Abstract
This study uses multilocus sequence typing (MLST) to investigate the epidemiology of Campylobacter coli in a continuous study of a population in Northwest England. All cases of Campylobacter identified in four Local Authorities (government administrative boundaries) between 2003 and 2006 were identified to species level and then typed, using MLST. Epidemiological information was collected for each of these cases, including food and recreational exposure variables, and the epidemiologies of C. jejuni and C. coli were compared using case-case methodology. Samples of surface water thought to represent possible points of exposure to the populations under study were also sampled, and campylobacters were typed with multilocus sequence typing. Patients with C. coli were more likely to be older and female than patients with C. jejuni. In logistic regression, C. coli infection was positively associated with patients eating undercooked eggs, eating out, and reporting problems with their water supply prior to illness. C. coli was less associated with consuming pork products. Most of the cases of C. coli yielded sequence types described elsewhere in both livestock and poultry, but several new sequence types were also identified in human cases and water samples. There was no overlap between types identified in humans and surface waters, and genetic analysis suggested three distinct clades but with several “intermediate” types from water that were convergent with the human clade. There is little evidence to suggest that epidemiological differences between human cases of C. coli and C. jejuni are a result of different food or behavioral exposures alone.
Campylobacterosis is a well-defined public health problem in many areas of the world, and several potential sources of infection are well characterized. However, in “industrialized” nations, the degree to which various transmission pathways contribute to human disease burden is still not certain. There has been some indication that the human epidemiology of Campylobacter coli differs from that of the more commonly reported Campylobacter jejuni (10), and the distributions of these species described in animal hosts also show some differences (29) (20). Although far less prevalent, infections with C. coli have consistently contributed around 7% of all human campylobacterosis in the United Kingdom (10, 26), corresponding to an approximate annual burden of 3,491 cases of severe gastroenteritis in 2008.
Campylobacter has been successfully isolated and cultured from surface freshwater in several countries (1) and both outbreaks and “sporadic” cases of campylobacterosis have been linked with exposure to drinking water (4, 18, 24). Specifically, C. coli has been isolated from surface and untreated drinking water and associated with an outbreak in drinking water in rural France, suggesting a potential risk of exposure from the environment (2, 9, 15, 23).
Multilocus sequence typing (MLST) has recently been adapted for C. coli (8), and we use this method to investigate the epidemiology of C. coli in a continuous population-based survey in Northwest (NW) England (the design of which has been described elsewhere [27]), comparing human strains of C. coli with those identified in local surface waters.
MATERIALS AND METHODS
Study population.
The study population was defined as all human cases of confirmed Campylobacter infection between April 2003 and March 2006 reported from residents in four Local Authorities (government administrative boundaries) in NW England, as previously described (27).
Data collection.
Confirmed cases of Campylobacter infection (using the United Kingdom's National Standard Method for diagnosis [http://www.hpa-standardmethods.org.uk/documents/bsopid/pdf/bsopid23.pdf]) are routinely reported to the NW Health Protection Agency (HPA) surveillance system by local National Health Service laboratories. Cases were identified as resident in the study area through available geographic information or by case names where this information was not available. Reports included basic demographic information such as age and sex, as well as date of onset. Where the date of onset was not available, the date of the report was used as a proxy. All cases were asked more detailed questions about their illness via a postal questionnaire. Information collected comprised basic demographics (including occupation), clinical data (including hospital admission and duration of illness), travel in the two weeks preceding symptoms (including destination and duration), detailed food and drink consumption, food habits (including diet type, consumption of rare-cooked food, and eating out), outdoor activity (including countryside and water sport exposures), and animal contact (including pet type and wild and farm animals).
Positive isolates of Campylobacter from confirmed cases resident in the study area were sent by the main diagnostic laboratories to the NW HPA Laboratory in Manchester for sequence typing. Cases were determined to be resident in the study area using the methods described above.
