Abstract
Ostreococcus spp. are extremely small unicellular eukaryotic green algae found worldwide in marine environments, and they are susceptible to attacks by a diverse group of large DNA viruses. Several biologically distinct species of Ostreococcus are known and differ in the ecological niches that they occupy: while O. tauri (representing clade C strains) is found in marine lagoons and coastal seas, strains belonging to clade A, exemplified by O. lucimarinus, are present in different oceans. We used laboratory cultures of clonal isolates of these two species to assay for the presence of viruses in seawater samples from diverse locations. In keeping with the distributions of their host strains, we found a decline in the abundance of O. tauri viruses from a lagoon in southwest France relative to the Mediterranean Sea, whereas in the ocean, no O. tauri viruses were detected. In contrast, viruses infecting O. lucimarinus were detected from distantly separated oceans. DNA sequencing, phylogenetic analyses using a conserved viral marker gene, and a Mantel test revealed no relationship between geographic and phylogenetic distances in viruses infecting O. lucimarinus.
Viruses are the most abundant and genetically diverse biological entities in marine environments (48). The three ways most often used to assess eukaryotic algal virus diversity are (i) using a functional host-virus system to quantify viruses specific to one host strain (i.e., culture-based studies) (4), (ii) using PCR amplification and sequencing a conserved gene (10, 12-14, 28), and (iii) using whole-community genome sequencing (i.e., metagenomics) (6, 8, 31). Recently, the advent of sequencing techniques like shotgun sequencing or pyrosequencing (38) has led to an increase in the number of metagenomics projects. The Global Ocean Sampling (GOS) Expedition has provided a unique opportunity to investigate viral diversity in different environments within the size fraction of 0.1 to 0.8 μm (39). The GOS data revealed highly abundant viral sequences (at least 3% of the predicted proteins had a viral origin) (53). In another study, the analysis of marine viromes from four oceanic regions suggested that the composition of viral assemblages depends on their geographic locations, but these authors conclude that similar viruses are widespread throughout the oceans (2). Despite these new methods and different ways to analyze viral diversity, we still do not really know if “everything is really everywhere” (7).
The present study addresses a specific part of this problem: are viruses infecting a single host strain present at geographically distant locations? If several viral strains are identified and characterized, how closely do these viruses resemble one another on a phylogenetic scale? In order to answer these questions, we focused on a microalgal (Prasinophyceae)-virus (Prasinovirus) system. The studied hosts belong to the genus Ostreococcus, a ubiquitous prasinophyte picoeukaryotic alga abundant throughout the oceanic euphotic zone (55, 56). Several strains from this genus were isolated and assigned to four distinct ecotypes according to their growth parameters under different light regimens (22, 36), which correspond to four well-defined phylogenetic clades in an internal transcribed spacer (ITS)-based phylogeny (clades A to D). The complete genome sequences of two Ostreococcus species have been described: O. tauri (19) and O. lucimarinus (35). In the present study, viruses infecting specific host species (Ostreococcus spp.) have been screened from a variety of locations around the world.
Among viruses infecting Ostreococcus, the genome of a single strain (OtV5) has been fully sequenced (18), and the phylogenetic relationships among several virus strains have been investigated (4). These viruses belong to the Prasinovirus group, a genus of the Phycodnaviridae family. Many viruses infecting phytoplankton are members of the Phycodnaviridae; they have double-stranded DNA genomes and large polyhedral capsids (20). In the prasinophyte-Prasinovirus system, the hosts and viruses can be grown on solid medium and are easily maintained in the laboratory. Ostreococcus viral strains have been isolated and characterized by phylogenetic analysis based on their B-family DNA polymerase (DNA pol) partial gene sequence (4). This DNA polymerase is a useful marker for phylogenetic analyses because its sequence is well conserved in all known members of nucleocytoplasmic large DNA viruses (NCLDVs) (26), including Phycodnaviridae. Furthermore, several previous studies have examined the abundance and the genetic diversity of marine eukaryotic viruses using environmental sequencing approaches and amplified DNA pol gene fragments (11, 12, 43-46), and Monier et al. (31) used this marker to describe the taxonomic distribution of large DNA viruses from the GOS data.
