Abstract
d-Galacturonic acid can be obtained by hydrolyzing pectin, which is an abundant and low value raw material. By means of metabolic engineering, we constructed fungal strains for the conversion of d-galacturonate to meso-galactarate (mucate). Galactarate has applications in food, cosmetics, and pharmaceuticals and as a platform chemical. In fungi d-galacturonate is catabolized through a reductive pathway with a d-galacturonate reductase as the first enzyme. Deleting the corresponding gene in the fungi Hypocrea jecorina and Aspergillus niger resulted in strains unable to grow on d-galacturonate. The genes of the pathway for d-galacturonate catabolism were upregulated in the presence of d-galacturonate in A. niger, even when the gene for d-galacturonate reductase was deleted, indicating that d-galacturonate itself is an inducer for the pathway. A bacterial gene coding for a d-galacturonate dehydrogenase catalyzing the NAD-dependent oxidation of d-galacturonate to galactarate was introduced to both strains with disrupted d-galacturonate catabolism. Both strains converted d-galacturonate to galactarate. The resulting H. jecorina strain produced galactarate at high yield. The A. niger strain regained the ability to grow on d-galacturonate when the d-galacturonate dehydrogenase was introduced, suggesting that it has a pathway for galactarate catabolism.
d-Galacturonate is the main component of pectin, an abundant and cheap raw material. Sugar beet pulp and citrus peel are both rich in pectin residues. At present, these residues are mainly used as cattle feed. However, since energy-consuming drying and pelletizing of the residues is required to prevent them from rotting, it is not always economical to process the residues, and it is desirable to find alternative uses.
Various microbes which live on decaying plant material have the ability to catabolize d-galacturonate using various, completely different pathways (19). Eukaryotic microorganisms use a reductive pathway in which d-galacturonate is first reduced to l-galactonate by an NAD(P)H-dependent reductase (12, 17). In the following steps a dehydratase, aldolase, and reductase convert the l-galactonate to pyruvate and glycerol (9, 11, 14).
In Hypocrea jecorina (anamorph Trichoderma reesei) the gar1 gene codes for a strictly NADPH-dependent d-galacturonate reductase. In Aspergillus niger a homologue gene sequence, gar2, exists; however, a different gene, gaaA, is upregulated during growth on d-galacturonate containing medium (16). The gaaA codes for a d-galacturonate reductase with different kinetic properties than the H. jecorina enzyme, having a higher affinity toward d-galacturonate and using either NADH or NADPH as cofactor. It is not known whether gar2 codes for an active protein.
Some bacteria, such as Agrobacterium tumefaciens or Pseudomonas syringae, have an oxidative pathway for d-galacturonate catabolism. In this pathway d-galacturonate is first oxidized to meso-galactarate (mucate) by an NAD-utilizing d-galacturonate dehydrogenase. Galactarate is then converted in the following steps to α-ketoglutarate. This route is sometimes called the α-ketoglutarate pathway (20). Galactarate can also be catabolized through the glycerate pathway (20). The products of this pathway are pyruvate and d-glycerate. These pathways have been described in prokaryotes, and it is not certain whether similar pathways also exist in fungi, some of which are able to metabolize galactarate.
d-Galacturonate dehydrogenase (EC 1.1.1.203) has been described in Agrobacterium tumefaciens and in Pseudomonas syringae, and the enzymes from these organisms have been purified and characterized (3, 6, 22). Recently, the corresponding genes were also identified (4, 24). Both enzymes are specific for NAD as a cofactor but are not specific for the substrate. They oxidize d-galacturonate and d-glucuronate to meso-galactarate (mucate) and d-glucarate (saccharate), respectively. The reaction product is probably the hexaro-lactone which spontaneously hydrolyzes. The reverse reaction can only be observed at acidic pH where some of the galactarate is in the lactone form (22).
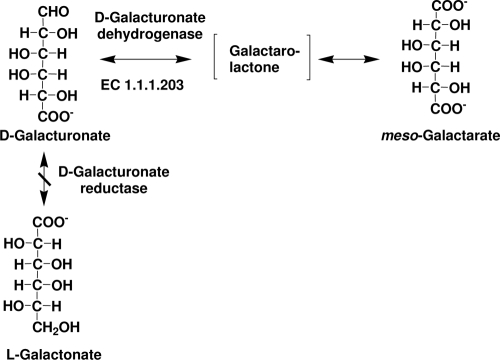
We describe here strains of filamentous fungi that have been genetically engineered to produce galactarate by disruption of d-galacturonate reductase and expression of d-galacturonate dehydrogenase (Fig. 1). Galactarate is currently commercially produced from d-galactose by oxidation with nitric acid (1) or from d-galacturonate by electrolytic oxidation (8). Oxidation with nitric acid is expensive and produces toxic wastes. Galactarate is used as a chelator and in skin care products. It was formerly used as a leavening agent in self-rising flour (2) and has potential applications in polymer synthesis (10) and as a platform chemical (for a review, see reference 13).