Water samples were collected approximately every 2 weeks from October 2003 until December 2005 as 2-liter grab samples from sampling points on the River Mersey (on a stretch located within Trafford Local Authority, Greater Manchester) and the River Wyre (on a stretch located within Wyre Local Authority, Lancashire). Water samples were transported under appropriate conditions to the Food and Environmental Microbiology Services laboratory (FEMS), Preston Microbiology Services, Royal Preston Hospital. Campylobacter species were isolated from the water samples by the addition of 10 ml of the water sample to 90 ml of warmed Campylobacter enrichment broth (product code CM0983; Oxoid Ltd., Basingstoke, United Kingdom) and incubated (37°C for 24 h followed by 42°C for 24 h). The enrichment broths were then subcultured onto Campylobacter blood-free selective agar (charcoal cefoperazone deoxycholate agar [CCDA], product code CM0739; Oxoid Ltd., Basingstoke, United Kingdom) and incubated (37°C for 48 h) microaerobically, using a microaerobic gas generating kit (product code CN0025; Oxoid Ltd., Basingstoke, United Kingdom). Campylobacter colonies were identified by morphology and confirmed by microaerobic and aerobic growth on blood agar, placed in Amies transport medium, and sent to the laboratory in Manchester for DNA extraction and characterization, as described below.
Characterization of Campylobacter isolates.
Campylobacter isolates were identified to species level using PCR as previously described (27). MLST for C. coli was performed as previously described (8). The amplification reactions were performed in a 50-μl volume containing approximately 1 μl of C. coli chromosomal DNA (10 ng/μl), 5 μl of each primer (10 pmol/μl), 10 μl of 1 mM deoxynucleoside triphosphates (dNTPs) (Roche), 5 μl 10× PCR buffer (Qiagen, United Kingdom), and 0.25 units of Taq DNA polymerase (Qiagen, United Kingdom). All other protocols for amplification and sequencing were as previously described for C. jejuni (27). All sequence assemblage and editing were performed using Sequencher 4.0 software (GeneCodes Corporation, MI).
MLST alleles, sequence types (STs), and clonal complexes were assigned using tools available through the Campylobacter PubMLST database (12) with sequences submitted for allele designation as appropriate.
Genetic analysis.
To analyze the genetic relationships between C. coli sequence types isolated from clinical cases and water samples, we used ClonalFrame software (7). Multi-FASTA files for each of the seven alleles used in MLST containing blocks of DNA sequence (separated into the sequence types compared) were prepared using a tool on the Campylobacter MLST database (12). The comparison of sequence data as separate allele blocks ensured that the software treated these data as noncontiguous sequence for a more accurate analysis. The tree was constructed using 50,000 burn-in cycles and 100,000 further iterations, and the final output was constructed using the “build consensus” capability from three independent analyses of the same data file. The convergence assessment of these three runs indicated that there was good convergence between them (data not shown), suggesting that the final tree was representative of the data. The analysis was exported as a “posterior sample” Newick file and presented as a “Consensus Network” using SplitsTree 4 software (11), enabling the representation of all sampled phylogenetic trees as a network. The branch points in the final network faithfully mapped onto those apparent in a basic tree analysis of the data.
Statistical analysis.
Case-case methodology was used in comparison of exposures between species and sequence types of Campylobacter (19). In all species analyses, cases of C. coli were considered “cases” and cases of C. jejuni were considered “controls.” In sequence type analyses, individual sequence types (cases) were compared with all other sequence types reported (controls). Statistical analysis was performed using STATA, version 10 (StataCorp, College Station, TX). The association of individual variables with C. coli was initially analyzed using two-way tabulations and Fisher's exact test. Logistic regression was performed to estimate the odds ratios (OR) of individual variables being associated with one species or the other and also to examine these relationships in a multivariable model. Data were collected and analyzed from two geographically distinct residential areas; therefore, area of residence was considered a stratified variable and controlled for in analysis. Likelihood ratio tests were used to assess the significance of including or excluding variables from multivariable analysis.
RESULTS
Incidence of reported disease.
Of the 1,594 cases of laboratory-confirmed Campylobacter spp. reported from the study area between 2003 (week 17) and 2006 (week 16), 1,142 (72%) were successfully identified to species level. Of these cases, 106 were C. coli (9.3%) and the remainder were C. jejuni, giving an approximate annual incidence of C. coli for this period of 5.75 cases per population of 100,000. Detailed epidemiological data were available for 88% (1,008/1,142) of the isolates that were identified, although completion rates of individual data fields varied. Both species were reported, with a higher incidence in the rural component of the study area, but beyond this, there was no significant geographical clustering of C. coli at the level of individual Local Authority area. However, the age and sex distributions differed between the species. The ratio of C. coli to C. jejuni cases increased with increased age, and cases infected with C. coli were significantly more likely to be over 65 years of age (OR = 3.23) and female (OR = 1.54) (Table 1) than those infected with C. jejuni. Age and sex were therefore controlled for in further analyses of association using logistic regression (in addition to the stratified variable of rural residence).