The first stage of this study was to isolate Ostreococcus viruses from different worldwide geographic locations, by culturing on various host strains. In a second stage, these viral strains were characterized via the sequencing of their pol sequence (encoding a part of their DNA polymerase gene), and their specificity toward different host strains was assessed in order to assess the potential host range of the viral strains isolated and to gain a better understanding of their population dynamics and distribution. Finally we compared these new Prasinovirus DNA sequences with metagenomic sequence data (obtained from sampling all around the world) and environmental sequence data identified using BLAST similarity to assess the global distribution of similar Ostreococcus viruses.
MATERIALS AND METHODS
Study sites.
Eight seawater samples were collected between September 2008 and February 2009 from different near surface depths from 3 sites within the Pacific, 3 from Atlantic, 1 from English Channel, and 1 from Mediterranean Sea (see Table S1 in the supplemental material). All samples were collected using Niskin bottles and were kept in the dark at room temperature until subsequent analysis was performed within 1 to 7 days after collection.
Culture of the algal host and production of viral lysis plaques.
Ostreococcus tauri strain RCC745 (Roscoff Culture Collection [51]) and O. lucimarinus CCMP2972 (Center for Culture of Marine Phytoplankton) were used as hosts for the infection experiments. Cultures were maintained in Keller's medium (27) prepared in autoclaved and 0.22-μm-pore-filtered seawater under a permanent irradiance of 100 μmol photon m−2 s−1 (Luminux cool white fluorescent tubes, type L36W/840). Routine analysis for plaques was done using a protocol modified from Derelle et al. (18). Seawater samples were prefiltered by gravity through membranes with a porosity of 3 μm (Millipore type SSWP) and then filtered again through a 0.45-μm filter (Millipore type SSWP). Ten milliliters of this filtrate was mixed with 7 ml of Keller's medium and 8 ml of 3.107 cells/ml of a growing host culture. A 1.5% solution of agarose (Euromedex type D-5, DNA grade) was dissolved in distilled water by heating to 100°C and held at 60°C in a water bath. Rapidly, 3 ml of the warm agarose solution was added to the seawater-medium mixture and poured immediately in a 9-cm-diameter petri dish. Collection and filtration of the water and plating out for visualization of lysis plaques were performed within 2 h. A minimum of 4 plates were prepared from each seawater sample and from each host tested. The plates were cultured (continuous light at 100 μmol photon m−2 s−1 at 20 ± 1°C) inside a transparent plastic box to maintain humidity for 1 to 2 weeks. A few days after plating, plaques often appeared inside the agarose gel. These were picked off, added to 400 μl of a solution of MgSO4 (SM buffer; CSH protocols, 2006; doi:10.1101/pdb.rec466) and conserved at 4°C. In order to obtain viral lysate for subsequent experiment on specificity, 200 μl of this lysis plaque was added to a host culture (in the exponential phase) of 6 ml, and few days later, this yielded 6 ml of cleared viral lysate.
Identification of viral sequences in environmental sequence databases.
First, we screened a metagenomics database (http://camera.calit2/net/) for potential DNA pol sequences from Ostreococcus viruses. A conservative approach was taken in order to distinguish only Ostreococcus viral sequences. A sequence was considered for further analysis only if its top BLASTN hit was clearly attributed to a viral DNA polymerase with a minimum size of 500 bp and an E value of <1e−20. Second, DNA pol viral environmental sequences amplified using the same primer set in this study were downloaded from GenBank. Many sequences from an “unknown host” were found, originating from several independent sampling series (12, 17, 45, 46). All of these sequences (from GenBank and metagenomics data) were used to generate an initial phylogenetic reconstruction, and then only one representative sequence from the clade closely related to Ostreococcus viruses was kept to build the final phylogenetic reconstruction.
PCR amplification.
Group-specific primers (AVS) already designed on the basis of viruses infecting Micromonas pusilla (MpV-Sp1), two Chlorovirus strains (PBCV-1 and NY-2A) (10, 11), and viruses infecting Ostreococcus (4) were used to amplify viral DNA polymerase fragments from lysis plaques. These primers AVS1 (5′-GARGGIGCIACIGTIYTIGAYGC-3′) and AVS2 (5′-GCIGCRTAICKYTTYTTISWRTA-3′) amplify a DNA polymerase fragment about 600 bp long. PCR conditions were described within a previous study (4). PCR bands were purified directly by using the NucleoSpin kit (Macheray-Nagel Company, Düren, Germany), and DNA fragments were sequenced (Macrogen, Inc., South Korea) using the amplification primers. To control for PCR or sequencing errors, fragments were sequenced in the reverse and forward senses, and all nucleotide differences were checked visually using the chromatograms. Special care was taken to avoid contamination or mislabeling: all experiments (from picking of viral lysis plaques to DNA amplification and purification) were independently run at least twice, by two different persons. The results were always similar.
