Abstract
Cyprinid herpesvirus 3 (CyHV-3), a lethal DNA virus that spreads in natural lakes and rivers, infects common carp and koi. We established a quantification method for CyHV-3 that includes a viral concentration method and quantitative PCR combined with an external standard virus. Viral concentration methods were compared using the cation-coated filter and ultrafiltration methods. The recovery of virus-like particles was similar for the two methods (cation-coated filter method, 44% ± 19%, n = 3; ultrafiltration method, 50% ± 3%, n = 3); however, the former method was faster and more suitable for routine determinations. The recovery of seeded CyHV-3 based on the cation-coated filter method varied by more than 3 orders of magnitude among the water samples. The recovery yield of CyHV-3 was significantly correlated with that of the seeded λ phage, and the average ratio of λ to the CyHV-3 recovery yield was 1.4, indicating that λ is useful as an external standard virus for determining the recovery yield of CyHV-3. Therefore, to quantify CyHV-3 in environmental water, a known amount of λ was added as an external standard virus to each water sample. Using this method, CyHV-3 DNA was detected in 6 of the 10 (60%) types of environmental water tested; the highest concentration of CyHV-3 DNA was 2 × 105 copies liter−1. The lowest recovery limit of CyHV-3 DNA was 60 copies liter−1. This method is practical for monitoring CyHV-3 abundance in environmental water.
Cyprinid herpesvirus 3 (CyHV-3) is a lethal DNA virus that infects the common carp (Cyprinus carpio L.) and koi carp (C. carpio koi). The occurrence of the disease in the United Kingdom has been dated to 1996, following outbreaks in the United States, Israel, Europe, and South Asia (10), and has afflicted cultured ornamental and common carps, causing severe losses to fish breeders, retailers, and hobbyists (28). Therefore, the characterization and diagnosis of the disease have been the subject of intensive research (15). In recent years, the mortality of wild carp has been reported in natural freshwater environments (11, 18, 23). In Lake Biwa in Japan, 60 to 80% of the wild carp population (>100,000) died in 2004, presumably due to CyHV-3 infection (Shiga Prefectural Government, http://www.pref.shiga.jp/g/suisan-s/seika/files/seikah1711.pdf [in Japanese]) (18). The mass mortality of wild carp can directly and indirectly affect community composition and environmental ecosystems (18). Nevertheless, the occurrence of the disease and the means of transmission of CyHV-3 in the natural environment are still not well understood.
CyHV-3 is present in several organs of infected fish, such as the intestines, kidneys (7), and gills (29). CyHV-3 is also detected in droppings (3); therefore, infected fish are suspected of releasing CyHV-3 into natural waters. Seasonal variation and the spatial distribution of CyHV-3 may be important for understanding the transmission routes and mechanisms by which CyHV-3 spreads. However, the lack of a reliable method for quantifying CyHV-3 in environmental water precludes our elucidation of how this disease spreads.
In general, the concentration of a pathogen in environmental water is considerably lower than that found in host bodies. Therefore, a CyHV-3 concentration method is required to detect and quantify the virus in environmental water. Several methods have been developed for determining concentrations of viruses in water samples. Ultrafiltration can concentrate a pathogen from a large volume of water in <100 liters (27, 35). An alternative method involving the use of electronegative or electropositive microporous adsorbent filters has also been used to concentrate viruses from environmental water (1, 8). The mechanism of concentration in this method is based on electrostatic interactions. Haramoto et al. established a cation-coated filter method in which viruses that had been trapped were eluted with NaOH solution (pH 10.8) instead of the conventional solution, beef extract, which inhibits the PCR (12, 13). The concentrated viruses can then be used for PCR-mediated identification. Using this method, they succeeded in the qualitative detection of CyHV-3 DNA from river water samples (13).
Viral recovery during concentration is influenced by soluble organic compounds (33, 34) and salts (31) in the water, which may vary in each sample. Therefore, quantification of the viral DNA from concentrated environmental water samples has been difficult. Because sediments contain many substances that influence DNA recovery, Mumy and Findlay developed a method for the routine determination of DNA extraction efficiency using an external DNA recovery standard, as follows: λ DNA was added to sediments, the total DNA was extracted, and the amount of target DNA recovered was determined by quantitative PCR (22).
In this study, we established a method for quantifying CyHV-3 in environmental water using a viral concentration method and TaqMan PCR combined with an external standard virus. To choose a suitable viral concentration method, we compared the viral recovery yields between the ultrafiltration and cation-coated filter methods, and the procedure was modified to increase sensitivity. We then confirmed that the recovery yields of CyHV-3 and the external standard virus λ from different environmental waters throughout the procedure were positively correlated. Finally, we applied this method to environmental water samples taken from Lake Biwa and Takaragaike Pond in Japan at 3 years and 1 month, respectively, after an outbreak of the disease for the detection and quantification of CyHV-3.
MATERIALS AND METHODS
Water samples.
Natural environmental surface water samples were used for all experiments (Table 1). Water temperature and pH were measured in the field. The specific conductivity (SpCond), dissolved oxygen (DO) content, oxidation-reduction potential (ORP), concentration of ammonium nitrogen and ammonia nitrogen, and turbidity in the samples were measured using a YSI multimeter (YSI, Yellow Springs, OH). Samples used for measurement of total phosphorus (TP) and total nitrogen (TN) were stored at −20°C until analysis. The water samples used for measuring the chlorophyll a concentration were filtered using GF/F glass fiber filters (47-mm diameter; Whatman, Tokyo, Japan), and the filters were stored at −20°C until analysis.
TABLE 1.