TABLE 1.
Odds ratios for age categories and sex in a multivariable model comparing cases of Campylobacter coli with C. jejuni, controlled for by area of residence (χ2 P = 0.003)a
| Variable | Odds ratio | 95% confidence interval |
|---|---|---|
| Age (yr) | ||
| 0-24 (baseline) | 1 | |
| 25-44 | 1.66 | 0.85-3.23 |
| 45-64 | 1.86 | 0.95-3.63 |
| >65 | 3.23 | 1.63-6.42 |
| Female sex | 1.54 | 1.03-2.32 |
| Rural residence | 1.06 | 0.70-1.60 |
Cases of C. coli were 3.23 times more likely to be over 65 years old compared with cases of C. jejuni (in relation to number of cases in the baseline category). Figures in bold show results statistically significant at a 95% level.
Sequence typing of human isolates.
From the human cases of C. coli identified, 96 of the 106 isolates were successfully typed using MLST. There were 36 distinct sequence types identified, all except six grouping to clonal complex 828 (Table 2). The remaining types are unassigned (UA) with respect to clonal complex until further identification of types allows the designation of new complexes. At the time of isolation and in reference to the PubMLST database and published literature, 26 of these sequence types had not previously been reported in human cases of campylobacterosis and 17 had never been reported (in any isolate). The most common sequence type isolated was ST-827 (27 cases, 28.1% of all typed cases), and 23 of the sequence types were represented by only one case.
TABLE 2.
MLST sequence types (ST) identified among C. coli isolates from human cases, showing the host and source of previous reportsa
| ST | No. of study cases | Host and source of previous report |
||||
|---|---|---|---|---|---|---|
| Swine | Chicken | Other livestock | Human | Not known | ||
| 825 | 12 | 17, 29 | 16, 20 | 8 | ||
| 827 | 27 | 20 | 20 | 8, 17 | ||
| 828 | 3 | 17, 29 | 17 | 17 | 8, 17 | |
| 829 | 2 | 20 | 8 | |||
| 854 | 1 | 17, 20, 29 | 8 | This study | ||
| 855 | 5 | 3, 8, 16, 17 | 17 | |||
| 872 | 5 | 8 | ||||
| 890 | 1 | 20, 29 | 20 | 8, 21 | This study | |
| 896 | 1 | This study | 8 | |||
| 901 | 1 | This study | 8 | |||
| 962 | 2 | 12 | This study | |||
| 1009* | 1 | 17 | ||||
| 1016 | 1 | 21 | 12 | |||
| 1055 | 3 | 20, 21, 29 | 3, 21 | This study | ||
| 1058 | 1 | 20, 21, 29 | This study | |||
| 1401* | 1 | 12 | This study | |||
| 1584* | 1 | 17 | 17 | |||
| 1614 | 6 | 12 | This study | |||
| 1688* | 1 | 12 | ||||
| 1761* | 1 | This study | ||||
| 1767 | 1 | This study | ||||
| 1768* | 1 | This study | ||||
| 1769 | 2 | This study | ||||
| 1770 | 2 | This study | ||||
| 1773 | 1 | This study | ||||
| 1774 | 2 | This study | ||||
| 1897 | 1 | This study | ||||
| 2003 | 1 | This study | ||||
| 2006 | 1 | This study | ||||
| 2008 | 1 | This study | ||||
| 2009 | 1 | This study | ||||
| 2014 | 1 | This study | ||||
| 2022 | 2 | This study | ||||
| 2177 | 1 | This study | ||||
| 2270 | 1 | This study | ||||
| 2273 | 1 | This study | ||||
| Total | 96 | |||||
All types belong to clonal complex 828, except those with an asterisk, which are unassigned.
Sequence typing of water isolates.
