Abstract
A gene encoding a glycoside hydrolase family 44 (GH44) protein from Clostridium acetobutylicum ATCC 824 was synthesized and transformed into Escherichia coli. The previously uncharacterized protein was expressed with a C-terminal His tag and purified by nickel-nitrilotriacetic acid affinity chromatography. Crystallization and X-ray diffraction to a 2.2-Å resolution revealed a triose phosphate isomerase (TIM) barrel-like structure with additional Greek key and β-sandwich folds, similar to other GH44 crystal structures. The enzyme hydrolyzes cellotetraose and larger cellooligosaccharides, yielding an unbalanced product distribution, including some glucose. It attacks carboxymethylcellulose and xylan at approximately the same rates. Its activity on carboxymethylcellulose is much higher than that of the isolated C. acetobutylicum cellulosome. It also extensively converts lichenan to oligosaccharides of intermediate size and attacks Avicel to a limited extent. The enzyme has an optimal temperature in a 10-min assay of 55°C and an optimal pH of 5.0.
Thirteen glycoside hydrolase (GH) families, each having members related to each other by amino acid sequence, contain enzymes that hydrolyze cellulose and/or cellooligosaccharides (4; http://www.cazy.org). Among them is GH family 44 (GH44), most of whose enzymes are endoglucanases (EGs). In general, EGs are more active on longer rather than on shorter chains and are more likely to attack bonds in the interiors of carbohydrate chains than those near their termini.
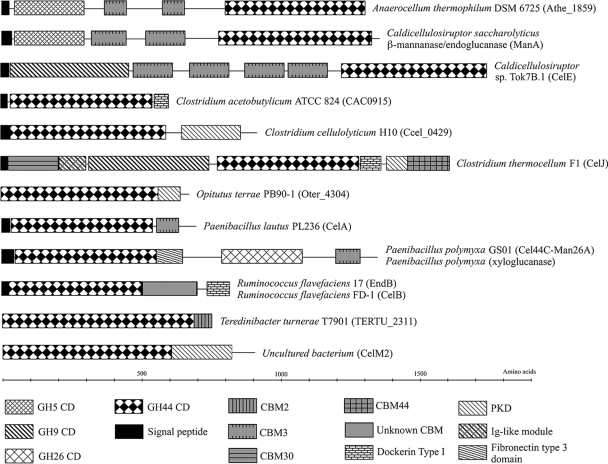
With one exception, GH44 enzymes are produced by bacteria, both aerobic and anaerobic. At present, 29 amino acid sequences of GH44 members have been determined (4). Often they are combined with other GHs in multienzyme proteins (Fig. 1).
FIG. 1.
Structural organization of genes coding for GH44 CDs, excluding GH44 members with only a signal peptide and a CD. The gene encoding O. terrae's GH44 member produces a 920-residue protein whose domain structure is unclassified. The sequence was searched against the Pfam library, and a low E-value, 1.22 × 10−22, was found for a PKD (polycystic kidney disease)-type domain. Ig, immunoglobulin.
Not all of these GH44 enzymes have been produced in vitro, and those that have been produced have only been partially characterized. Experimental results indicate that GH44 enzymes exclusively cleave β-1,4 bonds between glucosyl and xylosyl residues and that they have different abilities to attack xylan, lichenan, and different cellulose forms, such as Avicel, acid-swollen cellulose, and carboxymethyl cellulose (CMC), with the presence of a carbohydrate-binding module (CBM) allowing higher activity on solid cellulose. They appear to be inactive on short oligosaccharides, like p-nitrophenyl (PNP)-β-glucopyranoside, PNP-β-cellobioside, and PNP-β-xylopyranoside.
Most GH families containing cellulases have at least one member with a known tertiary structure. That was not true of GH44 until Kitago et al. (15) published six different crystal structures of an EG, CelJ, from Clostridium thermocellum. Three of the crystal structures are of the wild-type enzyme, and the other three are of the E186Q mutant, with each form being both unliganded and complexed with cellopentaose or cellohexaose. The enzyme uses a retaining mechanism, with Glu186 being the proton donor/acceptor and Glu359 being the nucleophile. Subsites −4 to −1 of the wild-type enzyme hold cellotetraose. When the E186Q mutant is soaked with cellopentaose or cellohexaose, different-length cellooligosaccharides are complexed in its subsites −4 to +5.
A second tertiary structure from an unidentified bacterium is similar to that from C. thermocellum (23). The enzyme, CelM2, is a triose phosphate isomerase (TIM)-like (β,α)8 barrel with a β-sandwich domain. It also has Glu221 and Glu393 as the catalytic proton donor/acceptor and nucleophile, respectively. These two residues are located approximately 4 Å apart from one another, similar to the catalytic residues of CelJ.
The present work concerns the GH44 putative EG from Clostridium acetobutylicum ATCC 824, a Gram-positive, mesophilic, anaerobic, solvent-producing bacterium. This organism and other solvent-producing Clostridium strains cannot grow on cellulose as a sole carbon source, but the first can produce EGs, mainly extracellular, when grown on glucose, xylose, mannose, and cellobiose (18). Nearly all of the same strains can grow on larch wood xylan, but C. acetobutylicum ATCC 824 can do this only when cultured in a chemostat, where it produces xylanase activity (19).
Genomic sequencing has found the gene CAC0915, which putatively encodes a fusion protein consisting of a signal peptide, a GH44 catalytic domain (CD), and a type I dockerin but no CBM in C. acetobutylicum ATCC 824 (25). This putative protein, CAC0915, has 606 amino acids for a calculated molecular mass of 66.8 kDa (25). The same project found genes for many other cellulases and xylanases. In fact, the complete coding for a cellulosome similar to those in the cellulolytic species Clostridium cellulovorans and Clostridium cellulolyticum appears to be present in C. acetobutylicum ATCC 824 (25), and a cellulosome is produced, but its cellulolytic activity is very low (28). Schwarz et al. (29) have hypothesized that C. acetobutylicum has repressed cellulosome expression and cellulolytic activity during evolution since it can grow on simpler substrates, including starch, oligosaccharides, and monosaccharides.