Phylogenetic reconstruction.
Sequences were read and corrected using BioEdit (version 7.0.0) (25). BLASTN (NCBI, http://www.ncbi.nlm.nih.gov/) was used to search for similar sequences in public databases (GenBank and CAMERA) (see Table S2 in the supplemental material). Amino acid sequences were then manually aligned using Se-Al (A. Rambaut, 1996; Se-Al: Sequence Alignment Editor [http://evolve.zoo.ox.ac.uk/]). Phylogenetic reconstructions based on amino acid sequences were carried out by Bayesian inference and maximum likelihood. Bayesian analysis was done with MrBayes 3.1.2 (37), with 4 chains of 2.106 generations and trees sampled every 200 generations, with the burn-in value set to 20% of the sampled trees. Using Bayesian inference, protein sequences were analyzed with a mixed-amino-acid model (37), and DNA sequences were considered with an evolutionary model designed for coding sequences taking the genetic code into account (21, 32, 42). Maximum likelihood reconstructions were carried out only on amino acid sequences, using PhyML (23, 24) with an evolutionary model selected via Akaike Information Criterion with ProtTest (1), and validated with 100 bootstrap replicates.
Statistical analysis.
To look for a biogeographic pattern, we used a Mantel test (30) to study the correlation between geographical distances and patristic distances computed from the phylogenic tree, using TreeEdit (Rambaut and Charleston, 2001; http://tree.bio.ed.ac.uk/software/treeedit/). The Mantel test was computed using the function “mantel” from the “vegan” library (J. Oksanen, R. Kindt, P. Legendre, and R. B. O'Hara, 2007; vegan: Community Ecology Package [http://cran.r-project.org/]) of the R statistical language v2.8.1 (R: a language and environment for statistical computing, 2008; R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org]). Significance was assessed using 1,000 permutations.
Specificity.
Virus susceptibilities were examined by using a plating technique on a range of prasinophytes. Plates with host cells were prepared (7 ml of Keller's medium, 8 ml of a 3.107 cells/ml of a growing host culture, and a 1.5% solution of agarose [Euromedex type D-5; DNA grade]) to which we added 2 μl of virus in duplicate. The plates were cultured (continuous light 100 μmol photon m−2 s−1, at 20 ± 1°C) inside a transparent plastic box to maintain humidity for 10 days. Plates that were not lysed 10 days after viral inoculation were considered to be resistant to the virus.
Nucleotide sequence accession numbers.
All sequences used in this study were deposited in GenBank. The accession numbers of virus DNA polymerase partial gene sequences are as follows: GQ412082, OlV349; GQ412083, OlV350; GQ412084, OlV359; GQ412085, OlV360; GQ412086, OlV364; GQ412087, OlV368; GQ412088, OlV402; GQ412089, OlV468; GQ412090, OlV470; GQ412091, OlV462; GQ412092, OlV464; GQ412093, OlV465; GQ412094, OlV466; GQ412095, OlV467; GQ412096, OlV536; GQ412097, OlV537; GQ412098, OlV458; GQ412099, OlV158; GQ412100, OlV164; and GQ412101, OlV155.
RESULTS
Isolation and characterization of prasinoviruses.