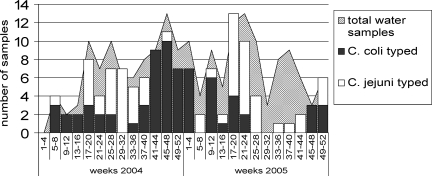
Campylobacter was isolated from surface river waters throughout the year, but there appeared to be some seasonal differences between C. coli and C. jejuni. C. coli was most frequently isolated in autumn and winter months, whereas C. jejuni was more likely to be isolated during spring and summer (Fig. 1). There was no corresponding seasonal pattern among human cases of C. coli (data not shown).
FIG. 1.
Prevalence of Campylobacter spp. isolated from surface waters at five sites on the River Wyre and River Mersey by 4-week period, 2004-2005. All isolates were successfully sequence typed. The total number of samples obtained in each 4-week period (hatched area) varied through the year, but all sampling sites are represented in the data, except for weeks 29 to 40 in 2005, when the River Wyre was not sampled.
Of all the water isolates collected from sites on the River Wyre and the River Mersey, 83% (140/169) were culture positive for Campylobacter spp., with samples from the River Wyre significantly more likely to be culture positive (95.6%, 66/69) than those from the River Mersey (74.0% [74/100]; chi-square, P = 0.0006) (Table 3). A higher proportion of isolates from the River Wyre than from the River Mersey were of C. coli (70% [48/69] compared with 31% [31/100]; chi-square, P < 0.0001). Only two sequence types were identified in water samples in any significant numbers (ST-1766 and ST-1764), and these were identified in both river systems. There was no strong evidence of any difference in the distribution of sequence types between the sampling sites on each river (data not shown). None of the C. coli sequence types identified in water samples were identified in clinical cases of infection in the study, and to date, none have not been identified in humans in the PubMLST database or published literature. All the sequence types identified in Table 3 represented newly characterized strains in the PubMLST database.
TABLE 3.
Isolation of Campylobacter from surface water samples from River Wyre (three sites) and River Mersey (two sites), 2004-2005, including sequence typesa
| Species and ST | No. of isolates at origin: |
Total no. of isolates | |
|---|---|---|---|
| River Wyre | River Mersey | ||
| C. coli | |||
| 1766 | 8 | 7 | 15 |
| 1764 | 7 | 3 | 10 |
| 1771 | 2 | 0 | 2 |
| 1976 | 0 | 2 | 2 |
| 1981 | 2 | 0 | 2 |
| 1988 | 1 | 1 | 2 |
| 1995 | 0 | 2 | 2 |
| 2005 | 0 | 2 | 2 |
| 2007 | 0 | 2 | 2 |
| 2017 | 2 | 0 | 2 |
| 2018 | 2 | 0 | 2 |
| 1762 | 1 | 0 | 1 |
| 1763 | 1 | 0 | 1 |
| 1765 | 1 | 0 | 1 |
| 1772 | 1 | 0 | 1 |
| 1975 | 1 | 0 | 1 |
| 1977 | 1 | 0 | 1 |
| 1978 | 1 | 0 | 1 |
| 1979 | 1 | 0 | 1 |
| 1980 | 0 | 1 | 1 |
| 1982 | 0 | 1 | 1 |
| 1983 | 1 | 0 | 1 |
| 1984 | 1 | 0 | 1 |
| 1985 | 0 | 1 | 1 |
| 1986 | 1 | 0 | 1 |
| 1989 | 1 | 0 | 1 |
| 1990 | 1 | 0 | 1 |
| 1991 | 1 | 0 | 1 |
| 1992 | 1 | 0 | 1 |
| 1996 | 1 | 0 | 1 |
| 1997 | 0 | 1 | 1 |
| 1998 | 0 | 1 | 1 |
| 1999 | 1 | 0 | 1 |
| 2000 | 0 | 1 | 1 |
| 2001 | 1 | 0 | 1 |
| 2002 | 0 | 1 | 1 |
| 2010 | 1 | 0 | 1 |
| 2011 | 1 | 0 | 1 |
| 2012 | 1 | 0 | 1 |
| 2013 | 1 | 0 | 1 |
| 2015 | 0 | 1 | 1 |
| 2016 | 0 | 1 | 1 |
| 2020 | 1 | 0 | 1 |
| 2021 | 0 | 1 | 1 |
| 2023 | 0 | 1 | 1 |
| 2024 | 0 | 1 | 1 |
| 2025 | 1 | 0 | 1 |
| All ST | 48 | 31 | 79 |
| C. jejuni, all ST | 18 | 43 | 61 |
| No Campylobacter | 3 | 26 | 29 |
| Grand total | 69 | 100 | 169 |
Sequence types for C. coli are shown in decreasing order of prevalence. None of these types were identified in human cases through the study. Sequence types of C. jejuni have been reported previously and are not shown here in detail. “No Campylobacter” refers to water samples where Campylobacter was not detected by the methods used.