FIG. 1.
Engineering the d-galacturonic acid pathway in fungi. Deletion of the gene encoding d-galacturonate reductase resulted in strains unable to utilize d-galacturonic acid as a carbon source. The expression of a bacterial udh gene, encoding an NAD-dependent d-galacturonate dehydrogenase, resulted in fungal strains, which were able to oxidize d-galacturonic acid to meso-galactaric acid (mucic acid). d-Galacturonate dehydrogenase forms a galactaro-lactone which spontaneously hydrolyzes.
MATERIALS AND METHODS
Strains and plasmids.
H. jecorina (anamorph T. reesei) QM6a and A. niger ATCC 1015 (CBS 113.46) and the strains derived from them were maintained as conidia in 20% (vol/vol) glycerol, 0.8% (wt/vol) NaCl, and 0.025% (vol/vol) Tween 20 at −80°C. Saccharomyces cerevisiae FY834 was obtained from Jay C. Dunlap (Dartmouth College, Hanover, NH) and stored at −80°C in 20% (vol/vol) glycerol with 0.9% (wt/vol) NaCl.
Plasmids pCSN44 (21) and pRS426 (ATCC 77107) were obtained from Jay C. Dunlap. pAN52-1NotI was obtained from P. J. Punt, TNO Nutrition and Food Research, Zeist, The Netherlands, and was modified as previously described (11). The selection plasmid pTOC202 contained the acetamidase gene (amdS) from A. nidulans. pRSET-A was obtained from Invitrogen. The pCL2-Amds vector was constructed from pAN52-1NotI by insertion of multiple cloning sites between the gpdA promoter and trpC terminator and by addition of the A. nidulans amdS gene as a selection marker.
Media.
Strains were grown on potato dextrose agar (Becton Dickinson) or on agar (1.5% [wt/vol]) containing 20% (vol/vol) carrot juice in 9-cm petri dishes to produce conidia.
H. jecorina strains were grown in medium containing 10 g of d-galacturonate liter−1 (pH 7.0), 0.5 g of proteose peptone liter−1, 15 g of KH2PO4 liter−1, 0.6 g of (NH4)2SO4 liter−1, 0.6 g of MgSO4·7H2O liter−1, 0.6 g of CaCl2·2H2O liter−1, and Mandels and Weber trace elements (15) to assess growth on d-galacturonate. A. niger strains were grown in medium containing 20 g of d-galacturonate liter−1 (pH 5.5), yeast nitrogen base (Becton Dickinson), synthetic complete amino acid mixture, and 20 g of agar liter−1 to assess growth on d-galacturonate.
For the transcriptional analysis, 500 ml of YP (20 g of yeast extract/liter, 40 g of peptone/liter) and 30 g of gelatin/liter was inoculated with 2 × 108 spores of the A. niger ATCC 1015 and the gaaA deletion strain, followed by cultivation overnight at 30°C (250 rpm). Then, 100 ml of each overnight culture was mixed with 400 ml of freshly made YP with d-galacturonate (final concentration, 20 g/liter; pH 6) and cultivated under the same conditions for different time intervals.
The defined medium of Vogel (23) was used for the production of galactarate. Precultures were grown in medium modified only by replacing sucrose with d-xylose as the carbon source and ammonium nitrate with (NH4)2SO4 as the nitrogen source. Medium for A. niger precultures also contained 0.4% (wt/vol) agar. Some media for precultures were also supplemented with 1 g of Bacto peptone liter−1. Production medium was further modified to reduce the concentration of phosphate and trisodium citrate, both of which interfere with high-pressure liquid chromatography (HPLC) analysis of galactaric acid. The modified production medium contained: 1.6 g of (NH4)2SO4 liter−1, 0.5 g of KH2PO4 liter−1, 0.1 g of MgSO4·7H2O liter−1, 0.05 g of CaCl2·2H2O liter−1, 2.63 mg of citric acid·H2O liter−1, 2.63 mg of ZnSO4·7H2O liter−1, 0.7 mg of FeSO4·7H2O liter−1, 50 μg of MnSO4·4H2O liter−1, 130 μg of CuSO4·5H2O liter−1, 50 μg of H3BO3 liter−1, 50 μg of Na2MoO4·2H2O liter−1, and 25 μg of biotin liter−1. Glucose (2 or 4 g liter−1) was provided as a carbon source. Production medium also contained d-galacturonate (9 or 17 g liter−1, provided as d-galacturonic acid monohydrate; Fluka). The pH of production media was adjusted to 4.0, 5.0, or 6.5 with NaOH.
