like gene fragments recovered from an agricultural soil that was treated with MCPA in 2002 were assigned to 11 clusters and were indicative of up to 122 distinct genotypes (at D < 3 [Table 3]) mainly related to alphaproteobacterial tfdA-like genes or two hitherto-unknown groups (Fig. 2; see also Fig. S2). Other studies of tfdA-like gene sequence diversity in soil without prior enrichment and/or isolation are still rather scarce and detect primarily group 1 tfdA-like genes indicative of Beta- and Gammaproteobacteria (3-5, 20). However, enrichments and analysis of isolates from soils revealed the presence of alphaproteobacterial PAA degraders in soils (29, 34) and are consistent with the detection of alphaproteobacterial tfdA-like genes in this study. Alphaproteobacteria of the genus Sphingomonas numerically dominate PAA degraders in certain soils (32), and those of the genera Sphingomonas and Bradyrhizobium are often detected by isolation (11, 27, 34, 41, 51). Since highly diverse tfdA-like genes indicative of Alphaproteobacteria were detected in this study, Alphaproteobacteria might represent a widely distributed, more diverse group of PAA degraders in soil than previously thought.
Bradyrhizobium-related PAA degraders of the Alphaproteobacteria are slow-growing oligotrophs (doubling times approximate 20 h), which are sensitive to high concentrations of organic nutrients (34, 59). Such organisms were also isolated from pristine soils that never received PAA treatment (34). Group 2 tfdA-like gene products are capable of PAA conversion (29), indicating that those genes are potentially involved in PAA conversion in soil, despite the occurrence of alternative potential PAA-degradative genes like cadAB among certain Bradyrhizobia (30, 35). Group 2 tfdA-like gene products display a higher activity for nonchlorinated PAA than for chlorinated ones (29). Group 1 tfdA-like gene products are capable of utilizing phenoxyacetic acid and cinnamic acid derivatives which occur in soil and are produced by plants (references 26 and 61 and references therein). Cinnamic acid-related flavonoids enhanced PAA utilization by a Bradyrhizobium sp. strain, indicating that such naturally occurring compounds might stimulate the expression of group 2 tfdA-like genes (58, 59). Since group 2 tfdA-like gene products are capable of converting PAA (29) and tfdA-like genes were abundant in the soil analyzed (Table 1), tfdA-like genes might have potential environmental relevance for (cometabolic) PAA degradation in soil. The primers developed in this study will facilitate a thorough evaluation of such a hypothesis in future studies.
The analysis of structural genes like tfdA might well be used for diversity studies of process-associated bacteria, but evidence for species-level diversity from such data might be obtained only if horizontal gene transfer of the structural gene in question is scarce. 16S rRNA gene similarities correlated with tfdA-like gene similarities for group 2 sequences but not for group 1 sequences (Fig. 4), indicating that horizontal gene transfer played a major role during evolution of group 1 rather than group 2 tfdA-like-gene-harboring organisms. Several lines of evidence support such a conclusion: (i) plasmid-encoded group 1 tfdA is common among PAA-degrading bacteria of the Beta- and Gammaproteobacteria (e.g., references 2, 14, and 18), (ii) such an encoded PAA-degrading function is transmissible (34, 46), and (iii) the degree of congruency between 16S rRNA and group 1 tfdA-like-gene-based phylogenetic trees is low (46). In contrast, the PAA-degrading function of group 2 and 3 tfdA-like gene-carrying organisms is not or only barely transmissible (34), the degree of congruency between 16S rRNA and group 2 tfdA-like-gene-based phylogenetic trees is high (30), and the GC contents of group 3 tfdA-like and housekeeping genes in Sphingomonas spp. are highly similar (30).