Comparison of isolates of C. coli from human and water sources.
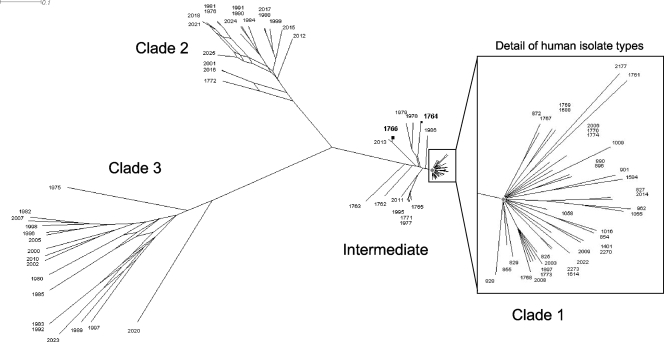
Genetic analysis of all C. coli sequence types generated in the study suggest three distinct clades of types: two identified from water samples (clades 2 and 3) and one from human samples (clade 1) (Fig. 2). In addition, there were several sequence types that did not group with any clade but showed close convergence to the tightly clustered clade 1 (labeled “Intermediate” in Fig. 2), and these included the sequence types most commonly identified from water samples, ST-1766 and ST-1764. Of the 11 other sequence types in this clade, 10 were solely identified in the River Wyre, but there were no consistent associations between clade of sample and its river source or the year or season in which it was taken. Several closely related types represent samples taken from the same river during the same few weeks (data not shown), but, in general, interpretation of the source of each clade was hampered by the very low numbers of each sequence type that were isolated.
FIG. 2.
ClonalFrame analysis of sequence types identified in human isolates and samples of surface waters from the rivers Mersey and Wyre (presented as a Consensus Network using SplitsTree). Genetic distance is indicated by the scale bar labeled 0.1. Most sequence types clustered into the three labeled clades, with the remainder labeled as “Intermediate.” Clade 1 (containing sequence types exclusively from human samples) is very tightly clustered at this scale, and detailed labeling is shown in the inset for clarity. All other sequence types shown were exclusively identified in water samples. The two most prevalent types in water samples grouped to the intermediate category and are shown in bold (ST-1766 and ST-1764).
Epidemiology of C. coli.
Duration of illness, frequency of hospitalization, and reporting of specific symptoms did not vary between cases of C. coli and C. jejuni (when controlled for age, sex, and residence in logistic regression), but there was weak evidence of a decreased likelihood of diarrhea among cases of C. coli (OR, 0.37; confidence interval [CI], 0.11 to 1.20; P = 0.097). Similarly, the frequency of travel outside the United Kingdom, participation in outdoor activities, and the food habits reported by cases did not vary between species groups isolated, except that eating undercooked eggs (OR, 6.28; CI, 1.05 to 37.63; P = 0.044; 839 observations) and eating out (OR, 1.58; CI, 0.95 to 2.64; P = 0.049; 839 observations) were more likely to be associated with cases of C. coli than with C. jejuni. Reported contact with any animal (including pets) was reported as frequently among cases of C. coli as among C. jejuni cases, and there were no discernible differences within categories of animals except for contact with wild animals (as opposed to farm or zoo). There was evidence that cases of C. coli were more likely than cases of C. jejuni to have had contact with wild animals (OR, 3.34; CI, 1.05 to 10.58; P = 0.041; 839 observations). The associations described remained unchanged when adjusted for travel outside the United Kingdom.