Sampling sites and quality of water samples
| Use of samples | Sample name | Sampling site (Japan) | Sampling date | Water temp (°C) | pH | Levels of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chlorophyll a (μg/liter) | TP (μg/liter) | TN (μg/liter) | SpCond (mS/cm) | DO (mg/liter) | ORP (mV) | Ammonium N (mg/liter) | Ammonia N (mg/liter) | Turbidity (NTU)a | ||||||
| Comparison of viral concn methods | 1 | Effluent river of Lake Biwa, Seta River | 9 May 2007 | 20.3 | 7.5 | 1.8 | 26 | 554 | ||||||
| 2 | Reed zone of Lake Biwa, Honmaru | 16 May 2007 | 18.8 | 6.5 | 6.0 | 193 | 3,275 | |||||||
| 3 | Sand beach of Lake Biwa, Yanagasaki | 30 May 2007 | 23.7 | 8.1 | 9.8 | 57 | 508 | |||||||
| Testing VLP recovery with sample vol | 4 | Sand beach of Lake Biwa, Minamihama | 7 June 2007 | 19.6 | 7.4 | 5.6 | 62 | 1,001 | ||||||
| Recovery testing of CyHV-3 and λ | RT1 | Sand beach of Lake Biwa, Kitakomatsu | 13 September 2007 | 27.2 | 7.6 | 1.2 | 23 | 270 | 0.15 | 7.5 | 212 | 0.07 | 0.00 | 0.9 |
| RT2 | Sand beach of Lake Biwa, Adogawacho-Yokoehama | 13 September 2007 | 27.1 | 8.0 | 2.1 | 14 | 236 | 0.13 | 7.8 | 206 | 0.04 | 0.00 | 0.5 | |
| RT3 | Reed zone of Lake Biwa, Shin'asahicho-Harie | 13 September 2007 | 23.9 | 6.8 | 8.9 | 204 | 1,525 | 0.23 | 5.4 | 88 | 0.29 | 0.00 | 14.9 | |
| RT4 | Harbor of Lake Biwa, Osue | 13 September 2007 | 26.7 | 8.2 | 0.6 | 52 | 2,447 | 0.47 | 13.7 | 200 | 0.26 | 0.03 | 0.6 | |
| RT5 | Harbor of Lake Biwa, Choumeiji | 13 September 2007 | 27.3 | 7.9 | 6.2 | 29 | 698 | 0.16 | 7.7 | 192 | 0.06 | 0.00 | 1.5 | |
| RT6 | Reed zone of Lake Biwa, Honmaru | 13 September 2007 | 27.6 | 6.8 | 9.3 | 147 | 2,528 | 0.19 | 6.3 | 125 | 0.10 | 0.00 | 15.0 | |
| RT7 | Sand beach of Lake Biwa, Yanagasaki | 13 September 2007 | 27.7 | 8.9 | 9.9 | 52 | 376 | 0.14 | 7.2 | 176 | 0.04 | 0.02 | 3.7 | |
| RT8 | Yamato River | 30 January 2008 | 9.0 | 6.4 | 5.0 | 0.32 | 11.8 | 297 | 0.59 | 0.00 | 8.2 | |||
| Application of newly established method | A | Sand beach of Lake Biwa, Matsunoura | 4 July 2007 | 24.3 | 7.5 | 1.6 | 35 | 332 | ||||||
| B1 | Reed zone of Komatsu-numa lagoon | 4 July 2007 | 25.2 | 6.3 | 6.4 | 105 | 437 | |||||||
| B2 | Offshore of Komatsu-numa lagoon | 4 July 2007 | 24.5 | 16.2 | 82 | 599 | ||||||||
| C1 | Shore of Katata lagoon | 5 July 2007 | 25.3 | 6.8 | 2.0 | 117 | 1,883 | |||||||
| C2 | Offshore of Katata lagoon | 5 July 2007 | 24.9 | 6.5 | 9.8 | 105 | 1,330 | |||||||
| C3 | Effluent channel of Katata lagoon | 5 July 2007 | 26.0 | 7.7 | 2.1 | 31 | 405 | |||||||
| D1 | Reed zone of Konohama lagoon | 6 July 2007 | 28.0 | 7.2 | 2.2 | 57 | 569 | |||||||
| D2 | Offshore of Konohama lagoon | 6 July 2007 | 26.9 | 7.1 | 4.1 | 38 | 544 | |||||||
| D3 | Effluent channel of Konohama lagoon | 6 July 2007 | 25.7 | 7.8 | 2.4 | 17 | 276 | |||||||
| E | Shore of Takaragaike pond | 30 June 2008 | 25.7 | 7.3 | 15.3 | |||||||||
NTU, nephelometric turbidity units.
Analytical measurements.
TP was measured according to the method of Menzel and Corwin (19). TN was converted to nitrate by alkaline persulfate oxidation (H3BO3 was omitted from the oxidizing reagent) and was determined using a UV spectrophotometric screening method (1). The chlorophyll a concentration was determined by standard fluorometric methods (38).
Comparison of viral concentration methods.
We compared two viral concentration methods, the cation-coated filter and the ultrafiltration methods, using water samples taken from three different sites at Lake Biwa (Table 1). Twenty-three liters of water was prefiltered through a 3-μm-pore-size low-protein-binding cellulose acetate membrane filter (142-mm diameter, C300A142C; Advantec, Tokyo, Japan) and a 0.8-μm-pore-size low-protein-binding cellulose acetate membrane filter (142-mm diameter, C080A142C; Advantec) using stainless steel filter holders (KS-142-US; Advantec) to remove large plankton and particles. These filters were separated using an interleaving mesh sheet (91221217; Advantec). Twenty liters of the prefiltrate was used for ultrafiltration, and 2.2 liters of the prefiltrate was used for the cation-coated filter method.
Ultrafiltration was performed using the spiral cartridge system S10Y30 (30-kDa cutoff, 540642; Millipore, Billerica, MA) (36); 20 liters of the prefiltrate was concentrated to a volume of 200 ml. The cartridge was cleaned after use by flushing it with 4 liters of autoclaved water, followed by circulation with 0.01 N NaOH for 20 min and rinsing with 4 liters of autoclaved water. The cartridge was stored in 0.01 N NaOH at 4°C, according to the manufacturer's recommendation.