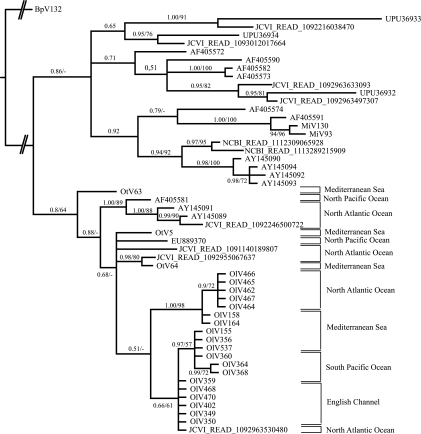
Water samples obtained from eight locations from different oceans (South and North Pacific, South and North Atlantic, Mediterranean Sea, and English Channel) were screened for the presence of viruses infecting two prasinophyte species, Ostreococcus tauri and O. lucimarinus, isolated from the Thau lagoon (Mediterranean Sea) and the North Pacific, respectively (see Table S1 in the supplemental material). Viruses of O. tauri were found only in samples from the Mediterranean Sea station, but viruses of O. lucimarinus were isolated from four locations (South Pacific, North Atlantic, English Channel, and Mediterranean Sea). We thus focused this study on O. lucimarinus viruses. Twenty viruses were isolated, and a fragment of the viral DNA pol gene was amplified and sequenced. The phylogeny of these 20 viral sequences (6 from the North Atlantic, 4 from the South Pacific, 5 from the English Channel, and 5 from the Mediterranean Sea) (Fig. 1) did not form clades that were associated with their geographic origins, except for North Atlantic sequences, which clustered together. We tested the correlation between geographical distances and phylogenetic distances via a Mantel test, which confirmed the absence of such a link (P = 0.338) and therefore supports the absence of a biogeographical pattern. Viral sequences from the Mediterranean Sea displayed more variability (mean nucleotide divergence of 10.84%), than those from the North Atlantic (0.18%) and South Pacific (3.54%) (Table 1). Sequences from the English Channel were almost all identical, with a mean divergence of 0.07% and a maximum divergence of 0.18%. The mean nucleotide divergence between sequences from South Pacific and English Channel was only 4.12%, but between North Atlantic and English Channel sequences, the mean nucleotide divergence was 20.12%. The nucleotide divergence of 10.84% reported above for 5 viruses from O. lucimarinus at the Mediterranean Sea site is higher than what was found in a previous study (L. Bellec, N. Grimsley, E. Derelle, H. Moreau, and Y. Desdevises, submitted for publication) at the same sample site, where 65 viruses of O. tauri were isolated, displaying a nucleotide divergence of only 1.61%.
FIG. 1.
Phylogenetic tree of prasinoviruses with environmental sequences from the partial DNA polymerase gene (for a final length of 195 amino acids [aa]) reconstructed by Bayesian inference (BI) and maximum likelihood (ML). Numbers are posterior probabilities (BI) and bootstrap proportions (ML) reflecting clade support (ML; values below 50 are indicated by dashes). Geographic origins of sequences are given on the right. GenBank accession numbers are as follows: MiV130, FJ267495; MiV93, FJ267493; OtV63, FJ267501; OtV64, FJ267502; BpV132, FJ267517; and OtV5, EU304328.
TABLE 1.
Nucleotide divergence among partial DNA polymerase gene sequences of viruses from different locations
| Virus source location(s) | % Nucleotide divergence |
||
|---|---|---|---|
| Maximum | Mean | SD | |
| English Channel | 0.18 | 0.07 | 0.09 |
| South Pacific | 4.86 | 3.54 | 1.86 |
| North Atlantic | 0.51 | 0.18 | 0.26 |
| Mediterranean Sea | 18.01 | 10.84 | 9.03 |
| English Channel and South Pacific | 5.04 | 4.12 | 1.35 |
| English Channel and North Atlantic | 20.36 | 20.12 | 2.1 |
| English Channel and Mediterranean Sea | 18.37 | 10.91 | 6.2 |
| South Pacific and North Atlantic | 20.72 | 20.49 | 2.56 |
| South Pacific and Mediterranean Sea | 18.91 | 9.57 | 7.7 |
| North Atlantic and Mediterranean Sea | 19.45 | 13.53 | 7.72 |
Host origin and specificity.
Eight prasinophyte host strains representing Ostreococcus sp. clades C (1 strain), D (1 strain), and A (3 strains), Bathycoccus sp., and Micromonas sp. were chosen (see Table S3 in the supplemental material) from various worldwide locations to test viral specificity. The same host specificity was observed for the 20 viruses tested (see Table S4 in the supplemental material). All viruses isolated using O. lucimarinus as a host were specific to this strain and did not infect any of the other 7 strains tested (5 Ostreococcus spp., 1 Micromonas sp., and 1 Bathycoccus sp.).
Environmental and metagenomic sequences.
Environmental DNA pol viral sequences were downloaded from GenBank and added to sequences extracted from metagenomic databases (Materials and Methods) and to the 20 new sequences obtained in this study from O. lucimarinus. We added some representative viral sequences of Ostreococcus and Micromonas isolated from a previous study (4) and added a Bathycoccus virus sequence as an outgroup to build a phylogenetic tree. This phylogeny clearly splits the sequences into two groups, one containing Ostreococcus viruses and the other containing Micromonas viruses (Fig. 1). Within the Ostreococcus virus clade, viruses from O. lucimarinus all cluster together, and 8 environmental sequences can be identified as probable Ostreococcus (4 from GenBank and 4 from metagenomes). The geographic origins of all sequences from the Ostreococcus virus group obtained in this study are heterogeneous, containing sequences from the North and South Pacific, North Atlantic (East and West), English Channel, and Mediterranean Sea (see Table S3 in the supplemental material).