Consumption of specific items of food and sources of water were reported almost equally between cases of C. coli and C. jejuni. The most striking exception was the less-frequent reporting of consumption of pork, ham, or bacon among cases of C. coli than among cases of C. jejuni (OR, 0.40; CI, 0.21 to 0.76; P = 0.005; 693 observations). Other differences in reported frequency were cases of C. coli more frequently having drunk from a drinking water fountain (OR, 2.60; CI, 1.02 to 6.63; P = 0.046; 839 observations) and having experienced water supply problems (OR, 2.25; CI, 0.99 to 5.09; P = 0.052; 839 observations). Problems listed mainly involved discolored water and were almost all reported by residents of Wyre Local Authority in the rural component of the study population. The study was not large enough to discern reliable associations between exposures and individual C. coli sequence types.
DISCUSSION
Although the organism is of less clinical importance than C. jejuni, the incidence of C. coli in the study population (5.75 cases per 100,000) is still a significant burden of disease. We present strong evidence that cases of C. coli were more likely to be older and female than cases of C. jejuni, and this has been described in other studies (10), but the mechanism for this differential age distribution remains unclear. In fact, the epidemiologies of C. coli and C. jejuni, in terms of exposures recorded, appear remarkably similar in the population described.
Although C. coli is prevalent in surface water samples in this study and there is some evidence that problems with drinking water supply are more associated with C. coli infection than with C. jejuni, sequence typing has demonstrated that the C. coli isolates represent not only a different set of designated MLST types but also a genetically distinct (and in most cases distant) population from that described in human cases of disease. This is in contrast to results of other studies, where overlap of types between water and human cases has been reported for C. coli (6) and C. jejuni (28). Environmental waters can be considered a reservoir for Campylobacter strains from both wildlife and livestock hosts and may also be contaminated with strains from human sewage effluent (13, 14, 30). There is no epidemiological or microbiological evidence from this study linking human disease from C. coli with direct exposure to environmental waters, but the population of bacteria identified in surface waters is of interest.
The separation of C. coli strains from this study into three clades is in concordance with previous genetic analysis of all known C. coli sequence types (25), and the individual sequence types presented here align with these clades (S. Sheppard and N. McCarthy, personal communication). The close clustering of types from human cases in clade 1 relative to the other types presented (from water samples) may result in part from the intense sampling to date of this population in poultry, livestock, and humans. In comparison, relatively few types from environmental samples and other species have been described and, as this data set expands, similar clustering may emerge in other clades. However, clade 1 also represents a population with abundant opportunity for horizontal gene transfer (through close proximity of livestock and their contact with and consumption by humans), and we do not yet know enough about the ecology of C. coli outside this domain to predict whether that emergence might happen.
It could be argued that the absence of human sequence types in clades 2 and 3 (water samples) in this study may simply reflect an incomplete picture of the full range of types identified in human samples and that as more sequence typing of C. coli from human isolates is done, we may see a more diverse representation of clades in the clinical data. However, of the 17 new types identified in human samples in this study, all align with clade 1, suggesting a true separation of human and water populations. Of interest in this discussion are the intermediate types presented, all isolated from surface waters (including those most commonly isolated, ST-1766 and ST-1764) but aligned genetically more closely with clade 1 than the remaining water isolates. All these types are newly reported in this study, and although there is no evidence at present that they contribute to the burden of human disease, 7 of the 13 sequence types possess between one and three alleles that are also found in human isolates, in contrast to the other water samples, which share no alleles (data not shown). They may therefore represent avian or wildlife strains prevalent in the environment that have had greater opportunity to recombine in a common host with human/livestock strains than the other water sample sequence types in clades 2 and 3 and may yet be identified in livestock or food animals with further sampling of these populations. Recombination may occur where ecological and adaptive barriers are breached (25), and types in clades 2 and 3 could represent populations of C. coli that either predominate in species with little contact with livestock, domestic poultry, or humans or have reduced virulence for these species. Since these types were isolated from surface waters that are easily accessible to humans, companion animals, and livestock in both rural and suburban settings, a virulence barrier is a reasonable hypothesis, making these strains an interesting subject of further study.
There has been a suggestion that C. coli infection may be associated with consumption of pâté and meat pies (10), and C. coli has been described as being found in offal in the United Kingdom with higher prevalence than C. jejuni (22). United Kingdom food survey data suggest that the average quantity of bacon, ham, and pork products, liver, and meat pies purchased per individual over 65 years old is greater than for younger adults whereas there is no age difference for pâté (5). Our data confirmed that study cases older than 65 years old, in addition to being more likely to have C. coli than C. jejuni, were significantly more likely (than all other age groups) to have consumed pork, ham, or bacon, offal, beef, lamb, and meat pies prior to illness (data not shown). Consumption of pâté, however, showed no association with either age or species of Campylobacter.