The virtual absence of horizontal gene transfer for group 2 and 3 tfdA-like genes allows for an estimation of species-level diversity based on group 2 and 3 tfdA-like gene analysis. Correlation analysis of tfdA-like and 16S rRNA gene similarity thus indicates that the number of tfdA-like genotypes that share less than 80% of translated protein sequences is a minimal estimate for species-level diversity of tfdA-like-gene-carrying bacteria (i.e., the true species-level diversity might be much higher). The calculated threshold similarities are well in agreement with those obtained for other structural genes (e.g., for narG, nosZ, dsrAB, and amoA gene fragments, they were 70 to 90% [25, 36, 37, 50]). The detected tfdA-like genes in this study consequently suggest that at least 48 distinct species of tfdA-like-gene-carrying bacteria occurred in agricultural soil (Table 3).
Such diverse tfdA-like genes were abundant in agricultural and pristine forest soils as indicated by up to 107 copy numbers per gram (dry weight) soil (Table 1). tfdA-like genes accounted for up to 3% of bacterial 16S rRNA genes. Copy numbers obtained in other studies with group 1 tfdA-like-gene-specific qPCR assays vary from 102 to 105 per gram (dry weight) soil prior to PAA incubation (3, 5, 20), indicating that group 2 and 3 genes outnumber group 1 genes in certain soils and highlighting a hitherto unknown PAA degradation potential in soils.
All qPCR assays in this study included a correction for inhibitory effects of environmental DNA extracts on the DNA polymerase used. Inhibition of the DNA polymerase during qPCR results in an underestimation of gene copy numbers (43). Such inhibitors include humic acids derived from plant material (53, 76). There are many approaches to overcome inhibition of qPCR, such as increase of Mg2+ concentration, addition of BSA, or dilution of template DNA (e.g., references 3, 23, and 38). DNA extraction and purification methods were also improved to reduce the amount of coextracted inhibitors (22, 75). However, all of these approaches failed in our study to avoid inhibition (Fig. 1 and data not shown). Thus, gene copy numbers were corrected for the inhibition caused by environmental DNA extracts. Since internal positive-control fragments are more suitable for TaqMan probe-based assays (12, 44), environmental DNA was spiked with an approximately 10-fold excess of the target gene to identify and correct for inhibition during qPCR. The quality of DNA extracts obtained with the same method from soils varies (63), necessitating the evaluation of inhibitory effects for every qPCR assay to minimize artificial variations in gene copy numbers.
Novel primers developed in this study enabled the detection of highly diverse group 2 and 3 tfdA-like gene sequences in soil (Fig. 2; see also Fig. S2 in the supplemental material). Such genes outnumber group 1 tfdA-like genes by several orders of magnitude in certain soils without prior enrichment on PAA (Table 1) (3, 4), indicating that PAA degradation is not the primary function of those genes. Inducers of gene expression of group 2 and 3 tfdA-like genes in soil and pure cultures need to be identified to shed light on the potential function of such genes. The present study provides a basis for such investigations.
Supplementary Material
Acknowledgments
We are grateful to Stephan Schulz, Michael Schloter, Jean Charles Munch, Daniela Degelmann, and Steffen Kolb for provision of soil samples.