DISCUSSION
In the present study, we used a host-virus system approach with the aim of isolating new viruses on one host using water samples from worldwide geographic locations. Five main results were obtained. First, viruses from O. tauri were only found within the Mediterranean Sea, whereas O. lucimarinus viruses were widely distributed (Atlantic, Pacific, Mediterranean Sea, and English Channel). Second, the DNA pol sequences of O. lucimarinus viruses exhibit a high genetic variability and they did not appear to cluster according to a geographical repartition. Third, the latter sequences recovered from distant oceans form a distinct clade of viruses within the genus Prasinovirus. Fourth, sequences with high similarities to Ostreococcus viruses can be found within metagenomics data and environmental sequences from different geographic locations. Five, viruses appeared to be highly specific to the host strain from which they were isolated.
In the present study, no O. tauri viruses were detectable in samples from 6 sites from different oceans or from one lagoon sample (Moorea, South Pacific). Viruses of O. tauri were isolated within the Gulf of Lion near the location where O. tauri was originally detected (16). In a previous study, we observed a high abundance of O. tauri viruses in a coastal Mediterranean lagoon (Bellec et al., submitted). These results suggest that O. tauri viruses may be limited to the Mediterranean Sea and preferentially within lagoons. Since the first isolation of O. tauri, isolates of Ostreococcus spp. have been cultured from many oceanic regions (22, 36), and their population density is high in coastal lagoons (3), but oceanic populations appear to be sporadic, with lower densities (100 cells ml−1) (33; Bellec et al., submitted), except during short bloom periods in the North Pacific or North Atlantic with high cell densities (105 cells ml−1) (15, 34). Ostreococcus cell density off the Spanish Mediterranean coast (100 km from the coastal lagoon) was close to zero, except for one period at 50 cells ml−1 (56). All of these studies used flow cytometry, denaturing gradient gel electrophoresis (DDGE), or quantitative PCR (qPCR) to assess cell density of the genus Ostreococcus but could not discriminate between strains such as O. tauri or O. lucimarinus. It would thus be informative to investigate coastal lagoons on a worldwide scale.
We found that viruses of O. lucimarinus were widely dispersed on a global scale. They were detected within a coastal lagoon in the Mediterranean Sea, displaying a high host specificity, but surprisingly strains similar to the host (O. lucimarinus) have not yet been found in cultures from Mediterranean samples. However, environmental sequences (52) show that diverse clade A strains (close to O. lucimarinus) are common in the western part of Mediterranean, i.e., in the nutrient-rich waters from the Morocco upwelling, the Strait of Gibraltar, and the Algerian Basin, in surface waters, although these authors did not specifically test water from lagoons. The simplest interpretation of these results is that clade A strains such as O. lucimarinus are ubiquitous (although more difficult to detect in cultures because the growth conditions for them may not be optimal), so their viruses are present everywhere, whereas clade C strains, such as O. tauri, may be easier to culture but more limited in their distribution to lagoons and coastal areas, so that their viruses in oceanic waters become diluted beyond the limits of detection by our method. This effect may be enhanced by the nature of the Mediterranean basin, with very limited contact with oceanic waters (the Suez Canal and Strait of Gibraltar), and its net water intake since its evaporation rate exceeds freshwater inputs.
Virion dispersal may also play a role in the wide distribution observed here for viruses of O. lucimarinus. Although the lifetime of viral particles in nature must be finite, we do not believe that virion viability plays a major role in reducing viral particle densities, since suspensions of viruses in the lab in seawater are stable for >3 years at 4°C and >1 year at 20°C (unpublished data). If these results could be confirmed in the field, that would suggest that viruses can move between biomes following oceanic currents without the need of a host (41). However, in the environment, other factors such as UV, grazing, or degradation by organics may act on viral decay (5, 54), which may be faster under natural conditions. In the sea, loss of viable virions by their erroneous nonspecific binding to related host phytoplankton thus seems a more plausible hypothesis. More information about the distribution of host species and experimental work on the specificity of viral particle binding is necessary to investigate the impact of nonspecific binding, but must await the availability of more complete metagenomic data since the above observations highlight that PCR-based and culture-based techniques give only an incomplete glimpse of species' diversity.