This article reports the phylogenetic tree of the GH44 enzymes and the production, purification, and subsequent structural and kinetic characterization of C. acetobutylicum GH44 EG. This protein apparently had not been observed in isolated form before this project.
MATERIALS AND METHODS
GH44 multiple sequence alignment and phylogenetic tree.
Primary amino acid sequences of GH44 CDs were obtained from the GenPept and UniProt databases via the CAZy database (4). An initial multiple sequence alignment was performed using ClustalX version 1.83 (32; http://bips.u-strasbg.fr/fr/Documentation/ClustalX/) using gap penalties of 30 for both pairwise and multiple alignments, with a delay for divergent species set at 40% and with a Gonnet series 250 protein weight matrix (7) to identify GH44 CDs in fusion proteins containing non-GH44 domains.
Following this, amino acid sequences of 23 of the 29 GH44 CDs were aligned using the same techniques. Two cellulase fragments from uncultured bacteria were omitted because their sequences were incomplete, Myxococcus xanthus sequence 15196 is the same as the M. xanthus DK 1622 sequence, and the sequences of CelJ in C. thermocellum ATCC 27405 and CelJ in C. thermocellum F1 are the same except for amino acid position 1394. The Paenibacillus pabuli EG sequence was excluded because of a segment of 36 unidentified amino acids.
A phylogenetic tree was constructed using Phylip 3.68 (http://evolution.genetics.washington.edu/phylip.html). The GH44 CD multiple sequence alignment was bootstrapped using Seqboot with molecular sequence and bootstrapping chosen, bootstrap block size of 1, input sequences interleaved, and 100 replicates generated. The output multiple sequence alignments were used as inputs for Protpars (parsimony), ProML (maximum likelihood), and ProtDist/Neighbor (neighbor joining) to find the best phylogenetic tree for each alignment by randomizing the input order of the sequences. A consensus tree was determined using Consense and majority-rule consensus type. Branch lengths were generated for the consensus tree by inputting the initial multiple sequence alignment and consensus tree into ProML and using the Jones-Taylor-Thornton probability model (12; http://xray.bmc.uu.se/alwyn/Distribution/distrib_frameset.html). Branch distances were given in terms of expected fraction of amino acids changed, such that 1.0 is the same as 100 point accepted mutations (PAMs).
Gene synthesis and transfer.
Conflicts in codon usage between source and host organisms can hinder successful protein expression (9, 21). Therefore, Protein2DNA (DNA 2.0, Menlo Park, CA) was used to adjust the codon bias of the first 1,643 nucleotides of CAC0915, coding for the signal peptide through the CD but not the dockerin domain, to that of Escherichia coli. This sequence was synthesized by Megabase Research Products (Lincoln, NE) and supplied as an E. coli XL1-Blue (Stratagene, La Jolla, CA) clone containing the synthesized gene in the pSTBlue-1 plasmid (Novagen, Madison, WI).
The DNA provided by Megabase was used as a template to produce a 1,544-base-pair gene fragment, yielding a mature protein of 511 amino acids, identical in sequence to that of the CD of the protein CAC0915. The nucleotides encoding the signal peptide were removed to eliminate expression problems. This was cloned into the Novagen pET-22b(+) vector, which codes for the fusion of a C-terminal histidine tag, and expressed in E. coli BL21(DE3) (Novagen).
Protein production and purification.
E. coli clones were grown in autoinduction medium [0.05% glucose, 0.5% glycerol, 0.2% lactose, 1.2% tryptone, 2.4% yeast extract, 25 mM succinate, 5 μM Fe2(SO4)3, 19 mM KH2PO4, 45 mM K2HPO4, 2 mM MgSO4, and 45 mM NaH2PO4] (31) supplemented with 50 μg/liter carbenicillin at room temperature and 250 rpm shaking until the absorbance at 600 nm was approximately 13, measured after dilution to bring the reading within the linear range. Harvested cells were resuspended in 20 ml of nickel-nitrilotriacetic acid (Ni-NTA) binding buffer (25 mM HEPES, pH 7.0, 300 mM NaCl, and 10 mM imidazole) (Novagen) and lysed four successive times in an SLM Aminco (Rochester, NY) French press at 125 MPa.
A 15-ml Ni-NTA His•Bind Superflow (Novagen) column resin was used to purify His-tagged proteins. The column was washed with Ni-NTA wash buffer (25 mM HEPES, pH 7.0, 300 mM NaCl, and 20 mM imidazole), and the enzyme was eluted with Ni-NTA elution buffer (25 mM HEPES, pH 7.0, 300 mM NaCl, and 250 mM imidazole). A 50-ml Sephadex G-25 (GE Healthcare, Piscataway, NJ) column was used to desalt the protein into 10 mM HEPES buffer, pH 7.0. If necessary, the protein was concentrated to 22 g/liter, using the Pierce (Rockford, IL) bicinchoninic acid assay (30) and bovine serum albumin standards, with a Vivaspin6 (Sartorius, Elk Grove, IL) polyethersulfone 5,000-Da MWCO spin filter at 8,000 × g. Based upon densitometry analysis of Pierce precast sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels using ImageJ, the protein in the elution fraction was ≫95% pure (data not shown). The contaminating band, seen only on silver staining, was of approximately the same molecular weight as the nickel-binding enzyme SlyD (26).
Crystallization and structure refinement.