For the cation-coated filter method (12), a nitrocellulose filter with a 0.45-μm pore size and a 142-mm diameter (HAWP14250; Millipore) was placed in a plastic container of 250 mM AlCl3 to form a cation (Al3+)-coated filter. Then, 2.2 liters of prefiltered sample water was passed through the filter. The filter containing trapped viruses was rinsed with 1 liter of 0.5 mM H2SO4 (pH 3.0) to remove aluminum ions, followed by elution of viruses with 200 ml of 1.0 mM NaOH (pH 10.8). To determine the relationship between the recovery yield of the viruses and the volume of the NaOH eluate, samples eluted with 1.0 mM NaOH (pH 10.8) were recovered four times in 50 ml, totaling 200 ml. The filtrates were neutralized by adding 1 ml of 100 mM H2SO4 (pH 1.0) and 2 ml of 100× Tris-EDTA buffer (pH 8.0).
To evaluate the recovery yields of native virus particles, virus-like particles (VLPs) in water samples before and after viral concentration were enumerated under an epifluorescence microscope using the SYBR green I staining method (25). Moreover, VLPs in filtrates that did not attach to the cation-coated filter or were not recovered using the spiral cartridge were collected; the VLPs in the rinse solution (0.5 mM H2SO4) were counted to estimate the amount of viral particles that did not bind during each concentration procedure.
To determine the relationship between VLP recovery yields and sample water volume, 0.3-, 1-, 3-, and 10-liter water samples were processed using the cation-coated filter method. VLP abundance in the prefiltrate was normalized to 100%. The experiments were conducted in duplicate.
Standard viruses.
CyHV-3 was propagated using the common carp brain (CCB) cell line, which was established from carp (Cyprinus carpio) brain tissues (24), and then purified via sucrose gradient ultrafiltration. Because this virus causes a disease recognized under the Law to Ensure Sustainable Aquaculture Protection in Japan, the viruses were inactivated with 0.3% formaldehyde to prevent their spread. No significant difference in recovery yields between raw and inactivated CyHV-3 has been confirmed in our preliminary experiment. The external standard virus λ (Promega, Tokyo, Japan) was propagated in Escherichia coli LE 392 cells (17) and purified by the polyethylene glycol method (30). The densities of the viruses were counted under an epifluorescence microscope using the SYBR green I staining method (25). CyHV-3 and λ stock solutions were stored at 4°C.
Sample preparation for CyHV-3 and λ recovery yield tests.
To determine the relationship between the recovery of CyHV-3 and λ after DNA extraction using the cation-coated filter method, CyHV-3 and λ were added directly to the water samples to a final concentration of 1 × 107 VLPs liter−1. We used 4-liter water samples taken from seven different sampling sites at Lake Biwa (no replicates) and from one sampling site at the Yamato River (five replicates) (Table 1). To evaluate native CyHV-3 and λ concentrations, we prepared water samples with no added CyHV-3 and λ. For λ, water samples were collected from sites RT1 and -3 on 22 September 2009 and from sites RT5 and -7 on 24 September 2009.
Sample preparation for application of the established method to environmental samples.
To determine the concentration of native CyHV-3 in environmental water, a known amount of external λ particles was added to each water sample to a final concentration of 1 × 107 VLPs liter−1. We used 4 liters of water samples taken from 10 different sampling sites (Table 1).
Viral concentration by the cation-coated filter method and polyethylene glycol precipitation.
The water samples were prefiltered, and the viral particles were concentrated using the cation-coated filter method to a final volume of 200 ml, as described above. The water samples were then reconcentrated to reduce the volume by precipitation with polyethylene glycol 6000 (PEG 6000) (16). Briefly, NaCl and PEG 6000 were added to a final concentration of 0.4 M and 8% (wt/vol), respectively. The resulting suspension was stirred for 1 h at 4°C and then incubated overnight at 4°C. The extract was centrifuged at 10,000 × g for 1 h, and the pellet was resuspended in 10 ml of SMC buffer (50 mM Tris-HCl [pH 7.0], 0.1 M NaCl, 8 mM MgSO4, 1 mM CaCl2, 0.01% [wt/vol] gelatin).
DNA extraction and purification.
Viral DNA was extracted using proteinase K and sodium dodecyl sulfate (SDS) (30), purified by phenol-chloroform extraction and ethanol precipitation (30), and further purified with the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The final volume of the DNA solution was 100 μl.
Real-time quantitative TaqMan PCR.
The extracted DNA samples were used as templates for TaqMan PCR amplification, with specific primer pairs and probe sets for CyHV-3 (7) and λ. λ-Specific primer pairs and a probe (Table 2) were commercially designed by Applied Biosystems (Tokyo, Japan) based on the enterobacterial λ phage complete genome sequence (GenBank accession no. J02459). The amplification and quantification of target DNA were performed with an ABI StepOnePlus real-time PCR system (Applied Biosystems). Each TaqMan reaction contained 900 nM primer and 125 nM TaqMan probe in a commercially available PCR mastermix (TaqMan gene expression mastermix; Applied Biosystems) and 2 μl of the extracted DNA sample (except for 5 μl of CyHV-3 in the application test of environmental samples) in a final volume of 20 μl. The PCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 60 s at 60°C. Three wells were used for the quantification of one sample. Standard genomic CyHV-3 and λ DNAs were extracted using the DNeasy blood and tissue kit (Qiagen). Quantification was performed using a NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). A dilution series of standard whole-genomic CyHV-3 DNA containing 1 × 100 to 3 × 107 copies per tube was amplified in triplicate using TaqMan PCR. Because one copy of the genomic DNA was detected in at least one well in the triplicate, we defined the detection limit of CyHV-3 DNA using TaqMan PCR as one copy.
TABLE 2.
