Abstract
Listeria monocytogenes is a food-borne human pathogen that causes listeriosis, a relatively rare infection with a high fatality rate. The regulation of virulence gene expression is influenced by several environmental factors, and the aim of the present study was to determine how disinfectants used routinely in the food industry affect the expression of different virulence genes in L. monocytogenes when added at sublethal concentrations. An agar-based assay was developed to screen the effect of disinfectants on virulence gene promoter expression and was validated at the transcriptional level by Northern blot analysis. Eleven disinfectants representing four different groups of active components were evaluated in this study. Disinfectants with the same active ingredients had a similar effect on gene expression. Peroxy and chlorine compounds reduced the expression of the virulence genes, and quaternary ammonium compounds (QAC) induced the expression of the virulence genes. In general, a disinfectant had similar effects on the expression of all four virulence genes examined. Northern blot analyses confirmed the downregulation of prfA and inlA expression by Incimaxx DES (a peroxy compound) and their upregulation by Triquart Super (a QAC) in L. monocytogenes EGD. Hence, sublethal concentrations of disinfectants routinely used in the food industry affect virulence gene expression in the human pathogen L. monocytogenes, and the effect depends on the active components of the disinfectant. From a practical perspective, the study underlines that disinfectants should be used at the lethal concentrations recommended by the manufacturers. Further studies are needed to elucidate whether the changes in virulence gene expression induced by the disinfectants have impact on virulence or other biological properties, such as antibiotic resistance.
Listeria monocytogenes is a food-borne, facultative intracellular pathogen that can cause invasive listeriosis in immunocompromised individuals, pregnant women, infants, and the elderly. The disease is relatively rare (0.3 cases per year per 100,000 inhabitants in the European Union), but it is associated with a high fatality rate (25 to 30%) (11).
L. monocytogenes can cross the intestinal barrier, the blood-brain barrier, and the placental barrier, and it invades and replicates in both phagocytic and nonphagocytic cells (47). The ability to invade and to survive and multiply within eukaryotic cells is determined by a number of chromosomal genes, most of which are found on the 9-kb pathogenic island known as Listeria pathogenicity island 1 (LIPI-1) but also on other places on the chromosome. These virulence genes encode products involved in adherence to and internalization by the host cell (inlA, inlB, and inlC), escape from the vacuoles (hly, plcA, and plcB), intracellular replication (htp), and cellular movement (actA). The major transcriptional factor which regulates the expression of these virulence genes, including its own transcription, is the 27-kDa polypeptide PrfA (5, 8, 12, 18, 27, 32).
The regulation of virulence gene expression by the PrfA protein is dependent on the concentration and activity of the protein but also on the configuration of the virulence gene promoters. The PrfA protein facilitates specific binding to its target site, the so-called PrfA box, which partially overlaps the promoter region, and affinity is weakened when this target sequence diverges from the perfect palindromic sequence (17, 44, 49).
The regulation of PrfA and virulence gene expression is influenced by several environmental factors. One example is the temperature-dependent control of translation of the prfA messenger, which is processed only at 37°C and not at 30°C (22, 26). Another example is the repression of virulence gene expression in response to high concentrations of fermentable carbohydrates (35), and a third example is the induction of virulence gene expression observed when L. monocytogenes is grown with activated charcoal, due to the absorption of an autorepressor (13, 40). Less-well-described physicochemical parameters also affect virulence gene expression. Nutrient stress conditions limiting growth induced gene expression of prfA, plcA, hly, and actA (3, 45), and high osmolarity induced expression of hly (37). On the other hand, growth at pH 6 to 6.8 repressed hly expression (2).
L. monocytogenes is frequently detected in the food processing environment, where it has a remarkable ability to persist (7, 29, 34, 38, 50). An efficient cleaning and disinfection process is essential in preventing contamination of food products with L. monocytogenes during processing. However, the disinfection process is not always adequately performed, or organic debris may inactivate the disinfectant; hence, the bacteria may be exposed to only sublethal concentrations and survive.
L. monocytogenes that resides in food processing environments may adapt to disinfectants after repeated exposure (1, 46); however, few studies have investigated how low concentrations of disinfectants affect the physiology of L. monocytogenes at the gene expression level. Recently, we found that exposure of L. monocytogenes to sublethal concentrations of a disinfectant stressed the cell, measured as a decrease in intracellular pH (24), and Ryan et al. (42) found that the sigB gene was upregulated in the presence of quaternary ammonium compounds (QAC) and sodium dodecyl sulfate (SDS), which are common components of industrial cleaning agents. Hence, an element in assessing the risk of L. monocytogenes contaminating a food product is to determine if food environment stress factors such as sublethal concentrations of disinfectant affect expression of virulence genes in L. monocytogenes.