Crystallization screening was performed using the hanging-drop vapor diffusion method with 500 μl of mother liquor in the reservoir and a 1:1 ratio of protein to mother liquor in a 4-μl drop and Hampton Research (Aliso Viejo, CA) Crystal Screen I and Crystal Screen II buffer kits. Initial crystals were obtained using Hampton's Crystal Screen buffer 20 [0.2 M (NH4)2SO4, 0.1 M sodium acetate (NaOAc), and 25% (wt/vol) polyethylene glycol 4000 (PEG 4000) at pH 4.6] at 23°C. The buffer composition was optimized to 0.2 M (NH4)2SO4, 0.1 M NaOAc, 10% (wt/vol) PEG 3350, and pH 5.4 for the native protein. The crystals were approximately 0.6 by 0.3 by 0.1 mm in size. They were soaked in 0.15 M (NH4)2SO4, 0.75 M NaOAc, 18.75% (wt/vol) PEG 3350, and 25% (wt/vol) glycerol at pH 5.4 and frozen before data collection. Diffraction data were collected at 100 K at the Iowa State University Macromolecular X-ray Crystallography Facility on a Rigaku/MSC home-source generator at a 1.54-Å wavelength and processed using d*TREK (27; http://www.px.nsls.bnl.gov/indexing.html#dtrek). The crystal belongs to space group P212121, and its unit cell parameters and relevant diffraction statistics are located in Table 1.
TABLE 1.
X-ray data collection and structure refinement
| Characteristic | Value(s)a |
|---|---|
| Space group | P212121 |
| Unit cell parameters | |
| a (Å) | 54.08 |
| b (Å) | 87.29 |
| c (Å) | 103.29 |
| α (°) | 90.0 |
| β (°) | 90.0 |
| γ (°) | 90.0 |
| Resolution (Å) | 66.67-2.20 (2.28-2.20) |
| Rmergeb (%) | 8.6 (20.9) |
| Completeness (%) | 99.5 (94.3) |
| Observed reflections | 123,672 |
| Unique reflections | 25,388 |
| Redundancy | 4.87 (4.19) |
| I/σ(I) | 11.4 (5.0) |
| Mosaicity | 0.5° |
| Refinement statistics | |
| No. of nonhydrogen atoms | 4,735 |
| No. of water molecules | 638 |
| Rworkc | 15.8 |
| Rfreed | 23.1 |
| Average B (Å2) | 14 |
| Ramachandran plot | |
| In most favored (%) | 88.7 |
| In additionally allowed (%) | 11.1 |
| In generously allowed (%) | 0.2 |
| RMSD from ideal geometry | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.3 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ∑hkl∑j|Ihkl,j − 〈Ihkl〉|/∑hkl∑jIhkl,j, where Ihkl,j is the intensity of the jth measurement of reflection hkl and 〈Ihkl〉 is the average over all j measurements of reflection hkl.
Rwork = ∑hkl(|Fhklobs| − |Fhklcalc|)/∑hkl|Fhklobs|, where Fhklobs and Fhklcalc are the structure-factor amplitudes.
Rfree was calculated for a random 5% of reflections.
Molecular replacement was used to solve enzyme structures using AMoRe from the CCP4 suite (5, 24; http://www.ccp4.ac.uk/ and http://www.ccp4.ac.uk/dist/ccp4i/help/modules/molrepl.html). The structure of C. thermocellum Cel44A (PDB ID 2e4t) (15) was used to solve the phase problem and to thread the amino acid sequence of the enzyme into the molecular replacement solution using Swiss-PdbViewer (8; http://www.expasy.org/spdbv/). Manual rebuilding of the model was performed using O (12; http://xray.bmc.uu.se/alwyn/Distribution/distrib_frameset.html), and the model was refined with REFMAC5 (22; http://www.ysbl.york.ac.uk/∼garib/refmac/docs/index.html). Structural calculations were performed with DSSP (14), and figures were created with PyMol (DeLano Scientific, Palo Alto, CA). The final model is a monomer consisting of 512 amino acid residues with 638 water molecules, 10 glycerol molecules, three acetate ions, one calcium ion, one chloride ion, and one sulfate ion, with structural refinement statistics shown in Table 1.
Products of carbohydrate hydrolysis.
The enzyme, at a concentration of 650 mg/liter, was incubated individually with the following substrates: 750 mg/liter of the cellooligosaccharides {[β-d-glucopyranosyl-(1→4)]n-β-d-glucose, n = 1-5} cellobiose (catalog no. C-7252; Sigma, St. Louis, MO), cellotriose (product no. 400400-1; Seikagaku, Tokyo, Japan), cellotetraose (product no. 400402-1; Seikagaku), cellopentaose (product no. 400404-1; Seikagaku), and cellohexaose (product no. 400406-1; Seikagaku), along with 20 g/liter Avicel (microcrystalline cellulose) (product no. 11363, lot no. 430118/1; Fluka, Buchs, Switzerland), 20 g/liter low-viscosity CMC (cellulose derivatized mainly with 2-O- and 6-O-linked carboxyl groups, averaging 0.6 to 0.95 groups per glucopyranosyl residue) (product no. C-5678, lot no. 065K0111; Sigma), 20 g/liter laminaran {primarily [β-d-glucopyranosyl-(1→3)]n-d-glucose, n = high} (product no. L-9634; Sigma), 10 g/liter lichenan {[β-d-glucopyranosyl-(1→3,1→4)]n-d-glucose, n = high} (product no. 155231, lot no. 9964F; Fisher), 10 g/liter mannan {[β-d-mannopyranosyl-(1→4)]n-d-mannose, n = high} (product no. M-7504, lot no 44C-1764; Sigma), 20 g/liter pullulan {[α-maltotriosyl-(1→6)-α-maltotriosyl]n-d-glucose, n = high} (product no. P0978, lot no. GA01; TCI America, Portland, OR), and 20 g/liter xylan {[β-d-xylopyranosyl-(1→4)]n-β-d-xylose, n = high, with significant branching initiated and terminated by other sugar residues} from birch wood (product no. X-0502, lot no. 129H0901; Sigma) or larch wood (product no. X-3875, lot no. 125C-00582; Sigma) at 25°C in 0.1 M NaOAc buffer, pH 5.0, for 16 h. The hydrolysis products were analyzed by thin-layer chromatography. A 60-Å silica gel plate (Whatman, Florham Park, NJ) was spotted with hydrolysate and developed using a single ascent of acetonitrile/ethyl acetate/1-propanol/water (1.7:0.4:1:1) mobile phase (10). The plate was dipped into a 5% (wt/vol) H2SO4, 0.5% (wt/vol) naphthol solution in ethanol and incubated at 95°C until the carbohydrate spots developed color.