Primers and probe specific for a newly developed real-time TaqMan PCR to detect the target λ
| Primer/probe | Sequence (5′→3′) | Length (bp) |
|---|---|---|
| Primers | ||
| Lambda-7184F | TTCTCTGTGGAGGAGTCCATGAC | 84 |
| Lambda-7267R | GCTGACATCACGGTTCAGTTGT | |
| Probe lambda-7210P | AGATGAACTGATTGCCCGTCTCCGCT |
Positive PCR products were excised from agarose gels and extracted using a QIAquick PCR purification kit (Qiagen). They were then cloned into the pGEM-T Easy vector (Promega). Sequences were determined commercially with M13F and M13R primers.
RESULTS
Comparison of VLP recovery between the cation-coated filter and ultrafiltration methods.
The recovery yield of VLPs using the cation-coated filter method (44% ± 19% of prefiltrate, n = 3) was similar to that of the ultrafiltration method (50% ± 3% of prefiltrate, n = 3) (Table 3). Some viruses were lost during cation-coated filtration due to failed adsorption to the filter or incomplete recovery from the filter. Similarly, viruses lost during ultrafiltration were due to incomplete recovery from the ultrafiltration unit (43% ± 3% of prefiltrate, n = 3).
TABLE 3.
Comparison of VLP recovery and loss yields in environmental waters using the cation-coated filter and ultrafiltration methodsc
| Sample name | Results obtained with indicated methods (%) |
||||||
|---|---|---|---|---|---|---|---|
| Cation-coated filter |
Ultrafiltration |
||||||
| Recovery of concentrate | Loss of: |
Recovery of concentrate | Loss of: |
||||
| Filtrate | H2SO4a | Unrecovered residue | Filtrate | Unrecovered residue | |||
| 1 | 64 | 48 | 0.00 | −12 | 46 | 10 | 44 |
| 2 | 43 | NAb | 0.04 | NA | 51 | 4 | 45 |
| 3 | 25 | 20 | 0.04 | 56 | 52 | 9 | 39 |
| Average results | 44 | 0.03 | 50 | 8 | 43 | ||
| SD | 19 | 0.02 | 3 | 3 | 3 | ||
H2SO4 indicates a rinse solution of 0.5 mM H2SO4.
NA, data are not available.
VLP abundance in prefiltrated water was set to 100%.
Relationship between the recovery yield of VLPs and elution water volume from cation-coated filters.
The initial 100 ml of NaOH solution eluted 37.3% of the VLPs in the prefiltrate, and an additional 100 ml of the solution eluted 1.5% of the VLPs (Fig. 1). The pH of the eluted NaOH solution was 10.8 (Fig. 1). Therefore, 200 ml of NaOH eluant was sufficient for the recovery of adsorbed viruses from the filter.
FIG. 1.
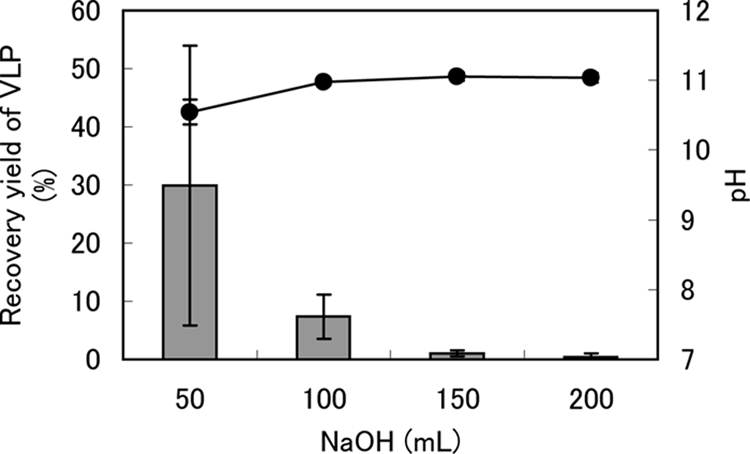
Relationship between the VLP recovery yield (%) and NaOH eluate volume using the cation-coated filter method. The gray bars and black circles indicate VLP recovery yields and pHs in NaOH eluant, respectively. VLP abundance in prefiltered water was normalized to 100%. The error bars indicate the standard errors of the means (n = 3).
Relationship between the VLP recovery yield and sample volume using the cation-coated filter method.
The highest recovery yield of VLPs was obtained from a 300-ml sample (Fig. 2A). Water samples ranging from 500 ml to 10 liters demonstrated reproducible recovery yields. The highest abundance of VLPs was recovered from the largest volume of samples (10 liters) (Fig. 2B).
FIG. 2.
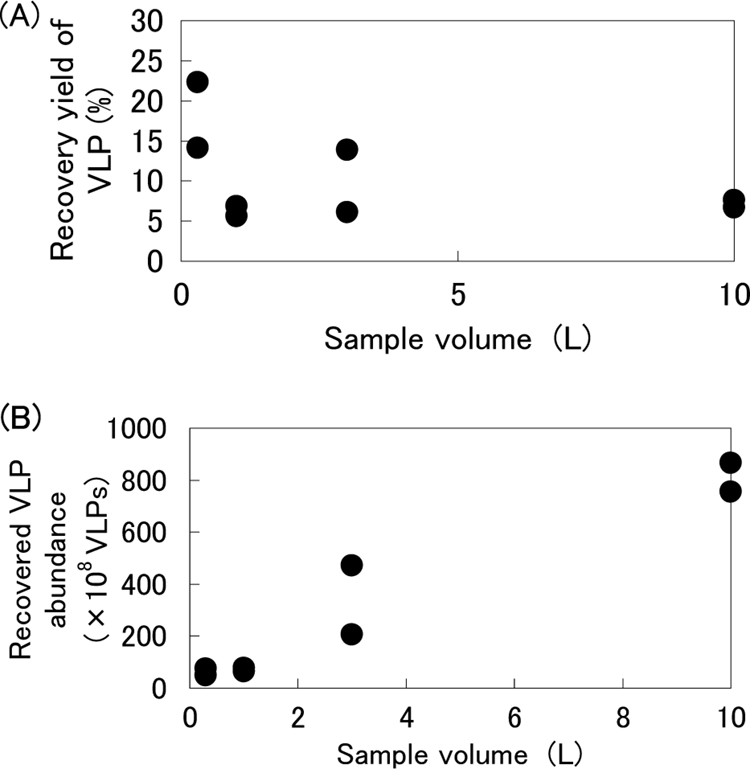
(A) Relationship between the VLP recovery yield (%) and sample water volume using the cation-coated filter method. VLPs in prefiltered water were normalized to 100%. L, liters. (B) Relationship between recovered VLP abundance and sample water volume using the cation-coated filter method.