However, despite C. coli commonly being reported as the most prevalent species in swine and pork meat (20, 21, 29, 31) and the associations described above, individuals with C. coli were less likely to have reported eating pork products prior to illness than those with C. jejuni. Although reliable associations between food consumption and individual sequence types were not possible, it is clear that the majority of types identified in humans in this study and previously identified in swine have also been identified in poultry (3, 8, 16, 20), and given the very high reporting of poultry consumption in this study (28), this route of infection seems the more likely. This is supported by a recent study of Danish isolates, demonstrating that only 9% of the C. coli sequence types described for humans were also described for pigs compared with 38% that were also described for poultry (17).
There is no strong evidence from this study of distinct exposures through food consumption or behavior reported by cases of C. jejuni and C. coli that could fully explain the different age and sex distribution of cases described. Where evidence of association is presented, the sample size of reported exposures cannot preclude association by chance, and the epidemiology and typing data together do not allow the confident proposal of a plausible hypothesis. The observed distribution may be mediated by factors related to host immunity or some mechanism of competitive colonization that is not yet understood.
New MLST sequence types of C. coli continue to be identified and published, and although the international MLST database is becoming more representative of the sources where types have been identified, it remains difficult to interpret epidemiology on the basis of sequence type alone. The demonstration in this study of the genetic difference between types commonly identified in humans and novel types in surface waters supports the continued application of genetic analysis to typing data in future studies as well as to existing data.
Acknowledgments
This work was supported by United Kingdom Food Standards Agency grant B14012. This publication made use of the Campylobacter jejuni MLST website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust.
We acknowledge the work of the local NHS laboratories in managing and making available the study area isolates from routine diagnostic work, colleagues from the United Kingdom Environment Agency for their collection of surface water samples, and Trudi Allen and Kevin Williamson from the Preston FEMS laboratory for the isolation of Campylobacter from these samples. In addition, we offer our thanks to Sam Sheppard and Noel McCarthy at the University of Oxford for comments on the manuscript.
There are no conflicts of interest.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Abulreesh, H. H., T. A. Paget, and R. Goulder. 2006. Campylobacter in waterfowl and aquatic environments: incidence and methods of detection. Environ. Sci. Technol. 40:7122-7131. [DOI] [PubMed] [Google Scholar]
- 2.Brown, P. E., O. F. Christensen, H. E. Clough, P. J. Diggle, C. A. Hart, S. Hazel, R. Kemp, A. J. Leatherbarrow, A. Moore, J. Sutherst, J. Turner, N. J. Williams, E. J. Wright, and N. P. French. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, S. A., V. M. Allen, G. Domingue, F. Jorgensen, J. A. Frost, R. Ure, R. Whyte, D. Tinker, J. E. Corry, J. Gillard-King, and T. J. Humphrey. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrique-Mas, J., Y. Andersson, M. Hjertqvist, A. Svensson, A. Torner, and J. Giesecke. 2005. Risk factors for domestic sporadic campylobacteriosis among young children in Sweden. Scand. J. Infect. Dis. 37:101-110. [DOI] [PubMed] [Google Scholar]
- 5.DEFRA. 5 December 2008, accession date. Expenditure and food survey. Purchased quantities of household food & drink by age of household reference person. https://statistics.defra.gov.uk/esg/publications/efs/datasets/ConsAGEHRPHH.xls.
- 6.Devane, M. L., C. Nicol, A. Ball, J. D. Klena, P. Scholes, J. A. Hudson, M. G. Baker, B. J. Gilpin, N. Garrett, and M. G. Savill. 2005. The occurrence of Campylobacter subtypes in environmental reservoirs and potential transmission routes. J. Appl. Microbiol. 98:980-990. [DOI] [PubMed] [Google Scholar]
- 7.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallay, A., H. De Valk, M. Cournot, B. Ladeuil, C. Hemery, C. Castor, F. Bon, F. Megraud, P. Le Cann, J. C. Desenclos, and the Outbreak Investigation Team. 2006. A large multi-pathogen waterborne community outbreak linked to faecal contamination of a groundwater system, France, 2000. Clin. Microbiol. Infect. 12:561-570. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, K. R. Neal, and Campylobacter Sentinel Surveillance Scheme. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huson, D., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 12.Jolley, K., and M.-S. Chan. 23 January 2009, accession date. Campylobacter jejuni and Campylobacter coli MLST home page. http://pubmlst.org/campylobacter/.