Assays for enzyme activity and thermostability.
Kinetic constants of the enzyme acting on CMC, birch wood xylan, and larch wood xylan were determined by measuring product-reducing sugars with a glucose standard curve using tetrazolium blue reagent (0.1% [wt/vol] tetrazolium blue, 0.05 M NaOH, and 0.5 M sodium potassium tartrate) (13). Standard assay conditions consisted of incubating enzyme (1.7 mg/liter) with 0.025 to 10 g/liter substrate in 0.1 M sodium acetate (NaOAc) buffer, pH 5.0, at 25°C. Samples were taken at 30-s to 5-min time intervals. Each sample was placed in a boiling water bath with 4 ml of tetrazolium blue reagent for 5 min to stop the reaction and develop reagent color. Specific activity for each substrate concentration was determined by a linear regression of the reducing sugar concentration liberated versus incubation time and dividing the slope by the mass of protein in the sample. Enzyme units are defined as μmol glucose liberated/min under the assay conditions. A plot of specific activity versus substrate concentration was generated for each substrate, and the maximal activities and Michaelis constants were determined by nonlinear regression. Activity on larch wood xylan decreased at high substrate concentrations, so an extra denominator term representing inhibitor concentration was included in the rate equation.
Optimal temperature and pH were determined with the tetrazolium blue assay. The former was found by reacting 2% (wt/vol) low-viscosity CMC with 1.7 mg/liter enzyme at 25 to 60°C and pH 5.0 in 0.1 M NaOAc buffer. Sampling was performed as described above, and linear regression was used to calculate activities at each temperature. The determination of optimal pH used 1.7 mg/liter enzyme incubated with 2% (wt/vol) low-viscosity CMC at 25°C. The reaction buffers were 0.1 M NaOAc buffer for pH 3.5 to 5.0 and 0.1 M sodium phosphate buffer for pH 5.5 to 8.0. Sampling and activity calculations were performed as described above.
Enzyme thermostability was determined by incubating the enzyme in 10 mM HEPES, pH 7.0, at various temperatures for five different incubation times. Each partially inactivated enzyme sample was reacted with 1.0% (wt/vol) CMC in 0.1 M NaOAc buffer, pH 7.0, and sampled as described above. A plot of ln(activity) versus incubation time was used to determine the first-order inactivation rate coefficient for each incubation temperature. Values of ln(rate coefficient) were plotted versus inverse temperature to determine the activation energy of enzyme inactivation.
Protein structure accession number.
The atomic coordinates and structure factors of this enzyme have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics (http://www.rcsb.org/), with the ID 3ik2.
RESULTS
GH44 multisequence alignment and phylogenetic tree.
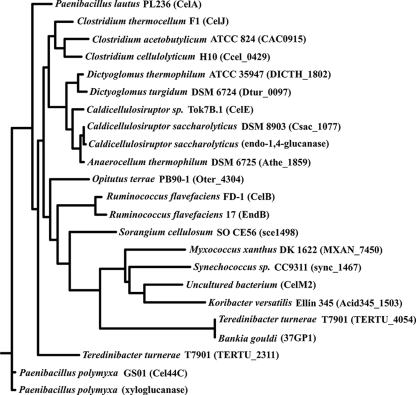
Although there are significant regions of sequence similarity in GH44 CDs, in general this family consists of enzymes with widely differing sequences (see Fig. S1 in the supplemental material). The neighbor-joining method produced the best consensus tree (Fig. 2), based upon frequency of branch occurrences. Two different groupings emerge, one comprising EGs from C. thermocellum, C. acetobutylicum, C. cellulolyticum, Dictyoglomus thermophilum, Dictyoglomus turgidum, Caldicellulosiruptor saccharolyticus and another Caldicellulosiruptor species, and Anaerocellum thermophilum and the other encompassing sequences from Opitutus terrae, Ruminococcus flavefaciens, Sorangium cellulosum, Myxococcus xanthus, Synechococcus sp., an uncultured strain, Candidatus Koribacter versatilis, Teredinibacter turnerae, and Bankia gouldi (a shipworm and the only nonbacterial species). The two Paenibacillus polymyxa xyloglucanases, the Paenibacillus lautus EG, and a second T. turnerae sequence are more distant from the other EGs. Specifically, the EG from C. thermocellum and the EG from C. acetobutylicum, which is the subject of this study, have very similar sequences.
FIG. 2.
Phylogenetic tree of GH44 CDs.
Enzyme crystal structure.