The aim of the present study was to determine how disinfectants used routinely in the food industry affect the expression of different virulence genes in L. monocytogenes when added at sublethal concentrations. An agar-based assay was developed and applied for screening the effect of disinfectants on virulence gene promoter expression and was validated at the transcriptional level by Northern blot analysis.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Listeria monocytogenes EGD and the ΔprfA mutant were obtained from Werner Goebel (Biozentrum, University of Würzburg). Strains of L. monocytogenes EGD with lacZ fusions to the hly, plcA, prfA, and inlA promoters in the vector pTCV-lac were from the laboratory collection at the Department of Veterinary Disease Biology, Faculty of Life Science, University of Copenhagen (25). Stock cultures were stored at −80°C in 15% (wt/vol) glycerol. The bacteria were cultivated on brain heart infusion (BHI) agar (CM0225 [Oxoid, Basingstoke, United Kingdom] supplemented with 1.5% agar) at 37°C for 24 to 48 h. One colony was cultured in BHI broth and incubated with shaking (180 rpm) at 37°C overnight, diluted 1,000-fold, and grown at 37°C and 180 rpm for 18 h. Kanamycin was added to growth media for the strains of L. monocytogenes EGD with lacZ fusions at a final concentration of 50 μg/ml.
Preparation of disinfectant solutions.
Eleven disinfectants commonly used in the food industry were tested in this study (Table 1). The disinfectants represent four different groups of active compounds. Disinfectants were prepared in 2-fold dilutions in sterilized, demineralized water to obtain concentrations at which inhibition zones with appropriate sizes (i.e., zones did not merge) were seen in the agar assay (described below).
TABLE 1.
Effect of different disinfectants on expression of different virulence genes of Listeria monocytogenes
| Type of disinfectant | Designation (pH) | Company | Concn (%) | Regulation of gene fusiona |
||||
|---|---|---|---|---|---|---|---|---|
| prfA-lacZ | plcA-lacZ | inlA-lacZ | ΔprfA inlA-lacZ | hly-lacZ | ||||
| Peroxy compounds | Incimaxx DES (acid) | Ecolab, Valby, Denmark | 1.25-40 | − | − | − | − | − |
| DES foam PAA (acid) | ITW Novadan APS, Kolding, Denmark | 0.31-10 | −/+ | −/+ | −/+ | −/+ | −/+ | |
| Chlorine | P3-hypochloran (alkaline) | Ecolab | 3.13-100 | − | − | − | − | − |
| Desinfect CL (alkaline) | ITW Novadan APS | 3.13-100 | − | − | −/+ | −/+ | − | |
| Chlorrengøring (alkaline) | Samson Enviro Industries, Kvistgård, Denmark | 3.13-100 | − | − | − | − | − | |
| Triclosan, ethanol | Overfladedesinfektion (neutral) | Pro-ren A/S, Holbaek, Denmark | 0.19-6 | + | None | + | + | + |
| Quaternary ammonium compounds | Triquart Super (alkaline) | Ecolab | 0.0063-0.2 | + | + | + | + | + |
| Multidesinfektion (neutral) | Samson Enviro Industries | 0.0063-0.2 | + | + | + | + | + | |
| Desinfect Maxi (neutral) | ITW Novadan APS | 0.0063-0.2 | + | + | + | + | + | |
| Desinfect Ultra (acid) | ITW Novadan APS | 0.0063-0.2 | + | + | + | + | + | |
| Desinfect Alka (alkaline) | ITW Novadan APS | 0.0063-0.2 | + | + | + | + | + | |
+, induction of expression of the virulence gene; −, reduction of expression of the virulence gene; −/+, reduction followed by induction of expression of the virulence gene; none, neither induction nor reduction of expression of the virulence gene.
Agar-based screening assay.
An agar-based assay was developed on the basis of an existing assay for Staphylococcus aureus (A. Nielsen, K. F. Nielsen, T. O. Larsen and H. Ingmer, unpublished data). BHI agar and charcoal-supplemented BHI (BHI-AC) (BHI with addition of 0.013% or 0.025% activated charcoal) was melted and tempered to 44°C. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (150 μg/ml) and kanamycin (50 μg/ml) were added. One milliliter of outgrown bacterial culture (i.e., grown overnight, diluted 1,000-fold, and grown for 18 h) with a promoter-lacZ fusion, diluted 1,000-fold in 0.9% NaCl, was mixed thoroughly with 25 ml of medium in a petri dish (diameter, 9 cm), and the plates were dried in a laminar air flow (LAF) bench for 45 min. Seven wells (4-mm diameter) were made in each plate with a sterilized drill, and 30 μl of disinfectant at different concentrations was added to each well. Water was used as control. The plates were incubated at 37°C for 48 h. Each disinfectant was tested in at least two independent experiments with all strains containing promoter-lacZ fusions. In each experiment, all strains were screened in BHI and BHI-AC. Strains containing plasmids pprfA-lacZ, phly-lacZ, and pinlA-lacZ were screened in 0.013% BHI-AC, while the strain containing plasmid pplcA-lacZ were screened in 0.025% BHI-AC in order to further induce promoter activity (see below).