- 13.Jones, I. G., and M. Roworth. 1996. An outbreak of Escherichia coli O157 and campylobacteriosis associated with contamination of a drinking water supply. Public Health 110:277-282. [DOI] [PubMed] [Google Scholar]
- 14.Jones, K. 2001. Campylobacters in water, sewage and the environment. Symp. Ser. Soc. Appl. Microbiol. 2001:68S-79S. [DOI] [PubMed] [Google Scholar]
- 15.Kemp, R., A. J. Leatherbarrow, N. J. Williams, C. A. Hart, H. E. Clough, J. Turner, E. J. Wright, and N. P. French. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 71:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinana, A. D., E. Cardinale, I. Bahsoun, F. Tall, J. M. Sire, S. Breurec, B. Garin, C. Saad-Bouh Boye, and J. D. Perrier-Gros-Claude. 2007. Campylobacter coli isolates derived from chickens in Senegal: diversity, genetic exchange with Campylobacter jejuni and quinolone resistance. Res. Microbiol. 158:138-142. [DOI] [PubMed] [Google Scholar]
- 17.Litrup, E., M. Torpdahl, and E. M. Nielsen. 2007. Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 103:210-218. [DOI] [PubMed] [Google Scholar]
- 18.Martin, S., P. Penttinen, G. Hedin, M. Ljungstrom, G. Allestam, Y. Andersson, and J. Giesecke. 2006. A case-cohort study to investigate concomitant waterborne outbreaks of Campylobacter and gastroenteritis in Soderhamn, Sweden, 2002-3. J. Water Health 4:417-424. [PubMed] [Google Scholar]
- 19.McCarthy, N., Y. Andersson, V. Jormanainen, O. Gustavsson, and J. Giesecke. 1999. The risk of Guillain-Barre syndrome following infection with Campylobacter jejuni. Epidemiol. Infect. 122:15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 21.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, J. E., and R. H. Madden. 1998. Occurrence of thermophilic Campylobacter spp. in porcine liver in Northern Ireland. J. Food Prot. 61:409-413. [DOI] [PubMed] [Google Scholar]
- 23.Rosef, O., G. Rettedal, and L. Lageide. 2001. Thermophilic campylobacters in surface water: a potential risk of campylobacteriosis. Int. J. Environ. Health Res. 11:321-327. [DOI] [PubMed] [Google Scholar]
- 24.Schonberg-Norio, D., J. Takkinen, M. L. Hanninen, M. L. Katila, S. S. Kaukoranta, L. Mattila, and H. Rautelin. 2004. Swimming and Campylobacter infections. Emerg. Infect. Dis. 10:1474-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard, S. K., N. D. McCarthy, D. Falush, and M. C. Maiden. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237-239. [DOI] [PubMed] [Google Scholar]
- 26.Sopwith, W., M. Ashton, J. A. Frost, K. Tocque, S. O'Brien, M. Regan, and Q. Syed. 2003. Enhanced surveillance of Campylobacter infection in the North West of England 1997-1999. J. Infect. 46:35-45. [DOI] [PubMed] [Google Scholar]
- 27.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2006. Campylobacter jejuni multilocus sequence types in humans, northwest England, 2003-2004. Emerg. Infect. Dis. 12:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2008. Identification of potential environmentally adapted Campylobacter jejuni strain, United Kingdom. Emerg. Infect. Dis. 14:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakur, S., W. E. Morrow, J. A. Funk, P. B. Bahnson, and W. A. Gebreyes. 2006. Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl. Environ Microbiol. 72:5666-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vereen, E. J., R. R. Lowrance, D. J. Cole, and E. K. Lipp. 2007. Distribution and ecology of campylobacters in coastal plain streams (Georgia, United States of America). Appl. Environ. Microbiol. 73:1395-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright, S. L., D. K. Carver, R. M. Siletzky, S. Romine, W. E. Morrow, and S. Kathariou. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J. Food Prot. 71:1791-1796. [DOI] [PubMed] [Google Scholar]