Recovery yield of CyHV-3 versus that of λ from environmental water.
Recovery yields of CyHV-3 and λ throughout the procedure were determined by TaqMan PCR. This test was conducted using environmental water samples (Table 1). The pH range of these water samples was 6.4 to 8.9. The native CyHV-3 and λ concentrations in environmental water samples were sufficiently low (0.1 to 8.1% and below 0.14% of the added concentration, respectively) (data not shown).
The recovery yields of CyHV-3 (0.03 to 5.5%) and λ (0.001 to 7.7%) varied by more than 3 orders of magnitude among the samples (Table 4). The average concentration efficiencies of CyHV-3 and λ were 2,182 and 3,097, respectively, and both viruses were concentrated effectively in almost all of the samples. However, in a sample taken from site RT6, the concentration efficiency of λ was below 1 (which corresponds to 0.002% of recovery yield), indicating that λ was not concentrated. When data except for those obtained from the RT6 sample were used, the recovery yields of CyHV-3 and λ were highly correlated (P < 0.01, r = 0.79) (Fig. 3), and the average ratio of the recovery yields of λ to CyHV-3 was 1.4 (Table 4). The variation in the recovery yield among sampling sites (0.03 to 5.5%; n = 7) was larger than that among the samples themselves (0.4 to 1.1%, n = 5) (Table 4). No correlations were found between the recoveries of CyHV-3 and any environmental factors, including water temperature, pH, chlorophyll a, TP, TN, SpCond, DO, ORP, ammonium nitrogen, ammonia nitrogen, and turbidity.
TABLE 4.
Recovery yields and concentration efficiencies of seeded CyHV-3 and λ in environmental waters and the λ/CyHV-3 ratio
| Sample name | Recovery yields (%) |
Concn efficiency |
λ/CyHV-3 ratio | ||
|---|---|---|---|---|---|
| CyHV-3 | λ | CyHV-3 | λ | ||
| RT1 | 5.5 | 2.8 | 2,182 | 1,124 | 0.5 |
| RT2 | 4.7 | 7.7 | 1,878 | 3,097 | 1.6 |
| RT3 | 0.6 | 0.2 | 253 | 94 | 0.4 |
| RT4 | 0.7 | 0.9 | 261 | 361 | 1.4 |
| RT5 | 0.03 | 0.01 | 13 | 5 | 0.4 |
| RT6 | 0.13 | 0.001 | 51 | 0.5 | 0.01 |
| RT7 | 0.4 | 0.1 | 150 | 31 | 0.2 |
| RT8-1a | 0.4 | 1.0 | 171 | 395 | 2.3 |
| RT8-2 | 0.7 | 1.3 | 290 | 521 | 1.8 |
| RT8-3 | 0.3 | 0.4 | 104 | 163 | 1.6 |
| RT8-4 | 1.1 | 1.7 | 440 | 695 | 1.6 |
| RT8-5 | 0.5 | 1.7 | 182 | 688 | 3.8 |
| Avg | 1.2 | 1.5 | 498 | 598 | 1.4b |
| SD | 1.8 | 2.1 | 728 | 859 | 1.1 |
RT8-1, -2, -3, -4, and -5 indicate 5 replicates of sample water at site RT8.
The average was calculated from data excluding those from site RT6, for which the λ concentration efficiency was below 1.
FIG. 3.
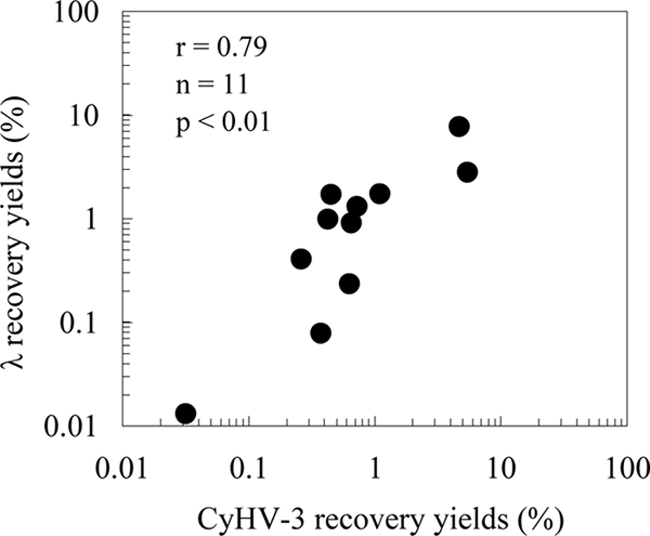
Relationship between the recovery yields (%) of seeded CyHV-3 and λ in environmental water. Data for sample RT6 were omitted because its concentration efficiency was below 1.
Application of the CyHV-3 quantification method to environmental water samples.