Construction of promoter-lacZ fusions in a ΔprfA strain.
To study if the responses of the different virulence gene promoters were influenced by the PrfA protein, the lacZ fusions were transformed into the ΔprfA L. monocytogenes EGD strain. Listerial plasmid DNA was purified from strains containing plasmid phly-lacZ, pinlA-lacZ, or pplcA-lacZ using the Qiaminiprep kit (Qiagen). The plasmid isolation procedure was modified by incubating the cell suspension in p1 buffer containing lysozyme (9 mg/ml) for 1 h at 37°C. Competent ΔprfA cells were prepared as described by Park and Stewart (39) with slight modifications. Briefly, 3 ml of an overnight culture was cultured in 150 ml BHI with 0.5 M sucrose at 37°C with shaking (200 rpm) until an optical density at 600 nm (OD600) of 0.2 was reached. Penicillin G (10 μg/ml) was added and the incubation continued for a further 2 h (100 rpm). Cells were harvested (3000 × g, 10 min, 4°C) and washed three times with sucrose electroporation buffer (1 mM HEPES [pH 7.0], 0.5 M sucrose). The final cell pellet was resuspended in 500 μl ice-cold glycerol solution (1 mM HEPES, 0.5 M sucrose, 15% glycerol) and stored at −80°C prior to use. Cells (40 μl) and 1 μl plasmid DNA were mixed, and electroporation was performed (25 μF, 2.5 kV, 200 Ω). After the pulse, 1 ml prewarmed BHI (37°C) was added, mixed with the cells, and transferred to 15 ml BHI. After 3 h of incubation at 30°C with shaking (150 rpm), cells were spread on BHI plates with kanamycin (5 μg/ml).
RNA extraction and Northern hybridization.
An outgrown culture (i.e., grown overnight, diluted 1.000-fold, and grown for 18 h) of L. monocytogenes EGD was diluted 100-fold in BHI and grown to an OD600 of 0.4. The culture was split and centrifuged at 3,000 × g for 10 min, and the pellet was resuspended in BHI-AC broth (0.2% charcoal) with 0.25% or 0.125% Incimaxx DES or in BHI broth with 0.0031% or 0.0016% Triquart Super. The bacterial cells were resuspended to an OD600 of 0.4, and 60 ml of each culture was transferred to a 300-ml Erlenmeyer flask. Demineralized water was used for the control cultures. Cells were grown at 37°C with shaking (130 rpm). Samples were taken after 15, 30, 60, and 180 min of growth with the disinfectant for RNA extraction. RNA was stabilized by the addition of 2 volumes RNA Protect (Qiagen), and bacterial cells were harvested by centrifugation at 5,000 × g for 10 min and stored at −80°C. Bacterial numbers were determined by surface plating on BHI agar plates after 0, 60, and 180 min of growth. BHI agar plates were incubated at 37°C overnight. To extract RNA, the bacterial cells were lysed using the Fast Prep FP120 instrument (BIO101, ThermoSavent) for 45 s at speed 6. The cells were kept on ice for 1 min and then lysed again. The treatment and cooling on ice were repeated three times. Total RNA was extracted from the cells using the RNeasy mini kit (Qiagen) according to the manufacturer's directions. Analysis of transcripts was performed as described by Frees et al. (15) with slight modifications. Briefly, total RNA was quantified with a Nanodrop 2000 (Thermo Fisher Scientific), and 5 μg of RNA of each preparation was loaded onto a 1% agarose gel and separated in 10 mM sodium phosphate buffer. RNA was transferred to a positively charged nylon membrane (Amersham HybondN; GE Health Care) by capillary blotting. Hybridization probes were generated by PCR from chromosomal DNA of L. monocytogenes EGD using specific primers for the prfA gene (1F, 5′TAA CCA ATG GGA TCC ACA AG-3′; 1R, 5′TGC TAA CAG CTG AGC TAT GTG-3′) and the inlA gene (1,5F, 5′ATC GAT GGA GTG GAA TAC TT-3′; 1,5R, 5′ GTG CCT ATA TCT TTT AAC TGG TTA C-3′). Probes were labeled with [32P]dCTP using Ready-to-Go DNA-labeling beads (Amersham Biosciences). RNA from at least two independent experiments was analyzed.
RESULTS
Screening of disinfectants in agar assay.