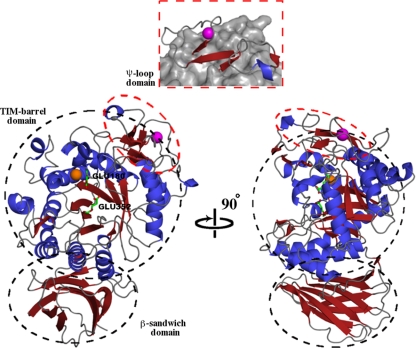
The C. acetobutylicum EG crystal structure, composed of 25 β-strands and 18 α-helices, was solved to a 2.2-Å resolution. It has a catalytic (β/α)8 TIM barrel-like structure (β3-β6, β11-β17, α1-α5, and α7-α18) with an additional ψ-loop motif (β7-β10 and α6) and β-sandwich (β1-β2 and β18-β25) of unknown function (Fig. 3). The catalytic proton donor/acceptor, Glu180, and the catalytic nucleophile, Glu352, are well defined in electron density and are located after the fourth β-strand (β11) and on the seventh β-strand (β16) of the TIM barrel core, respectively, with 5.4 Å separating their γ-carbon atoms and with Glu180 being part of an NEP motif. This structure indicates that the enzyme is part of clan GH-A and has a retaining mechanism. The catalytic proton donor/acceptor and nucleophile are the only two residues located in the generously allowed region of the Ramachandran plot. As with other EGs, C. acetobutylicum EG has a large, open binding cleft to accommodate bulky substrates.
FIG. 3.
Crystal structure of C. acetobutylicum ATCC 824 EG shown with a 90° rotation. The TIM barrel, a β-sandwich domain, and ψ-loop domains are shown. The catalytic acid/base, Glu180, and catalytic nucleophile, Glu352, are shown as sticks, as are amino acids involved in ligand binding. A chloride ion, orange, is located next to the catalytic acid in the TIM barrel. A cartoon diagram of the ψ-loop with respect to the rest of the protein is highlighted in the orange box. It contains a β-strand located between two antiparallel β-strands and hydrogen bonded to them. A calcium ion, magenta, appears to stabilize this domain.
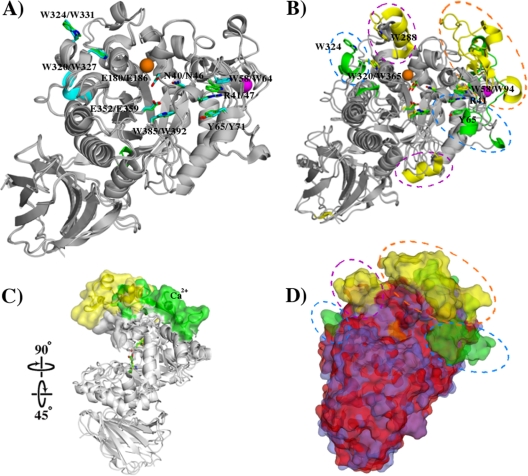
This structure and that of C. thermocellum EG (PDB ID 2e4t) have a root mean square deviation (RMSD) of 0.70 Å for 499 Cα atoms when superimposed. Figure 4A shows their alignment. Only two small secondary structure differences are observed: additional short helices exist in C. acetobutylicum EG (Pro18-Ile20) and C. thermocellum EG (Leu336-Ile338). Both EGs contain structural calcium ions to stabilize their ψ-loops. C. acetobutylicum EG has residues analogous to each of the ligand-binding amino acids of C. thermocellum EG: the catalytic proton donor/acceptor Glu180 (C. acetobutylicum EG)/Glu186 (C. thermocellum EG); the catalytic nucleophile Glu352/Glu359; hydrophobic platforms Trp58/Trp64, Tyr65/Tyr71, Trp320/Trp327, Trp324/Trp331, and Trp385/Trp392; and hydrogen bonders Asn40/Asn46 and Arg41/Arg47.
FIG. 4.
Structural alignments of C. acetobutylicum ATCC 824 EG with C. thermocellum EG or CelM2. (A) Alignment of cartoon representations of C. acetobutylicum EG and C. thermocellum EG. Ligand binding residues of C. thermocellum EG and their C. acetobutylicum EG analogs are shown as sticks, as are the catalytic residues. The chloride and calcium ions of both enzymes are orange and magenta spheres, respectively. Minimal structural differences are observed, with those additional secondary structure elements present in one enzyme but not the other shown in blue (C. thermocellum EG) or green (C. acetobutylicum EG). (B) Cartoon alignment of C. acetobutylicum EG and CelM2. Secondary structural features present in C. acetobutylicum EG but not in CelM2 are shown in green, and those found in CelM2 but not in C. acetobutylicum EG are shown in yellow. Key ligand binding amino acids and catalytic residues are shown as sticks and are colored like their secondary structure. The chloride and calcium ions of both enzymes are orange and magenta spheres, respectively. The ψ-loop of C. acetobutylicum EG and small domain of CelM2 that replaces it are shown in an orange circle. The blue ovals highlight both additional α-helices present in C. acetobutylicum EG, as well as its ligand binding residues (Arg41, Tyr65, and Trp324). (C) The alignment in panel B is rotated to show how the small domain of CelM2 extends the upper face of the ligand binding cleft relative to the ψ-loop of C. acetobutylicum EG. (D) A surface representation of the cartoon alignment in panel B, with C. acetobutylicum EG shown in blue and CelM2 shown in red. As in panel B, key differences are shown in yellow or green. Catalytic residues are shown in orange.
C. acetobutylicum EG and CelM2 (PDB ID 3fw6) have an RMSD of 1.19 Å for 407 Cα atoms. Figure 4B to D shows alignments and key structural differences between these two enzymes. CelM2 does not have a clear ψ-loop analogous to that of C. acetobutylicum EG. The amino acid residues that replace this small domain in CelM2 form three α-helices, three β-strands, and a twisted β-strand instead of the single α-helix, four β-strands, and one structural calcium ion that form the ψ-loop in C. acetobutylicum EG. The result is a difference in the shape of the binding cleft of the two EGs (Fig. 4C). The small CelM2 domain extends beyond the C. acetobutylicum EG ψ-loop, forming a deeper binding pocket.