The phylogenetic analysis indicated that all O. lucimarinus viruses formed a clade within Prasinovirus. There is not a clear geographical distribution, and it is remarkable that viral strains from the South Pacific are closely related to sequences from the English Channel (>95% identity) and from the Mediterranean Sea (>90% identical). However, these three samples were all taken from coastal sites, and their relatedness might reflect a similarity in their host communities. The 20 viruses of O. lucimarinus revealed that isolates from one sample site can be as diverse as between isolates from different oceans. For example, viruses from the Mediterranean Sea exhibit a nucleotide divergence of 10.84%, while the divergence between Mediterranean and South Pacific samples is 9.57%. Similar results have been observed for viruses from the prasinophyte Micromonas, in which some isolates of M. pusilla virus from widely separated geographical locations were more similar than isolates from a single location (12, 13). All of these results suggest that prasinoviruses (or at least Ostreococcus and Micromonas viruses) are diverse and widely distributed and that genetically distinct isolates can coexist at a single geographic location. Cottrell and Suttle (13) proposed the hypothesis that coexistence of genetically diverse viruses at the same site suggests that competition among these viral strains is limited. This hypothesis may be true for Micromonas viruses that exhibit different patterns of host specificity. A previous study showed that two Micromonas viruses display clear differences in their infection process. There was no relationship between susceptibility and geographic provenance of the host, nor with the season of collection of the host and the virus (57). In contrast, other studies on Micromonas pusilla (9, 40) reported susceptibility only in host strains from the same location of their virus (for DNA and RNA viruses). This pattern was also observed in Heterosigma akashiwo viruses (29, 49), different host isolates showing different viral infection characteristics can coexist during a bloom (50). The specificity of Phycodnaviridae was also investigated in several host-virus systems: in Emiliania huxleyi viruses that exhibited a similar host range for 10 virus isolates (42); in Chrysochromulina brevifilum viruses that lysed only 2 out of 10 Chrysochromulina host species (47); and, recently, in a virus of Ostreococcus tauri (OtV5), that showed strict host-strain specificity (18). Viral specificity is likely to play an important role in the host communities (14) because differences in host range imply different viral pressures on host genetic diversity.
This study presents primary results from a host-virus system on a global scale. However, the lack of knowledge about host ecology (cell density and susceptibility to viruses) and the few data currently available on their viruses emphasize the need for a deeper exploration of these associations in the future, including complete viral sequences to gain insight into those gene functions that may be involved in specificity and in-depth metagenomic analyses to form a more comprehensive nonbiased inventory of all the phycodnaviruses and unicellular eukaryotes present at various geographic sites. The lack of detection of O. tauri viruses in the oceans highlights that, even among life's smallest life forms, the spatial heterogeneity of oceanic phytoplankton requires further exploration.
Supplementary Material
Acknowledgments
We thank the following for providing us water samples from various worldwide locations: Pascal Romans (Mediterranean), Guglielmo Tita (North Atlantic), Pierre Sasal (Moorea), Jean-Lou Justine (Nouméa), Daniela Bottger (South Pacific), Station Biologique de Roscoff (English Channel), Alexandra Worden (Florida), and Graciela Salerno (Argentina). We also thank Gwenaël Piganeau and Hervé Moreau for stimulating discussions and Lucie Subirana and Camille Clerissi for technical assistance. We thank three anonymous reviewers for insightful comments.