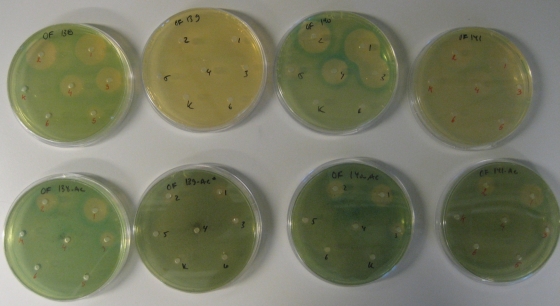
To determine how different disinfectants affect the expression of virulence genes in L. monocytogenes, we designed an agar-based screening assay in which virulence gene expression is monitored as β-galactosidase production from L. monocytogenes derivatives carrying lacZ fused to the promoters of each of the known virulence factor genes hly, plcA, prfA, and inlA (25). In order to be able to monitor changes in virulence gene expression, we chose to use a growth medium that supports a low level of virulence gene expression, as is the case with BHI and BHI-AC. When we incorporated the L. monocytogenes strains with lacZ fusions in the agar plates, a dense growth was observed. The L. monocytogenes prfA-lacZ strain turned a strong blue in BHI or BHI-AC agar plates, indicating that the prfA gene was expressed during growth in BHI and that charcoal did not visually induce the prfA promoter further (Fig. 1, two leftmost plates). The lacZ gene of the inlA- and hly-lacZ fusions was expressed in BHI, but addition of charcoal caused an even more intense blue color, indicating an induction of the promoters by charcoal (Fig. 1, four rightmost plates). In contrast, addition of activated charcoal was necessary to obtain visible lacZ expression of the plcA-lacZ fusion, and the amount of charcoal needed to obtain a sufficient blue color was 2-fold higher than that for the other strains (Fig. 1, two plates in the second column from the left).
FIG. 1.
Listeria monocytogenes with lacZ fusions with pprfA, pplcA, pinlA, and phly promoters (from left to right, respectively) diluted 1,000-fold and cast in BHI (upper row) and BHI-AC (lower row). Overfladedesinfektion was added at decreasing concentrations in wells 1 to 6 from right to left. Water was added as control (K) and did not alter the color of the agar. The results presented are representative of two independent experiments.
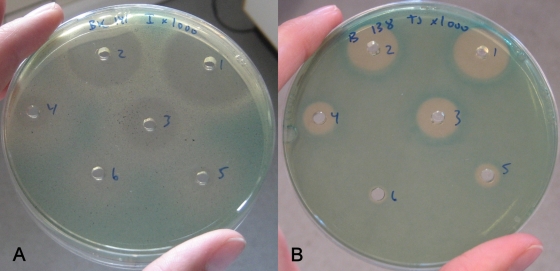
After having confirmed that differences in virulence gene expression could be monitored in agar plates, disinfectants were added to wells formed in the agar, and each of the lacZ fusion strains was examined in BHI and BHI-AC agar. Disinfectants were added at inhibitory concentrations, and for all strains the size of the inhibition zones decreased as expected with decreasing concentration of disinfectant (Fig. 2 and data not shown). Also, each well represents a screening of disinfectant concentrations as the disinfectant dilutes outward from each well. For some disinfectants, white colonies were seen outside of the inhibition zones, indicating that those disinfectants at sublethal concentrations reduced the expression of the virulence gene (Fig. 2A). For other disinfectants, colonies outside the inhibition zones were of a more intense blue color than the colonies further away from the inhibition zones, indicating that those disinfectants at sublethal concentrations induced the expression of the virulence gene (Fig. 2B). No inhibition or effect on gene expression was seen when water was added to the well (control) (Fig. 1).
FIG. 2.
(A) Listeria monocytogenes with a lacZ fusion with the phly promoter diluted 1,000-fold and cast in BHI-AC. Incimaxx DES was added at decreasing concentrations in wells 1 to 6. (B) L. monocytogenes with a lacZ fusion with the pprfA promoter diluted 1,000-fold and cast in BHI. Triquart Super was added at decreasing concentrations in well 1 to 6. The results are representative of two independent experiments.
In total, 11 disinfectants representing four groups of active components were evaluated in the present study (Table 1). They are all commonly used in the food industry and are supplied by different manufacturers. Disinfectants with the same active ingredients in general caused similar effects on gene expression. Peroxy and chlorine compounds reduced the expression of the virulence genes, and quaternary ammonium compounds (QAC) induced the expression of the virulence genes. However, for DES foam PAA, white colonies were seen around the inhibition zone and followed by an intense blue zone, which could indicate that the disinfectant at a sublethal concentration caused a reduction in the expression of the virulence gene but at a lower concentration caused induction of the virulence gene expression.
In general, a disinfectant compound had the same effect on the expression of all four virulence gene promoters (Table 1). However, Desinfect CL caused downregulation of three fusions (prfA, plcA, and hly), but DES foam PAA caused a combined down- and upregulation of inlA. Similarly, the compound Overfladedesinfektion induced only three promoters (prfA, inlA, and hly) and had no visible effect on the plcA promoter.
Screening of ΔprfA lacZ fusion strains.
Since many of the virulence genes in L. monocytogenes are controlled by PrfA, we determined if the effect of the different disinfectants on virulence gene expression was dependent on PrfA. To study this, the plasmids containing the reporter gene fusions (pplcA-lacZ, pinlA-lacZ, and phly-lacZ) were transformed into the prfA deletion mutant.