CyHV-3 DNA was detected successfully from 6 of the 10 (60%) environmental water samples; these samples were collected from sites B1, B2, C1, C2, D3, and E using our established method (Table 5). Positive PCR products were confirmed as the target sequence. Results from the calculation using recovery yields of λ and the average ratio of the recovery yields of λ to CyHV-3 (1.4) (Table 4) showed that the highest concentration was 2 × 105 copies liter−1 (Table 5). The lowest recovery limit for CyHV-3 DNA was 60 copies liter−1 in sample A, which was collected from Lake Biwa. Recovery yields of the external standard λ ranged from 0.005 to 11.5% in the samples. The recovery yields in 2 of the 10 samples were lower than 0.01%, which is below the recovery yield range in which the significant correlation between CyHV-3 and λ was confirmed (Fig. 3). Therefore, we did not calculate the detection limits or concentrations of CyHV-3 DNA in these samples.
TABLE 5.
Recovery yields of λ as the external standard virus, CyHV-3 concentration in environmental waters, and detection limits of CyHV-3 DNA
| Sampling site | Sample name | λ recovery yield (%) | CyHV-3 DNA concn in environmental water (copies/liter)a | Detection limit of CyHV-3 DNA (copies/liter)b |
|---|---|---|---|---|
| Lake Biwa | A | 11.5 | —c | 6 × 101 |
| Komatsu-numa | B1 | 0.05 | 2 × 105 | 1 × 104 |
| lagoon | B2 | 0.005 | NAd | NAe |
| Katata lagoon | C1 | 2.2 | >3 × 102f | 3 × 102 |
| C2 | 0.2 | 6 × 104 | 3 × 103 | |
| C3 | 4.3 | — | 2 × 102 | |
| Konohama lagoon | D1 | 0.001 | — | NAe |
| D2 | 0.04 | — | 2 × 104 | |
| D3 | 5.5 | >1 × 10 2f | 1 × 102 | |
| Takaragaike pond | E | 2.4 | 5 × 104 | 3 × 102 |
CyHV-3 concentration was calculated as follows: CyHV-3 DNA copy number in extracted DNA/[recovery yield of λ (%)/ratio (1.4) of recovery yield of λ to CyHV-3/100]/initial sample volume (liters).
We assumed that the detection limit by TaqMan PCR was 1 copy per well.
—, all three wells showed a negative signal by TaqMan PCR.
The result obtained from TaqMan PCR was positive, but the CyHV-3 concentration could not be calculated because the recovery yield of λ was below 0.01%.
The recovery yield of λ was below 0.01%.
One or two of the three wells showed a positive signal by TaqMan PCR.
DISCUSSION
In this study, we established a routine quantification method for CyHV-3 in environmental water. This was accomplished using a rapid viral concentration method and quantitative PCR combined with an external standard virus. The viral concentration method incorporated the use of a cation-coated filter that was modified to be more sensitive and suitable for highly turbid water samples. Addition of λ to water samples allowed us to determine the recovery yield of CyHV-3 and to calculate the abundance of CyHV-3 in environmental water samples. Using this method, we detected CyHV-3 DNA in 6 of the 10 environmental waters sampled; the highest concentration of viral DNA measured was 2 × 105 copies liter−1 (Table 5).
We compared the VLP recovery yields obtained with two major viral concentration methods, i.e., the cation-coated filter method and the ultrafiltration method. The average recovery yields obtained with the two methods were similar (Table 3). However, the ultrafiltration method had an increased propensity for losing CyHV-3 due to adherence to the ultrafiltration unit; 43% of the VLPs in the sample water were not recoverable from the ultrafiltration unit (Table 3). This could also increase the chances for contamination by CyHV-3 in the subsequent samples subjected to ultrafiltration. Conversely, the risk of cross-contamination among samples was minimized using the cation-coated filter method because the filter was changed after each sample application. Moreover, the ultrafiltration method is time-consuming due to the requirement of cleaning steps before the concentration of the sample (26). Therefore, we chose the cation-coated filter method for monitoring the concentration of CyHV-3 in environmental water.
We used a cation-coated filter method (12), with some modifications. At several points, the protocol was adapted for testing turbid samples and to increase the detection sensitivity; carp often exist in eutrophic areas containing large amounts of suspended solids. First, the samples were prefiltered to remove large particles (>0.8 μm) prior to filtration through the cation-coated filter. This effectively shortened the filtration time through the cation-coated filter and increased the maximum filterable water volume, especially for samples containing large amounts of suspended solids that can promote clogging. Second, the diameter of the filter and the volume of the sample were increased from 90 mm to 142 mm and from 500 ml to 4 liters, respectively. Sample water volumes could be extended from 4 to 10 liters without a decrease in viral recovery yield (Fig. 2). We also tested the relationship between elution efficiency and the volume of the NaOH eluant; 200 ml of NaOH eluant was sufficient for recovery of adsorbed viruses from the filter, and the initial 100 ml of elution solution had a pH of over 10.8 and contained almost all VLPs compared to those of the total 200 ml of elution solution (Fig. 1). Reconcentration of viruses by PEG precipitation instead of ultrafiltration was performed, because we used a filter with a larger diameter than that used by Haramoto et al. (12, 13) and the volume of the eluate increased correspondingly. We used a DNA solution equivalent to 200 ml of the original water sample, which was about 20 times larger than that used to detect CyHV-3 in the Tama River by Haramoto et al. (13), as a template for the PCRs. Additionally, we performed phenol-chloroform extraction and ethanol precipitation (30) before using the commercial DNA extraction kit; the recovery yield of DNA increased after this preparatory step (data not shown).
The total recovery yield of CyHV-3 (1.2%) (Table 4) obtained using our procedure was similar to the recovery yield (1.6%) obtained with a cation-coated filter concentration step, which was recently reported by Haramoto et al. (14). This suggested that loss of CyHV-3 throughout our whole procedure occurred mainly in the cation-coated filter concentration step, not in following steps of reconcentration by PEG precipitation and DNA extraction.