Support for this study was provided by the Deutsche Forschungsgemeinschaft (grant HO4020/1-1, DFG Priority Program 1315 “Biogeochemical interfaces in soil”), by the University of Bayreuth, and by a grant from the German Academic Exchange Service (DAAD) to Y.-J. Liu.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amy, P. S., J. W. Schulke, L. M. Frazier, and R. J. Seidler. 1985. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 49:1237-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bælum, J., T. Henriksen, H. C. B. Hansen, and C. S. Jacobsen. 2006. Degradation of 4-chloro-2-methylphenoxyacetic acid in top- and subsoil is quantitatively linked to the class III tfdA gene. Appl. Environ. Microbiol. 72:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bælum, J., and C. S. Jacobsen. 2009. TaqMan probe-based real-time PCR assay for detection and discrimination of class I, II, and III tfdA genes in soils treated with phenoxy acid herbicides. Appl. Environ. Microbiol. 75:2969-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bælum, J., M. H. Nicolaisen, W. E. Holben, B. W. Strobel, J. Sørensen, and C. S. Jacobsen. 2008. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2:677-687. [DOI] [PubMed] [Google Scholar]
- 6.Bollag, J.-M., C. S. Helling, and M. Alexander. 1967. Metabolism of 4-chloro-2-methylphenoxyacetic acid by soil bacteria. Appl. Environ. Microbiol. 15:1393-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borken, W., A. Muhs, and F. Beese. 2002. Changes in microbial and soil properties following compost treatment of degraded temperate forest soils. Soil Biol. Biochem. 34:403-412. [Google Scholar]
- 8.Borken, W., Y. J. Xu, and F. Beese. 2003. Conversion of hardwood forests to spruce and pine plantations strongly reduced soil methane sink in Germany. Global Change Biol. 9:956-966. [Google Scholar]
- 9.Braid, M. D., L. M. Daniels, and C. L. Kitts. 2003. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 52:389-393. [DOI] [PubMed] [Google Scholar]
- 10.Bryant, F. O. 1992. Biodegradation of 2,4-dichlorophenoxyacetic acid and 2,4,5-trichlorophenoxyacetic acid by dichlorophenol-adapted microorganisms from fresh-water, anaerobic sediments. Appl. Microbiol. Biotechnol. 38:276-281. [Google Scholar]
- 11.Chung, M. J., and J. O. Ka. 1998. Isolation and characterization of 2,4-dichlorophenoxyacetic acid-degrading bacteria from paddy soils. J. Microbiol. 36:256-261. [Google Scholar]
- 12.Courtney, B. C., M. M. Smith, and E. A. Henchal. 1999. Development of internal controls for probe-based nucleic acid diagnostic assays. Anal. Biochem. 270:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Cupples, A. M., and G. K. Sims. 2007. Identification of in situ 2,4-dichlorophenoxyacetic acid-degrading soil microorganisms using DNA-stable isotope probing. Soil Biol. Biochem. 39:232-238. [Google Scholar]
- 14.Don, R. H., and J. M. Pemberton. 1981. Properties of 6 pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, J. C. 1980. Enumeration of the soil microorganisms able to degrade 2,4-D by metabolism or co-metabolism. Chemosphere 9:169-174. [Google Scholar]
- 16.Fukumori, F., and R. P. Hausinger. 1993. Alcaligenes eutrophus JMP134 2,4-dichlorophenoxyacetate monooxygenase is an alpha-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukumori, F., and R. P. Hausinger. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311-24317. [PubMed] [Google Scholar]
- 18.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. E. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gintautas, P. A., S. R. Daniel, and D. L. Macalady. 1992. Phenoxyalkanoic acid herbicides in municipal landfill leachates. Environ. Sci. Technol. 26:517-521. [Google Scholar]
- 20.Gonod, L. V., F. Martin-Laurent, and C. Chenu. 2006. 2,4-D impact on bacterial communities, and the activity and genetic potential of 2,4-D degrading communities in soil. FEMS Microbiol. Ecol. 58:529-537. [DOI] [PubMed] [Google Scholar]
- 21.Good, I. J. 1958. The population frequency of species and the estimation of the population parameters. Biometrics 40:237-246. [Google Scholar]
- 22.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Baumann, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 24.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Horn, M. A., H. L. Drake, and A. Schramm. 2006. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl. Environ. Microbiol. 72:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotopp, J. C. D., and R. P. Hausinger. 2001. Alternative substrates of 2,4-dichlorophenoxyacetate/alpha-ketoglutarate dioxygenase. J. Mol. Catal. B Enzymatic 15:155-162. [Google Scholar]
- 27.Huong, N. L., K. Itoh, and K. Suyama. 2007. Diversity of 2,4-dichlorophenoxyacetic acid (2,4-D) and 2,4,5-trichlorophenoxyacetic acid (2,4,5-T)-degrading bacteria in Vietnamese soils. Microbes Environ. 22:243-256. [DOI] [PubMed] [Google Scholar]
- 28.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 29.Itoh, K., R. Kand, Y. Sumita, H. Kim, Y. Kamagata, K. Suyama, H. Yamamoto, R. P. Hausinger, and J. M. Tiedje. 2002. tfdA-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in Alphaproteobacteria. Appl. Environ. Microbiol. 68:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh, K., Y. Tashiro, K. Uobe, Y. Kamagata, K. Suyama, and H. Yamamoto. 2004. Root nodule Bradyrhizobium spp. harbor tfdA alpha and cadA, homologous with genes encoding 2,4-dichlorophenoxyacetic acid-degrading proteins. Appl. Environ. Microbiol. 70:2110-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Analysis of competition in soil among 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 60:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Use of gene probes to aid in recovery and identification of functionally dominant 2,4-dichlorophenoxyacetic acid-degrading populations in soil. Appl. Environ. Microbiol. 60:1116-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitagawa, W., S. Takami, K. Miyauchi, E. Masai, Y. Kamagata, J. M. Tiedje, and M. Fukuda. 2002. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 37.Koops, H.-P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2004. The lithoautotrophic ammonia-oxidizing bacteria, p. 2625-2637. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes—an evolving electronic resource, release 3.13 ed. Springer Verlag, New York, NY.