The three ΔprfA promoter-lacZ transformants were all screened in the agar assay. Expression of inlA was induced in BHI, and addition of charcoal increased also the expression in the absence of PrfA. However, there was no visible expression of plcA or hly in the prfA deletion strains during growth in BHI or BHI-AC, nor could growth of the two strains in diluted BHI (1:2, 1:4, and 1:8) agar (1.5%) or diluted tryptic soy (1:7) agar (1.5%) with or without 0.013% charcoal induce the promoters. Hence, only the ΔprfA pinlA-lacZ strain could be screened with the disinfectants in our agar assay.
The influence of the 11 disinfectants on gene expression in the ΔprfA pinlA-lacZ strain was determined. All disinfectants had the same effect on this strain as on the wild-type pinlA-lacZ strain, which contains the prfA gene (Table 1). When comparing the intensity of the background color as well as the blue induction zones or the white reduction zones, no clear difference was seen between the ΔprfA pinlA-lacZ strain and the pinlA-lacZ strain. This indicates that the up- or downregulation of the inlA promoter by disinfectants was not primarily caused by PrfA.
Northern blot analysis of virulence gene expression in the presence of disinfectant.
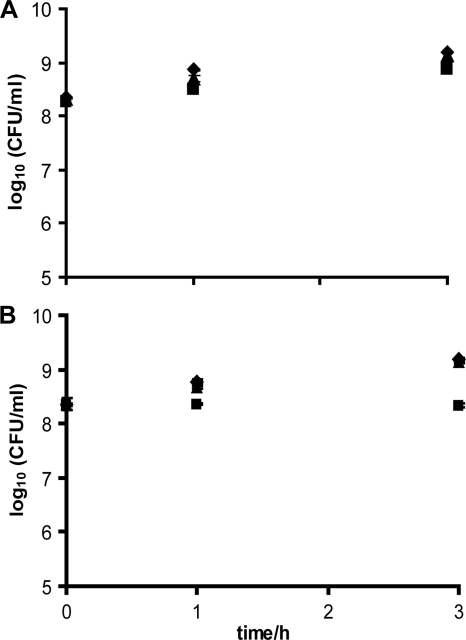
To study whether the effect of disinfectants on the virulence gene promoter was indeed a true gene induction or repression effect, we determined the effect of sublethal concentrations of disinfectants on the transcript levels of the prfA and inlA genes. Northern blot analyses were performed with the wild-type L. monocytogenes EGD and the two disinfectants Incimaxx DES and Triquart Super, which down- and upregulate gene expression in the agar assay, respectively. Both disinfectants were tested at sublethal and noninhibiting concentrations (Fig. 3) to ensure that the growth rates were similar so the total RNA concentrations per bacterial cell were comparable.
FIG. 3.
(A) Growth of Listeria monocytogenes EGD in BHI-AC (0.2% charcoal) broth with water (control) (⧫) or 0.25% (▪) or 0.125% (▴) Incimaxx DES. (B) Growth of Listeria monocytogenes EGD in BHI broth with water (control) (⧫) or 0.0031% (▪) or 0.0016% (▴) Triquart Super. The results are representative of two independent experiments.
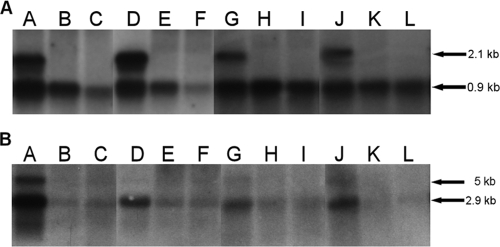
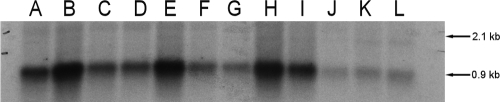
A marked downregulation of transcript levels was seen for prfA and inlA compared to EGD grown with water when L. monocytogenes EGD was grown with 0.25% and 0.125% Incimaxx DES (Fig. 4). The plcA-prfA transcript of 2.1 kb was downregulated at 15, 30, 60, and 180 min of growth with both 0.125% (lanes B, E, H, and K) and 0.250% (lanes C, F, I, and L) Incimaxx DES compared to the control (lanes A, D, G, and J) (Fig. 4A). Similarly, the monocistronic prfA transcripts of 0.8 and 0.9 kb were downregulated compared to controls with water. The downregulation was more pronounced in trials with 0.25% Incimaxx DES than in those with 0.125% Incimaxx DES. Both inlA (2.9 kb) and inlAB (5 kb) transcripts were detected after 15 min of growth in the control, whereas only the inlA transcript was detected during further growth (Fig. 4B). However, only very weak transcripts of inlA were detected in L. monocytogenes EGD grown with both concentrations of Incimaxx DES.
FIG. 4.