D-galacturonate solutions for production of galactarate contained 17 g of d-galacturonate liter−1, adjusted to pH 5.0 with NaOH. d-Galacturonate solutions contained sodium d-galacturonate and d-galacturonic acid, but no other mineral salts.
Deletion of the d-galacturonate reductase gene gar1 in H. jecorina.
The gar1 deletion cassette was constructed by cloning 1.5-kb areas of genomic DNA from both sides of the d-galacturonate reductase gene from H. jecorina genomic DNA by PCR, using the primers gar1 5f, gar1 5r, gar1 3f, and gar1 3r (Table 1). Other elements in the deletion cassette were the hph (hygromycin B phosphotransferase) gene under the Aspergillus trpC promoter and elements for replication and selection in yeast and Escherichia coli. The hph-containing fragment was obtained by PCR using pCSN44 as a template and the primers hphF and hphR (Table 1) to obtain a PCR fragment of 1,447 bp.
TABLE 1.
Primers used for PCR
| Primer | Sequence |
|---|---|
| gar1 5f | GTAACGCCAGGGTTTTCCCAGTCACGACGAAGCTTATATCCACCGTGTCCCAG |
| gar1 5r | ATCCACTTAACGTTACTGAAATCTCCAACGACGCAGTTGTTTGAGCAAC |
| gar1 3f | CTCCTTCAATATCATCTTCTGTCTCCGACTTGCATTGGTCAGAGCGGTA |
| gar1 3r | GCGGATAACAATTTCACACAGGAAACAGCAAGCTTAAGCAGTGGATGACTTGCTG |
| hphF | GTCGGAGACAGAAGATGATATTGAAGGAGC |
| hphR | GTTGGAGATTTCAGTAACGTTAAGTGGAT |
| pyrG-5-F | TATAGAATTCTGCGGGCATGATGTTTCAAC |
| pyrG-5-R | TATACCCGGGAAATCATCGCGAGCCCCT |
| pyrG-3-F_n | TATACCCGGGATGTCGAGCACGGGTAGTCA |
| pyrG-3-R_n | TATAGAATTCATGCAAGCGGCAGAGTACTTT |
| gaaA-5-F | TATACTCGAGTGAATTGCACTCTTCGTACCG |
| gaaA-5-R | TATACCATGGTGTGATTGCTGTGGTGTAAAT |
| gaaA-3-F | TATACCATGGCCGTTTATGATTCTGGTCCATC |
| gaaA-3-R | TATAGAATTCTCGAGTTAATTCCCTTAGCG |
| pyrG-del-F_n | TATACCCGGGTGATTGAGGTGATTGGCGAT |
| pyrG-del-R_n | TATACCCGGGTTATCACGCGACGGACAT |
A fragment for replication and selection in S. cerevisiae and E. coli was obtained by digesting pRS426 with restriction enzymes EcoRI and XhoI to release a 5.7-kb fragment. The two PCR fragments containing the flanking regions for homologous recombination were combined with the hph and replication fragments, and all four DNA fragments were transformed to S. cerevisiae FY834. The single plasmid obtained through homologous recombination was isolated from S. cerevisiae and amplified in E. coli. The deletion cassette, 4,520 bp containing the gar1 flanking regions on either side of the hygromycin B resistance gene, was released by HindIII digestion of the combined plasmid and transformed to H. jecorina QM6a. Transformants were selected by resistance to hygromycin B (240 μg liter−1). Strains with a successful deletion were identified by colony PCR. Integration of only a single copy of the deletion cassette was verified by Southern hybridization using hph as a probe.
Expression of the udh gene in the H. jecorina gar1 deletion strain.