- 38.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, T. H., S. Kurata, C. H. Nakatsu, and Y. Kamagata. 2005. Molecular analysis of bacterial community based on 16S rDNA and functional genes in activated sludge enriched with 2,4-dichlorophenoxyacetic acid (2,4-D) under different cultural conditions. Microb. Ecol. 49:151-162. [DOI] [PubMed] [Google Scholar]
- 40.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macur, R. E., J. T. Wheeler, M. D. Burr, and W. P. Inskeep. 2007. Impacts of 2,4-D application on soil microbial community structure and on populations associated with 2,4-D degradation. Microbiol. Res. 162:37-45. [DOI] [PubMed] [Google Scholar]
- 42.Maurer, D., S. Kolb, L. Haumaier, and W. Borken. 2008. Inhibition of atmospheric methane oxidation by monoterpenes in Norway spruce and European beech soils. Soil Biol. Biochem. 40:3014-3020. [Google Scholar]
- 43.McDevitt, J. J., P. S. J. Lees, W. G. Merz, and K. J. Schwab. 2004. Development of a method to detect and quantify Aspergillus fumigatus conidia by quantitative PCR for environmental air samples. Mycopathologia 158:325-335. [DOI] [PubMed] [Google Scholar]
- 44.McDevitt, J. J., P. S. J. Lees, W. G. Merz, and K. J. Schwab. 2007. Inhibition of quantitative PCR analysis of fungal conidia associated with indoor air particulate matter. Aerobiologia 23:35-45. [Google Scholar]
- 45.McGhee, I., and R. G. Burns. 1995. Biodegradation of 2,4-dichlorophenoxyacetic acid (2,4-D) and 2-methyl-4-chlorophenoxyacetic acid (MCPA) in contaminated soil. Appl. Soil Ecol. 2:143-154. [Google Scholar]
- 46.McGowan, C., R. R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortensen, S. K., and C. S. Jacobsen. 2004. Influence of frozen storage on herbicide degradation capacity in surface and subsurface sandy soils. Environ. Sci. Technol. 38:6625-6632. [DOI] [PubMed] [Google Scholar]
- 48.Mrkonjic Fuka, M., M. Engel, A. Gattinger, U. Bausenwein, M. Sommer, J. C. Munch, and M. Schloter. 2008. Factors influencing variability of proteolytic genes and activities in arable soils. Soil Biol. Biochem. 40:1646-1653. [Google Scholar]
- 49.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer, K., H. L. Drake, and M. A. Horn. 2009. Genome-derived criteria for assigning environmental narG and nosZ sequences to operational taxonomic units of nitrate reducers. Appl. Environ. Microbiol. 75:5170-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, I. H., and J. O. Ka. 2003. Isolation and characterization of 4-(2,4-dichlorophenoxy)butyric acid degrading bacteria from agricultural soils. J. Microbiol. Biotechnol. 13:243-250. [Google Scholar]
- 52.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. G. Ludwig, J. Peplies, and F. O. Glöckner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radstrom, P., R. Knutsson, P. Wolffs, M. Lovenklev, and C. Lofstrom. 2004. Pre-PCR processing—strategies to generate PCR-compatible samples. Mol. Biotechnol. 26:133-146. [DOI] [PubMed] [Google Scholar]
- 54.Robertson, B. K., and M. Alexander. 1994. Growth-linked and cometabolic biodegradation—possible reason for occurrence or absence of accelerated pesticide biodegradation. Pest. Sci. 41:311-318. [Google Scholar]
- 55.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 56.Scheidleder, A. 2000. Groundwater quality and quantity in Europe. European Environment Agency, Copenhagen, Denmark.