Listeria monocytogenes prfA (A) and inlA (B) transcription measured by Northern blotting using RNA isolated from L. monocytogenes EGD grown in BHI-AC (0.2% charcoal) for 15 min (lanes A, B, and C), 30 min (lanes D, E, and F), 60 min (lanes G, H, and I), or 180 min (lanes J, K, and L) with water (control) (lanes A, D, G, and J) or 0.125% (lanes B, E, H, and K) or 0.250% (lanes C, F, I, and L) Incimaxx DES. The arrows show the plcA-prfA (2.1-kb), prfA (0.8- and 0.9-kb), inlAB (5-kb), and inlA (2.9-kb) transcripts. The results are representative of two independent experiments.
Upregulation of prfA and inlA in L. monocytogenes EGD grown in BHI with Triquart Super was seen on the transcript level with the Northern blot analysis (Fig. 5 and data not shown). The monocistronic prfA transcripts were upregulated after 15, 30, and 60 min of growth with 0.0031% Triquart Super (lanes B, E, and H) compared to the control (lane A, D, and G). Growth with 0.0016% Triquart Super also upregulated prfA after 60 min. The transcript at 2.1 kb was only weakly expressed, since L. monocytogenes was grown in BHI broth without addition of charcoal. Only a weakly monocistronic inlA transcript was detected. However induction could be seen after 30 min of growth with 0.0031% Triquart Super (data not shown).
FIG. 5.
prfA transcription measured by Northern blotting using RNA isolated from Listeria monocytogenes EGD grown for 15 min (lanes A, B, and C), 30 min (lanes D, E, and F), 60 min (lanes G, H, and I), or 180 min (lanes J, K, and L) with water (control) (lanes A, D, G, and J) or 0.0031% (lanes B, E, H, and K) or 0.0016% (lanes C, F, I, and L) Triquart Super. The arrows show the sizes of the plcA-prfA (2.1-kb) and prfA (0.8- and 0.9-kb) transcripts. The results are representative of two independent experiments.
Together, the Northern blot analyses confirmed the observations obtained with the agar assay.
DISCUSSION
In the present study, we have developed an agar-based assay based on an existing assay intended to isolate fungal compounds that reduce virulence gene expression in S. aureus (Nielsen et al., unpublished data). Similarly, our assay can be used to study how different compounds affect virulence gene expression in L. monocytogenes. In the screening assay, the expression is determined as β-galactosidase activity of virulence gene promoters fused to a lacZ gene. However, this relates very well to the transcript level as confirmed by Northern blot analyses. The assay has high sensitivity, since it detects differences in expression on the transcript level, but it is not quantitative. In the present study, we used the assay to determine the effects of different disinfectants routinely used in the food industry on the gene expression of four virulence genes, but the assay could also be used for screening of other compounds and their influence on other (virulence) genes in L. monocytogenes.
In general, disinfectants with the same active ingredients had similar effects on virulence gene expression. Peroxy and chlorine compounds reduced the virulence gene expression, and QAC induced the expression of the virulence. Ryan et al. (42) found that two QAC, benzalkoniumchloride (BC) and cetylpyridinium chloride (CPC), at sublethal concentrations, induced expression of sigB in L. monocytogenes as measured by reverse transcriptase PCR; hence, QAC may affect a number of bacterial behaviors. No housekeeping genes were included in our study. However, we find it unlikely that QAC induce the expression of all genes in L. monocytogenes, as BC and CPC did not induce 16S RNA in the study by Ryan et al. (42). The difference in the effect on gene expression is not due to differences in pH per se, since disinfectants with peroxy compounds have a low pH, whereas the disinfectants with chlorine and QAC compounds generally have a high pH. Others have determined the effect of subinhibitory concentrations of commercial disinfectants on virulence in other pathogens. Disinfectants including QAC reduced virulence factors in Pseudomonas aeruginosa and Streptococcus agalactiae when measured on the phenotypic level (19, 20, 30), benzyl alcohol induced expression of an operon involved in biofilm formation in Staphylococcus epidermidis, and peracetic acid induced some and downregulated other virulence-related genes in S. aureus when studied by transcriptomics (6, 36).
PrfA is transcribed both as monocistronic prfA from its own two promoters and as a prfA-plcA bicistron from pplcA (4, 16, 32). Basal amounts of PrfA are made from the monocistronic prfA transcripts, and these stimulate the transcription of the prfA-plcA bicistron. In our Northern blot analysis, no transcript was detected at 2.1 kb in trials with Incimaxx DES and when L. monocytogenes EGD was grown in BHI, which could be due to a too-low basal amount of monocistronic prfA. The increase caused by Triquart Super was not sufficient to induce the transcription of the prfA-plcA bicistron. Interestingly, the effect on promoter activity and transcription level indicates that the disinfectants, even at levels not inhibiting growth, interact with the transcription process directly or indirectly. This could be through interaction with the promoter region or the RNA polymerase. The disinfectant also could act in the allosteric transition of PrfA (e.g., by interacting with the so-far-unidentified cofactor) that is necessary for the protein to stimulate the virulence promoters (41, 48, 49).