This work was supported by an “Agence Nationale de Recherche” grant “PICOVIR” (coordinator N.G.; no. BLAN07-1_200218). Laure Bellec benefited from a doctoral fellowship from the French Ministry of Education and Research.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bec, B., J. Husseini-Ratrema, Y. Collos, P. Souchu, and A. Vaquer. 2005. Phytoplankton seasonal dynamics in a Mediterranean coastal lagoon: emphasis on the picoeukaryote community. J. Plankton Res. 27:881-894. [Google Scholar]
- 4.Bellec, L., N. Grimsley, H. Moreau, and Y. Desdevises. 2009. Phylogenetic analysis of new Prasinoviruses (Phycodnaviridae) that infect the green unicellular algae Ostreococcus, Bathycoccus and Micromonas. Environ. Microbiol. Rep. 1:114-129. [DOI] [PubMed] [Google Scholar]
- 5.Bongiorni, L., M. Magagnini, M. Armeni, R. Noble, and R. Danovaro. 2005. Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 71:6644-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. Lond. B Biol. Sci. 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., P. Salamo, and B. Andresen. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brussaard, C. P., A. A. Noordeloos, R. A. Sandaa, M. Heldal, and G. Bratbak. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280-291. [DOI] [PubMed] [Google Scholar]
- 10.Chen, F., and C. A. Suttle. 1995. Amplification of DNA-polymerase gene fragments from viruses infecting microalgae. Appl. Environ. Microbiol. 61:1274-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, F., and C. A. Suttle. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170-178. [DOI] [PubMed] [Google Scholar]
- 12.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cottrell, M. T., and C. A. Suttle. 1995. Genetic diversity of algal viruses which lyse the photosynthetic picoflagellate Micromona pusilla (Prasinophyceae). Appl. Environ. Microbiol. 61:3088-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cottrell, M. T., and C. A. Suttle. 1991. Wide-spread occurrence and clonal variation in viruses which cause lysis of a cosmopolitan, eukaryotic marine phytoplankter, Micromonas pusilla. Mar. Ecol. Prog. Ser. 78:1-9. [Google Scholar]
- 15.Countway, P. D., and D. A. Caron. 2006. Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl. Environ. Microbiol. 72:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courties, C., A. Vaquer, M. Troussellier, J. Lautier, M. J. Chretiennot-Dinet, J. Neveux, C. Machado, and H. Claustre. 1994. Smallest eukaryotic organism. Nature 370:255. [Google Scholar]
- 17.Culley, A. I., B. F. Asuncion, and G. F. Steward. 2008. Detection of inteins among diverse DNA polymerase genes of uncultivated members of the Phycodnaviridae. ISME J. 3:409-418. [DOI] [PubMed] [Google Scholar]
- 18.Derelle, E., C. Ferraz, M. L. Escande, S. Eychenie, R. Cooke, G. Piganeau, Y. Desdevises, L. Bellec, H. Moreau, and N. Grimsley. 2008. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One 3:e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derelle, E., C. Ferraz, S. Rombauts, P. Rouze, A. Z. Worden, S. Robbens, F. Partensky, S. Degroeve, S. Echeynie, R. Cooke, Y. Saeys, J. Wuyts, K. Jabbari, C. Bowler, O. Panaud, B. Piegu, S. G. Ball, J.-P. Ral, F.-Y. Bouget, G. Piganeau, B. De Baets, A. Picard, M. Delseny, J. Demaille, Y. Van de Peer, and H. Moreau. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647-11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 21.Goldman, N., and Z. H. Yang. 1994. Codon-based model of nucleotide substitution for protein-coding DNA-sequences. Mol. Biol. Evol. 11:725-736. [DOI] [PubMed] [Google Scholar]
- 22.Guillou, L., W. Eikrem, M. J. Chretiennot-Dinet, F. Le Gall, R. Massana, K. Romari, C. Pedros-Alio, and D. Vaulot. 2004. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155:193-214. [DOI] [PubMed] [Google Scholar]
- 23.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 24.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 26.Iyer, L. M., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 27.Keller, M. D., R. C. Selvin, W. Claus, and R. R. L. Guillard. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23:633-638. [Google Scholar]
- 28.Larsen, J. B., A. Larsen, G. Bratbak, and R. A. Sandaa. 2008. Phylogenetic analysis of members of the Phycodnaviridae virus family, using amplified fragments of the major capsid protein gene. Appl. Environ. Microbiol. 74:3048-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence, J. E., A. M. Chan, and C. Suttle. 2001. A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae). J. Phycol. 37:216-222. [Google Scholar]