The recovery yield of CyHV-3 from samples taken from different sites differed by at least 3 orders of magnitude (Table 4 and Fig. 3). Moreover, the variation among replicates from the same water sample was smaller than that among different water samples (Table 4), suggesting that water quality affected the recovery yield. In this study, recovery yield depended on recovery at each step of the procedure, including cation-coated filtration, reconcentration by PEG precipitation, and DNA extraction. Many factors influence viral adsorption to the filter during viral concentration and DNA extraction, including pH, salinity, soluble organic compounds, and particulate matter (9, 33, 34). These factors may explain why we observed no relationship between CyHV-3 recovery yield and environmental factors in the water samples.
In a sample taken from site RT6, the concentration efficiency of λ was below 1, indicating that λ was not concentrated. Although aggregation of the virus was observed via epifluorescence microscopy, we are uncertain as to what caused aggregation in this sample. Moreover, in water samples taken from the reed zone at this site, turbidity, chlorophyll a concentration, and TP and TN concentrations tended to be higher than those in samples collected at the sand beach and harbor (Table 1). Therefore, our method was less effective in concentrating the virus in water samples that contained substances with an aggregating effect or many suspended particles.
Despite the variability of CyHV-3 recovery among the water samples, the CyHV-3 recovery yield was correlated with the external λ standard (Fig. 3). Therefore, we conclude that λ is a suitable external standard virus for the estimation of CyHV-3 recovery yield. The approach of adding a known amount of λ as an external standard prior to viral concentration and DNA extraction provided an efficient and reliable way to evaluate the concentration efficiency of each individual sample in a single tube, and quantification of CyHV-3 in environmental water was accomplished using this approach, even though the concentration efficiencies varied among samples. There is no need to increase the number of samples and no risk of contamination of the added target virus to the sample. Many studies have examined the detection of waterborne pathogenic viruses in environmental water through viral concentration (2, 4, 6, 12), and the recovery yields were assumed to be consistent. The application of the external standard method to quantify the viruses could provide more accurate estimations, especially in the case of a high variability in recovery yield.
Using our established method, we have succeeded in detecting CyHV-3 not only in Takaragaike Pond, where an outbreak occurred 1 month before sampling, but also in lagoons of Lake Biwa, where an outbreak occurred 3 years before sampling. In Lake Biwa and its lagoons, the outbreak of CyHV-3 disease occurred in 2004 and killed about 100,000 carp. Since then, no further mass mortality has been observed. However, surprisingly high concentrations of CyHV-3 DNA were detected in samples from lagoons in 2007 (Table 5) and were comparable to concentrations (5 × 104 copies liter−1) (Table 5) in Takaragaike Pond, where an outbreak occurred 1 month before sampling. The quantitative method established in this study revealed that a similar level (average level, 103 to 104 copies liter−1) of CyHV-3 DNA existed in a river where mass mortality occurred due to the disease (20). These data suggest that the disease caused by CyHV-3 occurred in and around Lake Biwa in 2007. Indeed, several carp dead from the disease have been found every year in Lake Biwa and its lagoons (Shiga Prefectural Fisheries Experiment Station, unpublished data). Therefore, with the endemic occurrence of CyHV-3 disease in a population, infectious CyHV-3 released into the water by diseased carp may cause subsequent infection. A possible explanation for the fact that no subsequent acute outbreak occurred in the host population in Lake Biwa is that most of the population had likely acquired resistance to CyHV-3 disease, although CyHV-3 was abundant in the water. The fact that 54% of fish larger than 300 mm were positive for the antibody (37) supported this explanation. Another explanation is that the abundance or virulence of CyHV-3 released from infected carp during the endemic occurrence of an infectious disease in a population was lower or weaker, respectively, than that from infected carp during an acute clinical outbreak in the host population. Moreover, our results show that CyHV-3 DNA was distributed heterogeneously (Table 5), suggesting the possibility that viral “hot spots” can exist. Quantitative monitoring of CyHV-3 DNA in environmental water could be helpful in investigating the occurrence of this disease in natural environmental ecosystems (21).
The infectivity of CyHV-3 has been reported to decline within 3 days in intact environmental water (32). Although genetic methods can also detect inactive virus, such methods correlate with the detection of viable viruses when used with samples in natural waters and poliovirus (5). The transmission mechanisms of CyHV-3 infection among wild carp in environmental ecosystems are not clear. Investigations concerning the distribution and seasonal dynamics of CyHV-3 in environmental waters using the method described herein would be helpful for understanding when, where, and how CyHV-3 spreads and could be used to predict viral outbreaks.
Acknowledgments
We thank T. Itayama and N. Tanaka for valuable comments and N. Ibuki and the members of the C-06 research project at the Research Institute for Humanity and Nature (RIHN) for their assistance. We thank the Shiga Prefectural Fisheries Experiment Station for providing information on carp that had died from CyHV-3 disease in Lake Biwa and its lagoons. We also thank the Center for Ecological Research for allowing us to use their laboratory and equipment as affiliated scientists.
This work was supported by the RIHN (C-06 project). M.N.H. was supported by a Grant-in-Aid for Young Scientists (B) (grant 20710013) from the Ministry of Education, Science, Sports, and Culture, Japan.
Footnotes
Published ahead of print on 13 November 2009.
REFERENCES
- 1.American Public Health Association. 1998. Detection of enteric viruses. In Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 2.Brooks, H. A., R. M. Gersberg, and A. K. Dhar. 2005. Detection and quantification of hepatitis A virus in seawater via real-time RT-PCR. J. Virol. Methods 127:109-118. [DOI] [PubMed] [Google Scholar]
- 3.Dishon, A., A. Perelberg, J. Bishara-Shieban, M. Ilouze, M. Davidovich, S. Werker, and M. Kotler. 2005. Detection of carp interstitial nephritis and gill necrosis virus in fish droppings. Appl. Environ. Microbiol. 71:7285-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 5.Enriquez, C. E., M. Abbaszadegan, I. L. Pepper, K. J. Richardson, and C. P. Gerba. 1993. Poliovirus detection in water by cell culture and nucleic acid hybridization. Water Res. 27:1113-1118. [Google Scholar]
- 6.Gersberg, R. M., M. A. Rose, R. Robles-Sikisaka, and A. K. Dhar. 2006. Quantitative detection of hepatitis A virus and enteroviruses near the United States-Mexico border and correlation with levels of fecal indicator bacteria. Appl. Environ. Microbiol. 72:7438-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilad, O., S. Yun, F. J. Zagmutt-Vergara, C. M. Leutenegger, H. Bercovier, and R. P. Hedrick. 2004. Concentrations of a koi herpesvirus (CyHV-3) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Organ. 60:179-187. [DOI] [PubMed] [Google Scholar]
- 8.Goyal, S. M., and C. P. Gerba. 1982. Concentration of viruses from water by membrane filters, p. 59-119. In C. P. Gerba and S. M. Goyal (ed.), Methods of environmental virology. Marcel Dekker, New York, NY.