It is well known that PrfA regulates the expression of several virulence genes in L. monocytogenes, including hly, inlA, plcA, and prfA. However, activation by PrfA is more efficient at promoters which posses a perfectly symmetrical PrfA box, such as pplcA or phly, than at promoters which have substitutions, such as pinlAB (44). Also, some virulence genes have more than one promoter. plcA is transcribed from one PrfA-dependent promoter, and hly and inlA are transcribed from three and four promoters, respectively. Two hly promoters are PrfA dependent and one is PrfA independent, whereas only one of the inlA promoters is PrfA dependent (9, 10, 28, 33). This might lead to the different dependency on PrfA seen in this study, where only pinlA was sufficiently induced during growth in the ΔprfA strain to produce a visible amount of β-galactosidase. In the screening assay, the presence of PrfA did not visibly affect the effects of the different disinfectants on pinlA. This indicates that the modulation of inlA expression by disinfectants is not dependent on PrfA concentration. However, an interaction of PrfA on pinlA cannot be excluded by this assay.
To our knowledge, this is the first study showing that at sublethal concentrations disinfectants routinely used in the food industry affect virulence gene expression in the human pathogen L. monocytogenes and that the effect depends on the active component in the disinfectant. From a practical perspective, the study underlines that disinfectants should be used at the lethal concentrations recommended by the manufacturers based on data obtained according to accepted disinfectant test standards. Further studies are necessary to elucidate whether these differences have biological importance. One may hypothesize that sublethal concentrations of disinfectants could affect the stress response or virulence of the bacteria. This would be highly relevant in risk analysis of the use of disinfectants, especially in settings where the route of the pathogen from disinfectant exposure to the host is shorter, as it is in food processing settings.
Recently concerns have been expressed about the potential effect of biocide exposure on development of resistance to disinfectants or antibiotics. Resistance to disinfectants is believed to be a relatively rare event because most disinfectants are often complexes of antimicrobials that inactivate multiple cell targets (31). However, sublethal concentrations of disinfectants can select for antibiotic resistance. In Salmonella enterica serovar Typhimurium, a QAC showed the highest selectivity for variants with reduced susceptibility to different antibiotics (23). This variant had increased level of acrB, a marker for efflux, leading to decreased sensitivity to antibiotics. Also, induction of efflux pumps caused by biocides has been detected in other pathogens (14, 21, 43). Similarly, one may speculate that the increase in virulence gene expression detected with some disinfectants in the present study could be generalized to more genes in L. monocytogenes and thereby cause an increase in antibiotic resistance in this bacterium.
Acknowledgments
This work was financed by the European Commission within the VI Framework Program, contract no. 007081, “Pathogen Combat: control and prevention of emerging and future pathogens at cellular and molecular level throughout the food chain.”
Footnotes
Published ahead of print on 6 November 2009.
REFERENCES
- 1.Aase, B., G. Sundheim, S. Langsrud, and L. M. Rørvik. 2000. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62:57-63. [DOI] [PubMed] [Google Scholar]
- 2.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 4.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty, T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, W., F. Toghrol, and W. E. Bentley. 2006. Toxicogenomic response of Staphylococcus aureus to peracetic acid. Environ. Sci. Technol. 40:5124-5131. [DOI] [PubMed] [Google Scholar]
- 7.Chasseignaux, E., M.-T. Toquin, C. Ragimbeau, G. Salvat, P. Colin, and G. Ermel. 2001. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry- and pork-processing plants. J. Appl. Microbiol. 91:888-899. [DOI] [PubMed] [Google Scholar]
- 8.Chico-Calero, I., M. Suárez, B. González-Zorn, M. Scortti, J. Slaghuis, W. Goebel, the European Listeria Genome Consortium, and J. A. Vázquez-Boland. 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U. S. A. 99:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domann, E., J. Wehland, K. Niebuhr, C. Haffner, M. Leimeister-Wächter, and T. Chakraborty. 1993. Detection of a prfA-independent promoter responsible for listeriolysin gene expression in mutant Listeria monocytogenes strains lacking the PrfA regulator. Infect. Immun. 61:3073-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dramsi, S., C. Kocks, C. Forestier, and P. Cossart. 1993. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotrophic activator prfA. Mol. Microbiol. 9:931-941. [DOI] [PubMed] [Google Scholar]
- 11.EFSA. 2009. The community summary report on trends and sources of zoonoses and zoonotic agents in the European Union in 2007. EFSA J. 223:1-141. [Google Scholar]
- 12.Engelbrecht, F., S.-K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 13.Ermolaeva, S., S. Novella, Y. Vega, M.-T. Ripio, M. Scortti, and J. A. Vázquez-Boland. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible repressor. Mol. Microbiol. 52:601-611. [DOI] [PubMed] [Google Scholar]