C. acetobutylicum EG has three residues, Arg41 in subsite −3, Tyr65 in subsite +3, and Trp324 in subsite +5 (encircled by blue ovals in Fig. 4B), that are involved in substrate binding and that do not have structural analogs in CelM2. It also has two hydrophobic residues, Trp58 and Tyr65, both in subsite −4, on opposite faces of the active site that can bind a substrate. Conversely, CelM2 has two α-helices where only coils are present in C. acetobutylicum EG (Fig. 4B). The helix on top of CelM2, on which Trp288 is found, is at the end of the binding cleft and forms a protrusion that points toward its opposite face, holding Trp365, another hydrophobic residue, and they could potentially form stacking interactions with a substrate. These two residues are located at the opposite end of the binding cleft from the two opposing hydrophobic residues in C. acetobutylicum EG.
Carbohydrate hydrolysis products.
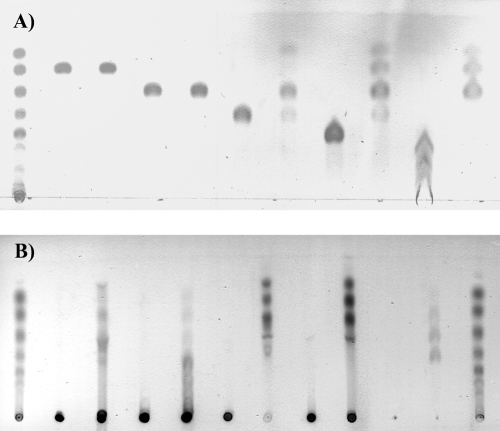
Thin-layer chromatography shows that C. acetobutylicum EG attacks cellotetraose, cellopentaose, and cellohexaose but not cellobiose and cellotriose (Fig. 5A). Cellotetraose yields mainly cellotriose and glucose, with some unreacted cellotetraose and, perhaps, some cellobiose. Cellotriose, cellobiose, glucose, and cellotetraose are produced from cellopentaose. Cellohexaose yields cellotriose, cellobiose, and glucose, larger products presumably being completely hydrolyzed because of the long incubation times and high enzyme concentrations used here.
FIG. 5.
Thin-layer chromatography of hydrolysis products when enzyme was incubated with 10 g/liter cellooligosaccharides or 10 g/liter various polysaccharides in 0.1 M NaOAc buffer, pH 5.0, at 25°C for 16 h. (A) Left-hand lane: glucose, cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose standards (from top to bottom). Lanes 2 to 11: cellobiose control (without enzyme), cellobiose incubation (with enzyme), cellotriose control, cellotriose incubation, cellotetraose control, cellotetraose incubation, cellopentaose control, cellopentaose incubation, cellohexaose control, and cellohexaose incubation, respectively. (B) Left- and right-hand lanes: glucose, cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose standards (from top to bottom). Lanes 2 to 11: CMC control (without enzyme), CMC incubation, lichenan control, lichenan incubation, birch wood xylan control, birch wood xylan incubation, larch wood xylan control, larch wood xylan incubation, Avicel control and Avicel incubation, respectively.
C. acetobutylicum EG attacks CMC, birch wood and larch wood xylan, lichenan, and to a limited extent, Avicel (Fig. 5B), but not laminaran, mannan, and pullulan (data not shown). The hydrolysis products of CMC, birch wood xylan, and larch wood xylan are mainly mono- to tetrasaccharides, while those of lichenan are in general longer. Avicel yields small amounts of cellobiose, cellotriose, and cellotetraose.
Enzyme kinetic and thermostability properties.
Enzyme activity on CMC and the two xylans increases with increasing substrate concentrations (see Fig. S2 in the supplemental material), leading to the kinetic values shown in Table 2. Activity decreases at high larch wood xylan concentrations, perhaps because of an inhibitory material in the xylan. It is noteworthy that this EG has higher kcat values on the two xylans than on CMC, even though most characterized GH44 members are classified as either EGs or xyloglucanases.
TABLE 2.
Kinetic constants of C. acetobutylicum GH44 endoglucanase on different substrates in 0.1 M NaOAc buffer at pH 5.0
| Substrate | kcat (s−1)a | Km (g/liter)a | kcat/Km (liter/g·s) |
|---|---|---|---|
| CMC | 18.9 ± 0.5 | 0.263 ± 0.027 | 65 |
| Birch wood xylan | 31.6 ± 1.0 | 0.412 ± 0.045 | 77 |
| Larch wood xylan | 29.5 ± 1.7 | 0.278 ± 0.035 | 106 |
Values are means ± standard errors.
The enzyme has an optimal temperature near 55°C on CMC in a 10-min assay at pH 5 and has an activation energy for activity of 26.9 ± 3.0 kJ/mol, where the second value is the standard error (see Fig. S3 in the supplemental material). It has an optimal pH on CMC of 5.0 (see Fig. S4 in the supplemental material). The thermostability of the enzyme at pH 7 is shown in Fig. S5 in the supplemental material. The activation energy of thermoinactivation is 230 ± 42 kJ/mol, much higher than the activation energy for activity, as is expected.
DISCUSSION
C. acetobutylicum GH44 EG has been isolated for the first time. It is the third and most thoroughly characterized EG from this organism, after work by Zappe et al. (33) on an EG from an unidentified GH family and by López-Contreras et al. (20) on a GH9 EG.
The similarity in tertiary structures of C. acetobutylicum and C. thermocellum EGs and their more significant differences from the crystal structure of CelM2 is not unexpected, considering the proximity of the first two enzymes on the GH44 phylogenetic tree and their distance from the third (Fig. 2).