The udh gene, coding for the A. tumefaciens d-galacturonic acid dehydrogenase, was expressed in an H. jecorina gar1 deletion strain, obtained as described above. The open reading frame of the udh gene (GenBank no. BK006462.1 [24]) was cloned in a pCR2.1-TOPO vector, as previously described (4), and released as a BamHI fragment. This fragment was ligated to the BamHI site of the modified pAN52-1NotI vector (11) that contained the Aspergillus nidulans gpdA promoter and trpC terminator. The orientation of the udh in the resulting plasmids was checked to be suitable for transcription. This plasmid was then cotransformed to the gar1 deletion strain with selection plasmid pTOC202, which has the A. nidulans amdS gene coding for acetamidase as a marker. Transformants were selected for growth in the presence of acetamide. Transformants were verified by PCR, and crude extracts of the mycelia were tested for d-galacturonic acid dehydrogenase activity.
Deletion of the d-galacturonate reductase gene, gaaA, in A. niger ATCC 1015 ΔpyrG.
ATCC 1015 ΔpyrG was generated by using a deletion cassette containing 1,500 bp from the A. niger pyrG (orotidine-5′-phosphate decarboxylase) promoter and 1,560 bp from the A. niger pyrG terminator. These fragments were obtained by PCR of A. niger ATCC 1015 genomic DNA using the primers pyrG-5-F, pyrG-5-R, pyrG-3-F_n, and pyrG-3-R_n (Table 1) and the proofreading DNA polymerase Phusion (Finnzymes). After digestion of plasmid pRSET-A (Invitrogen) with EcoRI and PvuII (both NEB) and the terminator fragment (pyrG-3) with EcoRI, an intermediary construct was obtained by ligation using T4 DNA ligase (NEB). This intermediary construct was digested with XmaI (NEB) and Ecl136II (Fermentas) and ligated to the XmaI-digested promoter fragment (pyrG-5). After digestion with EcoRI, this construct was introduced to A. niger ATCC 1015 using standard fungal transformation methods. Transformants were screened for integration of the deletion cassette at the pyrG locus by growth in the presence of 5-fluoroorotic acid and the deletion of the pyrG gene was confirmed by PCR.
The cassette for the deletion of gaaA from A. niger ATCC 1015 was generated by using a deletion cassette containing 1,550 bp from the A. niger gaaA promoter, 1,468 bp from the A. niger gaaA terminator, and a 1,927-bp fragment containing the pyrG gene flanked with its native promoter and terminator. These fragments were obtained by PCR of A. niger ATCC 1015 genomic DNA using the primers gaaA-5-F, gaaA-5-R, gaaA-3-F, gaaA-3-R, pyrG-del-F_n, and pyrG-del-R_n (Table 1) and the proofreading DNA polymerase Phusion. Plasmid pRSET-A (Invitrogen) was digested with EcoRI and PvuII (both NEB) and the terminator fragment (gaaA-3) with EcoRI to produce an intermediary construct by ligation using T4 DNA ligase (NEB). This intermediary construct was digested with NcoI (NEB) and Ecl136II (Fermentas) and ligated to the NcoI-digested promoter fragment (gaaA-5). The resulting vector was digested with NcoI (NEB) and treated with Klenow polymerase and phosphatase. The pyrG DNA fragment, after digestion with SmaI, was inserted between the two gaaA flanking regions. The deletion cassette, 4,854 bp, containing the gaaA flanking regions and the pyrG gene, was released by NheI digestion and transformed into A. niger ATCC 1015 ΔpyrG. Transformants were selected by ability to grow in the absence of uracil. Strains with a successful deletion were verified by PCR.
Expression of the udh gene in the A. niger gaaA deletion strain.
A codon-optimized udh gene, for expression in A. niger, was custom synthesized by Geneart (Regensburg, Germany). A fragment obtained by ApaI and SpeI restriction digestion, containing the udh open reading frame, was ligated to the ApaI-SpeI sites of pCL2-Amds. The resulting plasmid, containing the udh gene under the control of the A. nidulans gpdA promoter and trpC terminator, was transformed to the A. niger gaaA deletion strain. Transformants were selected by ability to grow on acetamide as the sole nitrogen source and verified by PCR.
Transcription analysis.