- 14.Fraud, S., A. J. Campigotto, Z. Chen, and K. Poole. 2008. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob. Agents Chemother. 52:4478-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frees, D., A. Chastanet, S. Qazi, K. Sørensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 16.Freitag, N. E., L. Rong, and G. C. Port. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag, N. E., P. Youngman, and D. A. Portnoy. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard, J.-L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 19.Galice, D. M., C. Bonacorsi, V. C. G. Soares, M. S. G. Raddi, and L. M. de Fonseca. 2006. Effect of subinhibitory concentration of chlorhexidine on Streptococcus agalactiae virulence factor expression. Int. J. Antimicrob. Agents 28:143-146. [DOI] [PubMed] [Google Scholar]
- 20.Hostacká, A. 1997. Influencing virulence factors of Pseudomonas aeruginosa by commercially manufactured disinfectants. Pharmazie 52:317-319. [PubMed] [Google Scholar]
- 21.Huet, A. A., J. L. Raygada, K. Mendiratta, S. M. Seo, and G. W. Kaatz. 2008. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 154:3144-3153. [DOI] [PubMed] [Google Scholar]
- 22.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 23.Karatzas, K. A. G., L. P. Randall, M. Webber, L. J. V. Piddock, T. J. Humphrey, M. J. Woodward, and N. G. Coldham. 2008. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 74:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastbjerg, V. G., D. S. Nielsen, N. Arneborg, and L. Gram. 2009. Response of Listeria monocytogenes to disinfection stress at the single-cell and population levels as monitored by intracellular pH measurements and viable-cell counts. Appl. Environ. Microbiol. 75:4550-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen, M. H., B. H. Kallipolitis, J. N. Christiansen, J. E. Olsen, and H. Ingmer. 2006. The response regulator ResD modulates virulence gene expression in response to carbohydrates in Listeria monocytogenes. Mol. Microbiol. 61:1622-1635. [DOI] [PubMed] [Google Scholar]
- 26.Leimeister-Wächter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundén, J. M., T. J. Autio, A.-M. Sjöberg, and H. J. Korkeala. 2003. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 66:2062-2069. [DOI] [PubMed] [Google Scholar]
- 30.Majtán, V., and L. Majtánova. 2002. Influence of subinhibitory concentrations of disinfectants on hydrophobicity, alginate production, and motility of Pseudomonas aeruginosa. Chem. Pap. 56:345-350. [Google Scholar]
- 31.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vazquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 33.Mengaud, J., M. F. Vicente, and P. Cossart. 1989. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect. Immun. 57:3695-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miettinen, M. K., K. J. Björkroth, and H. J. Korkeala. 1999. Characterization of Listeria monocytogenes from an ice cream plant by serotyping and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 46:187-192. [DOI] [PubMed] [Google Scholar]
- 35.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 36.Milisavljevic, V., L. P. Tran, C. Batmalle, and H. J. Bootsma. 2008. Benzyl alcohol and ethanol can enhance the pathogenic potential of clinical Staphylococcus epidermis strains. Am. J. Infect. Control 36:552-558. [DOI] [PubMed] [Google Scholar]
- 37.Myers, E. R., A. W. Dallmier, and S. E. Martin. 1993. Sodium chloride, potassium chloride, and virulence in Listeria monocytogenes. Appl. Environ. Microbiol. 59:2082-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norton, D. M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, S. F., and G. S. A. B. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 40.Ripio, M.-T., G. Domínguez-Bernal, M. Suárez, K. Brehm, P. Berche, and J. A. Vázquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to change in the extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 41.Ripio, M.-T., G. Domínguez-Bernal, M. Lara, M. Suárez, and J. A. Vázquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan, E. M., C. G. Gahan, and C. Hill. 2008. A significant role for Sigma B in the detergent stress response of Listeria monocytogenes. Lett. Appl. Microbiol. 46:148-154. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, P., E. Moreno, and J. L. Martinez. 2005. The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob. Agents Chemother. 49:781-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokolovic, Z., J. Riedel, M. Wuenscher, and W. Goebel. 1993. Surface-associated, PfrA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol. Microbiol. 8:219-227. [DOI] [PubMed] [Google Scholar]
- 46.To, M. S., S. Favrin, N. Romanova, and M. W. Griffiths. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5258-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vega, Y., C. Dickneite, M.-T. Ripio, R. Böckmann, B. González-Zorn, S. Novella, G. Domínguez-Bernal, W. Goebel, and J. A. Vázquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vega, Y., M. Rauch, M. J. Banfield, S. Ermolaeva, M. Scortti, W. Goebel, and J. A. Vázquez-Boland. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulated PrfA. Mol. Microbiol. 52:1553-1565. [DOI] [PubMed] [Google Scholar]
- 50.Wulff, G., L. Gram, P. Ahrens, and B. F. Vogel. 2006. One group of genetically similar Listeria monocytogenes strains frequently dominate and persist in several fish slaughter- and smokehouses. Appl. Environ. Microbiol. 72:4313-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]