- 30.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 31.Monier, A., J. M. Claverie, and H. Ogata. 2008. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 9:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muse, S. V., and B. S. Gaut. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11:715-724. [DOI] [PubMed] [Google Scholar]
- 33.Not, F., M. Latasa, D. Marie, T. Cariou, D. Vaulot, and N. Simon. 2004. A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Appl. Environ. Microbiol. 70:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Kelly, C. J., M. E. Sieracki, E. C. Thier, and I. C. Hobson. 2003. A transient bloom of Ostreococcus (Chlorophyta, Prasinophyceae) in West Neck Bay, Long Island, New York. J. Phycol. 39:850-854. [Google Scholar]
- 35.Palenik, B., J. Grimwood, A. Aerts, P. Rouze, A. Salamov, N. Putnam, C. Dupont, R. Jorgensen, E. Derelle, S. Rombauts, K. Zhou, R. Otillar, S. S. Merchant, S. Podell, T. Gaasterland, C. Napoli, K. Gendler, A. Manuell, V. Tai, O. Vallon, G. Piganeau, S. Jancek, M. Heijde, K. Jabbari, C. Bowler, M. Lohr, S. Robbens, G. Werner, I. Dubchak, G. J. Pazour, Q. Ren, I. Paulsen, C. Delwiche, J. Schmutz, D. Rokhsar, Y. Van de Peer, H. Moreau, and I. V. Grigoriev. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 104:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez, F., E. Derelle, L. Guillou, F. Le Gall, D. Vaulot, and H. Moreau. 2005. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7:853-859. [DOI] [PubMed] [Google Scholar]
- 37.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 38.Rothberg, J. M., and J. H. Leamon. 2008. The development and impact of 454 sequencing. Nat. Biotechnol. 26:1117-1124. [DOI] [PubMed] [Google Scholar]
- 39.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahlsten, E. 1998. Seasonal abundance in Skagerrak-Kattegat coastal waters and host specificity of viruses infecting the marine photosynthetic flagellate Micromonas pusilla. Aquat. Microb. Ecol. 16:103-108. [Google Scholar]
- 41.Sano, E., S. Carlson, L. Wegley, and F. Rohwer. 2004. Movement of viruses between biomes. Appl. Environ. Microbiol. 70:5842-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder, D., and A. H. Perera. 2002. A comparison of large-scale spatial vegetation patterns following clearcuts and fires in Ontario's boreal forests. For. Ecol. Manag. 159:217-230. [Google Scholar]
- 43.Shapiro, B., A. Rambaut, and A. Drummond. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7-9. [DOI] [PubMed] [Google Scholar]
- 44.Short, S. M., and C. M. Short. 2008. Diversity of algal viruses in various North American freshwater environments. Aquat. Microb. Ecol. 51:13-21. [Google Scholar]
- 45.Short, S. M., and C. A. Suttle. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Short, S. M., and C. A. Suttle. 2003. Temporal dynamics of natural communities of marine algal viruses and eukaryotes. Aquat. Microb. Ecol. 32:107-119. [Google Scholar]
- 47.Suttle, C., and A. M. Chan. 1995. Viruses infecting the marine Prymnesiophte Chrysochromulina spp.: isolation, preliminary characterization and natural abundance. Mar. Ecol. Prog. Ser. 118:275-282. [Google Scholar]
- 48.Suttle, C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801-812. [DOI] [PubMed] [Google Scholar]
- 49.Tai, V., J. E. Lawrence, A. S. Lang, A. I. Culley, and C. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akahiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 50.Tarutani, K., K. Nagasaki, and M. Yamaguchi. 2000. Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo. Appl. Environ. Microbiol. 66:4916-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaulot, D., F. Le Gall, D. Marie, L. Guillou, and F. Partensky. 2004. The Roscoff Culture Collection (RCC): a collection dedicated to marine picoplankton. Nova Hedwigia 79:49-70. [Google Scholar]
- 52.Viprey, M., L. Guillou, M. Ferreol, and D. Vaulot. 2008. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ. Microbiol. 10:1804-1822. [DOI] [PubMed] [Google Scholar]
- 53.Williamson, S. J., D. B. Rusch, S. Yooseph, A. L. Halpern, K. B. Heidelberg, J. I. Glass, C. Andrews-Pfannkoch, D. Fadrosh, C. S. Miller, G. Sutton, M. Frazier, and J. C. Venter. 2008. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worden, A. Z., J. K. Nolan, and B. Palenik. 2004. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol. Oceanogr. 49:168-179. [Google Scholar]
- 56.Zhu, F., R. Massana, F. Not, D. Marie, and D. Vaulot. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79-92. [DOI] [PubMed] [Google Scholar]
- 57.Zingone, A., F. Natale, E. Biffali, M. Borra, G. Forlani, and D. Sarno. 2006. Diversity in morphology, infectivity, molecular characteristics and induced host resistance between two viruses infecting Micromonas pusilla. Aquat. Microb. Ecol. 45:1-14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.