C. acetobutylicum EG is active on a variety of β-1,4-linked glucans, with somewhat higher activity on xylans than on CMC, the opposite of the case with C. thermocellum EG. Both studies used low-viscosity CMC from Sigma. The differences in relative activity on xylan versus CMC for these two enzymes may be due to the use of oat spelt xylan in the C. thermocellum EG study, whereas birch wood and larch wood xylans were used in this study. The impact of plant source on differences in xylanase activity is a more likely explanation than structural differences between the enzymes, given the low RMSD between their structures and the lack of obvious structural differences in their active sites.
Activity on CMC and xylan is consistent with the crystal structure of C. acetobutylicum EG, where a broad binding cleft allows entry of bulky side chains. The unbalanced nature of the products of cellotetraose hydrolysis can be explained by subsites to one side of the cleavage point having a higher affinity for substrate residues than those on the other side. If this is the case, then subsites −4 to −1 should bind substrate residues more strongly than subsites +1 to +5, since (i) Kitago et al. (15) found the hydrolysis product cellotetraose in subsites −4 to −1 when the crystals of the closely related C. thermocellum wild-type EG had been soaked with longer substrates and (ii) C. acetobutylicum EG subsites +1 to +3 lack any amino acid residues that can hydrogen-bind substrates, while Trp58 in subsite −4; Arg41 and Tyr65 in subsite −3; and Asn40, Glu352, and Trp385 in subsite −1 can do so.
The cellooligosaccharide hydrolysis results (Fig. 5A) indicate that C. acetobutylicum EG reacts faster on longer substrates than on shorter ones, as no cellopentaose and cellohexaose but some cellotetraose remain when they are hydrolyzed over long periods by a very high concentration of enzyme. Furthermore, the enzyme does not attack cellobiose and cellotriose at all. This behavior is caused by the progressive loss of ability, due to a progressively less negative binding free energy, of the enzyme to bind substrates as their chain length decreases. This is a common trait of endo-acting enzymes like EGs, which have long active sites with many subsites binding carbohydrate residues, most with negative binding energies.
Inactivity against mannan is likely due to an inability of the catalytic nucleophile to hydrogen bond with the 2-OH group of the mannopyranosyl ring in subsite −1, therefore leaving it in a 4C1 nondistorted conformation. Inactivity on laminarin is due to the substrate's β-1,3-glycosidic bonds, which would require the hydrogen-bond donors in the binding cleft to be on opposite, equatorial sides of the residue in subsite −1 to be able to distort it.
C. acetobutylicum GH44 EG is optimally active on CMC at pH 5.0 and 55°C. This is comparable to the optimal pH of 5.2 observed for the extracellular unidentified EGs produced by C. acetobutylicum (18). Its optimal temperature is slightly higher than the 50°C optimal temperature of the C. acetobutylicum EG from an unknown GH family studied by Zappe et al. (33) and is much lower than the 70°C optimal temperature of C. thermocellum GH44 EG (1). Although the activation energy of thermoinactivation was not determined for the latter enzyme, it is stable for 10 min up to 80°C, whereas C. acetobutylicum EG is stable for <2 min at 60°C.
The causes of high enzyme thermostability include a more charged surface, higher aliphaticity, and higher hydrophobicity (11). C. thermocellum EG has a higher aliphatic index than C. acetobutylicum EG with the His tag attached, 73.2 versus 61.5, calculated by ProtParam (6; http://www.expasy.org/tools/protparam.html). It also has a less negative grand average of hydrophobicity (GRAVY) score (16) than C. acetobutylicum EG, −0.495 versus −0.683, meaning that it is more hydrophobic. Furthermore, it has a more acidic surface, which potentially would be more charged. Thus, the difference in thermostability of these two enzymes agrees with the results of previous work correlating differences in structural features of thermophiles and mesophiles to thermostability.
The activity of the purified C. acetobutylicum cellulosome against CMC, 0.115 U/mg (28), is about 0.6% of the activity of the GH44 C. acetobutylicum EG against CMC, 18.9 s−1 (Table 2) or 20.0 U/mg, these values measured at a lower temperature and pH. C. acetobutylicum ATCC 824 Cel9G, a GH9 EG, whose encoding gene was found in the same cellulosomal gene cluster as the gene encoding the GH44 EG studied here (25), has a specific activity of 7.4 U/mg against CMC (20). The experiments were conducted at different pHs, and the cellulosome does contain noncatalytic proteins, which may contribute to some of its lower observed activity, since activity is based upon mass instead of molarity. However, the synergistic increase in activity of a cellulosome due to the proximity effect should offset much or all of a decrease in activity due to noncatalytic proteins. Another likely contributing factor was the use of cellobiose as the carbon source to produce the C. acetobutylicum cellulosome by Sabathé et al. (28), as they were unable to grow the organism on cellulose. Other cellulosomes had higher cellulase activity when the production organism was grown on cellulose (2, 3, 17). This effect is likely due to a difference in cellulosome enzymatic composition caused by a different carbon source.
The dramatically higher activities of C. acetobutylicum Cel9G and GH44 EGs than that of its cellulosome against CMC would suggest that these enzymes were absent from the cellulosome when its activity was characterized. However, two bands on an SDS-PAGE gel of the cellulosome components, not identified by N-terminal sequencing (28), are the correct molecular masses for C. acetobutylicum GH44 EG, 66 kDa, and Cel9G, 76 kDa. Furthermore, the genes encoding these proteins, CAC0915 and CAC0916, are both present in the cellulosomal gene cluster, and CAC0915 is the only gene in the cluster that produces a protein with a molecular mass between 60 and 76 kDa. It is therefore likely that both enzymes were incorporated into the cellulosome. Thus, the difference in CMCase activity is not due to the absence of these two EGs from the cellulosome, and the differences in reaction conditions, substrate source, and/or enzymatic composition of the cellulosome seem to be incomplete explanations for the discrepancy in activities. The fact that the recombinant forms of these two EGs are so much more active on CMC than the cellulosome in which they are normally found suggests that the latter might be engineered to yield higher activities or that different conditions may activate it. Two other possibilities are (i) that the cellulosome components in the Sabathé et al. study (28) were improperly folded and (ii) that the absence of a signal peptide in our enzyme led to an increase in its activity.