- 9.Guttman-Bass, N., and J. Catalano-Sherman. 1985. Effects of humic materials on virus recovery from water. Appl. Environ. Microbiol. 49:1260-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haenen, O. L. M., K. Way, S. M. Bergmann, and E. Ariel. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24:293-307. [Google Scholar]
- 11.Hara, H., H. Aikawa, K. Usui, and T. Nakanishi. 2006. Outbreaks of koi herpesvirus disease in rivers of Kanagawa Prefecture. Fish Pathol. 41:81-83. (In Japanese.) [Google Scholar]
- 12.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2005. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 71:2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haramoto, E., M. Kitajima, H. Katayama, and S. Ohgaki. 2007. Detection of koi herpesvirus DNA in river water in Japan. J. Fish Dis. 30:59-61. [DOI] [PubMed] [Google Scholar]
- 14.Haramoto, E., M. Kitajima, H. Katayama, T. Ito, and S. Ohgaki. 2009. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 32:297-300. [DOI] [PubMed] [Google Scholar]
- 15.Ilouze, M., A. Dishon, and M. Kotler. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol. Mol. Biol. Rev. 70:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, G. D., and T. G. Metcalf. 1988. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 54:1983-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 18.Matsui, K., M. Honjo, Y. Kohmatsu, K. Uchii, R. Yonekura, and Z. Kawabata. 2008. Detection and significance of koi herpesvirus (CyHV-3) in freshwater environments. Freshw. Biol. 53:1262-1272. [Google Scholar]
- 19.Menzel, D. W., and N. Corwin. 1965. The measurement of total phosphorus in seawater based on the liberation of organically bound fractions by persulfate oxidation. Limnol. Oceanogr. 10:280-282. [Google Scholar]
- 20.Minamoto, T., M. N. Honjo, K. Uchii, H. Yamanaka, A. A. Suzuki, Y. Kohmatsu, T. Iida, and Z. Kawabata. 2009. Detection of cyprinid herpesvirus 3 DNA in river water during and after an outbreak. Vet. Microbiol. 135:261-266. [DOI] [PubMed] [Google Scholar]
- 21.Minamoto, T., M. N. Honjo, and Z. Kawabata. 2009. Seasonal distribution of cyprinid herpesvirus 3 in Lake Biwa, Japan. Appl. Environ. Microbiol. 75:6900-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mumy, K. L., and R. H. Findlay. 2004. Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. J. Microbiol. Methods 57:259-268. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi, N., M. Furuno, and S. Tanaka. 2007. Outbreaks of koi herpesvirus disease in Mie Prefecture. Bull. Fish. Res. Div. Mie Pref. 15:23-27. (In Japanese.) [Google Scholar]
- 24.Neukirch, M., K. Bottcher, and S. Bunnajirakul. 1999. Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19:221-224. [Google Scholar]
- 25.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 26.Olszewski, J., L. Winona, and K. H. Oshima. 2005. Comparison of 2 ultrafiltration systems for the concentration of seeded viruses from environmental waters. Can. J. Microbiol. 51:295-303. [DOI] [PubMed] [Google Scholar]
- 27.Paul, J. H., S. C. Jiang, and J. B. Rose. 1991. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, Y. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Israeli J. Aquaculture-Bamidgeh 55:5-12. [Google Scholar]
- 29.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steinitz, A. Perelberg, D. Soffer, and M. Kotler. 2004. Pathogenesis of acute viral disease induced in fish by carp interstitial nephritis and gill necrosis virus. J. Virol. 78:9544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shields, P. A., and S. R. Farrah. 1983. Influence of salts on electrostatic interactions between poliovirus and membrane filters. Appl. Environ. Microbiol. 45:526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu, T., N. Yoshida, H. Kasai, and M. Yoshimizu. 2006. Survival of koi herpesvirus (KHV) in environmental water. Fish Pathol. 41:153-157. [Google Scholar]
- 33.Sobsey, M. D., and J. S. Glass. 1984. Influence of water quality on enteric virus concentration by microporous filter methods. Appl. Environ. Microbiol. 47:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobsey, M. D., and A. R. Hickey. 1985. Effects of humic and fulvic acids on poliovirus concentration from water by microporous filtration. Appl. Environ. Microbiol. 49:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soule, H., O. Genoulaz, B. Gratacap-Cavallier, P. Chevallier, J.-X. Liu, and J.-M. Seigneurin. 2000. Ultrafiltration and reverse transcription-polymerase chain reaction: an efficient process for poliovirus, rotavirus, and hepatitis A virus detection in water. Water Res. 34:1063-1067. [Google Scholar]
- 36.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1991. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 57:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchii, K., K. Matsui, T. Iida, and Z. Kawabata. 2009. Distribution of the introduced cyprinid herpesvirus 3 in a wild population of common carp, Cyprinus carpio L. J. Fish Dis. 32:857-864. [DOI] [PubMed] [Google Scholar]
- 38.Wetzel, R. G., and G. E. Likens. 2000. Limnological analysis, 3rd ed. Springer Verlag, Berlin, Germany.