- 57.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw, L. J., and R. G. Burns. 2004. Enhanced mineralization of [U-14C]2,4-dichlorophenoxyacetic acid in soil from the rhizosphere of Trifolium pratense. Appl. Environ. Microbiol. 70:4766-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw, L. J., and R. G. Burns. 2005. Rhizodeposition and the enhanced mineralization of 2,4-dichlorophenoxyacetic acid in soil from the Trifolium pratense rhizosphere. Environ. Microbiol. 7:191-202. [DOI] [PubMed] [Google Scholar]
- 60.Shelton, D. R., and J. S. Karns. 1998. Pesticide bioremediation: genetic and ecological considerations, p. 181-216. In P. C. Kearny and T. Roberts (ed.), Pesticide remediation in soils and water. John Wiley & Sons, Chichester, United Kingdom.
- 61.Smejkal, C. W., T. Vallaeys, S. K. Burton, and H. M. Lappin-Scott. 2001. Substrate specificity of chlorophenoxyalkanoic acid-degrading bacteria is not dependent upon phylogenetically related tfdA gene types. Biol. Fertil. Soils 33:507-513. [Google Scholar]
- 62.Smith, A. E. 1985. Identification of 4-chloro-2-methylphenol as a soil degradation product of ring-labeled [C-14] mecoprop. Bull. Environ. Contam. Toxicol. 34:656-660. [DOI] [PubMed] [Google Scholar]
- 63.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 64.Sorensen, S. R., A. Schultz, O. S. Jacobsen, and J. Aamand. 2006. Sorption, desorption and mineralization of the herbicides glyphosate and MCPA in samples from two Danish soil and subsurface profiles. Environ. Pollut. 141:184-194. [DOI] [PubMed] [Google Scholar]
- 65.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 66.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44:846-849. [Google Scholar]
- 67.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suwa, Y., A. D. Wright, F. Fukumori, K. A. Nummy, R. P. Hausinger, W. E. Holben, and L. J. Forney. 1996. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 62:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 70.Thompson, D. G., G. R. Stephenson, K. R. Solomon, and A. V. Skepasts. 1984. Persistence of (2,4-dichlorophenoxy)acetic acid and 2-(2,4-dichlorophenoxy)propionic acid in agricultural and forest soils of Northern and Southern Ontario. J. Agric. Food Chem. 32:578-581. [Google Scholar]
- 71.Tonso, N. L., V. G. Matheson, and W. E. Holben. 1995. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30:3-24. [DOI] [PubMed] [Google Scholar]
- 72.Tsai, Y. L., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol. Ecol. 20:163-172. [Google Scholar]
- 74.van der Ploeg, J. R., E. Eichhorn, and T. Leisinger. 2001. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch. Microbiol. 176:1-8. [DOI] [PubMed] [Google Scholar]
- 75.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 43:633-642. [DOI] [PubMed] [Google Scholar]
- 76.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Worthing, C. R., and R. J. Hance. 1991. The pesticide manual—a world compendium. The British Crop Protection Council, Farnham, United Kingdom.
- 78.Zakaria, D., H. Lappin-Scott, S. Burton, and C. Whitby. 2007. Bacterial diversity in soil enrichment cultures amended with 2 (2-methyl-4-chlorophenoxy) propionic acid (mecoprop). Environ. Microbiol. 9:2575-2587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.