In conclusion, GH44 C. acetobutylicum EG has been produced, purified, and characterized, and its crystal structure has been solved. This is the first experimental work ever reported on this enzyme from this source. It is phylogenetically similar to other EGs produced by Clostridium species (Fig. 2), although, despite close similarity in amino acid sequences and crystal structures, differences in relative activities, pH and temperature optima, and thermostabilities have still occurred. It is active on cellotetraose and longer cellooligosaccharides, soluble cellulose, xylan, and lichenan, and slightly active on crystalline cellulose.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the U.S. Department of Agriculture through the Biotechnology Byproducts Consortium and through the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant number 2007-35504-18252. The Iowa State University Macromolecular X-ray Crystallography Facility is supported by the Office of Biotechnology, the College of Agriculture and Life Sciences, the College of Liberal Arts and Sciences, and the Plant Sciences Institute.
We thank chemical engineering undergraduates Theresa Russo, Paul Low, Christopher Setina, and Waddah Moghram for their help during this project, and we thank Erica Fuchs for her outstanding work in keeping the laboratory functioning.
Footnotes
Published ahead of print on 13 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahsan, M. M., M. Matsumoto, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Purification and characterization of the family J catalytic domain derived from the Clostridium thermocellum endoglucanase CelJ. Biosci. Biotechnol. Biochem. 61:427-431. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat, K. M., P. W. Goodenough, E. Owen, and T. M. Wood. 1993. Cellobiose: a true inducer of cellulosome in different strains of Clostridium thermocellum. FEMS Microbiol. Lett. 111:73-78. [Google Scholar]
- 4.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 6.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonnet, G. H., M. A. Cohen, and S. A. Benner. 1992. Exhaustive matching of the entire protein sequence database. Science 256:1443-1445. [DOI] [PubMed] [Google Scholar]
- 8.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 9.Gustafsson, C., S. Govindarajan, and J. Minshull. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22:346-353. [DOI] [PubMed] [Google Scholar]
- 10.Han, N. S., and J. F. Robyt. 1998. Separation and detection of sugars and alditols on thin layer chromatograms. Carbohydr. Res. 313:135-137. [Google Scholar]
- 11.Haney, P. J., J. H. Badger, G. L. Buldak, C. I. Reich, C. R. Woese, and G. J. Olsen. 1999. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. U. S. A. 96:3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 13.Jue, C., and P. N. Lipke. 1985. Determination of reducing sugars in the nanomole range with tetrazolium blue. J. Biochem. Biophys. Methods 11:109-115. [DOI] [PubMed] [Google Scholar]
- 14.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 15.Kitago, Y., S. Karita, N. Watanabe, M. Kamiya, T. Aizawa, K. Sakka, and I. Tanaka. 2007. Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum. J. Biol. Chem. 282:35703-35711. [DOI] [PubMed] [Google Scholar]
- 16.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydrophatic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 17.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Technol. 7:37-41. [Google Scholar]
- 18.Lee, S. F., C. W. Forsberg, and L. N. Gibbins. 1985. Cellulolytic activity of Clostridium acetobutylicum. Appl. Environ. Microbiol. 50:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S. F., C. W. Forsberg, and L. N. Gibbins. 1985. Xylanolytic activity of Clostridium acetobutylicum. Appl. Environ. Microbiol. 50:1068-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-Contreras, A. M., A. A. Martens, N. Szijarto, H. Mooibroek, P. A. M. Claassen, J. van der Oost, and W. M. de Vos. 2003. Production by Clostridium acetobutylicum ATCC 824 of CelG, a cellulosomal glycoside hydrolase belonging to family 9. Appl. Environ. Microbiol. 69:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makoff, A. J., M. D. Oxer, M. A. Romanos, N. F. Fairweather, and S. Ballantine. 1989. Expression of tetanus toxin fragment C in E. coli: high level expression by removing rare codons. Nucleic Acids Res. 17:10191-10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 23.Nam, K. H., S.-J. Kim, and K. Y. Hwang. 2009. Crystal structure of CelM2, a bifunctional glucanase-xylanase protein from a metagenomic library. Biochem. Biophys. Res. Commun. 383:183-186. [DOI] [PubMed] [Google Scholar]
- 24.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 25.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsy, C. B., C. J. Chapman, A. C. Barnes, J. F. Robertson, and A. Murray. 2007. Two-step method to isolate target recombinant protein from co-purified bacterial contaminant SlyD after immobilised metal affinity chromatography. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 853:314-319. [DOI] [PubMed] [Google Scholar]
- 27.Pflugrath, J. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55:1718-1725. [DOI] [PubMed] [Google Scholar]
- 28.Sabathé, F., A. Bélaïch, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz, W. H., V. V. Zverlov, and H. Bahl. 2004. Extracellular glycosyl hydrolases from clostridia. Adv. Appl. Microbiol. 56:215-261. [DOI] [PubMed] [Google Scholar]
- 30.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 31.Studier, F. W. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed]
- 33.Zappe, H., D. T. Jones, and D. R. Woods. 1986. Cloning and expression of Clostridium acetobutylicum endoglucanase, cellobiase and amino acid biosynthesis genes in Escherichia coli. J. Gen. Microbiol. 132:1367-1372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.