To quantify the transcription of gaaA, gaaB, and gaaC after induction with d-galacturonate, mycelia were grown in complex medium and then transferred to medium containing d-galacturonate (pH 6) as described above. The mycelium was removed by filtration at 0, 0.5, 1, 2, 4, 8, 16, and 24 h after transfer to d-galacturonate containing medium. The mycelium was mixed with 500 μl of RNA extraction buffer (1 mM EDTA, 0.1 M LiCl, 0.1 M Tris-HCl [pH 7.5], 1% sodium dodecyl sulfate), 500 μl of acid-washed glass beads (Sigma), and 500 μl of PCI (citrate-buffered, water-equilibrated phenol [pH 4.2]; chloroform; and isoamyl alcohol at a volume ratio 25:24:1). The cells were disrupted by Fast Prep (Bio 101), and the total RNA was precipitated in the presence of ethanol and potassium acetate. Next, 100 μg of the total RNA was purified by using an RNeasy minikit (Qiagen). Finally, 6 μg of purified total RNA was used in the reverse transcription reaction (SuperScript; Invitrogen).
cDNA produced in the reverse transcription reaction was diluted 1:6 with water and 5 μl of diluted solution was used in quantitative PCR (qPCR) reactions (LightCycler 480 SYBR green I Master; Roche, Switzerland). The primers are listed in Table 2. The expression of gaaA, gaaB, gaaC, and actin were analyzed. The reactions were carried out in a LightCycler 480 Instrument II (Roche, Switzerland), and the analysis was performed with the accompanying software (Advance Relative Quantification tool). The signal was normalized to that of actin.
TABLE 2.
Primers used for qPCR
| Primer | Sequence |
|---|---|
| gaaA_qPCR_F | AGGACACGATTACTCTACTTGTG |
| gaaA_qPCR_R | GAGCCCATATAATGGAAGTACTG |
| gaaB_qPCR_F | GTGGTCAGTGGAAAGAAGAG |
| gaaB_qPCR_R | CCTTCATCTCAACACTATACCC |
| gaaC_qPCR_F | AGTTGTTGAGTAGGGCTGAC |
| gaaC_qPCR_R | GGCATCCTTGATCTTCTCAG |
| act_qPCR_F | CAACATTGTCATGTCTGGTGG |
| act_qPCR_R | GGAGGAGCAATGATCTTGAC |
Culture conditions.
Flask cultures were grown in 250-ml Erlenmeyer flasks containing 50 ml of medium or substrate and incubated at 28 or 30°C and at 200 rpm. Flasks were inoculated with conidial suspensions to give final concentrations of 2 × 105 conidia ml−1 or with mycelia grown in modified Vogel's medium.
Precultures to obtain mycelium were allowed to grow for 24 (A. niger) or 45 (H. jecorina) h before being harvested by centrifugation (A. niger) or vacuum filtration (H. jecorina) through disks of sterile disposable cleaning cloth (X-tra, 100% viscose household cleaning cloth; Inex Partners Oy, Helsinki, Finland) and rinsed with equal or greater volumes of sterile H2O to remove residual d-xylose and peptone (when present). The mycelium was aseptically transferred to d-galacturonate-containing solution or medium to provide initial biomass concentrations of 1.6 to 2.4 g liter−1.
Bioreactor cultures were grown in 1 liter of medium in B. Braun Biotech International (Sartorius AG, Germany) Biostat CT (2.5 liters, maximum working volume) bioreactors. Cultures were inoculated with an initial biomass of 1.8 ± 0.1 g liter−1 and maintained at 30°C and 500 rpm, with a 1.0 volume gas (volume culture)−1 min−1 (vvm). Culture pH was kept constant at pH 4.0 or 6.5 by the addition of sterile 1 M KOH or 1 M H3PO4. The gas concentration (CO2, O2, N2, and Ar) was analyzed continuously in an Omnistar quadrupole mass spectrometer (Balzers AG, Liechtenstein), calibrated with 3% CO2 in Ar.
Chemical analyses.
The concentrations of d-glucose, d-galacturonic acid, citric acid, and galactaric acid were determined by HPLC using a Fast Acid Analysis column (100 mm by 7.8 mm; Bio-Rad Laboratories, Hercules, CA) linked to an Aminex HPX-87H organic acid analysis column (300 mm by 7.8 mm; Bio-Rad Laboratories) with 2.5 mM H2SO4 as eluant and a flow rate of 0.5 ml min−1. The column was maintained at 55°C. Peaks were detected by using a Waters 410 differential refractometer and a Waters 2487 dual wavelength UV (210 nm) detector. The yield of galactarate from d-galacturonate was calculated from the concentrations of the corresponding acids measured by HPLC.
Enzyme activity measurements.
Crude cell extract of the mycelia was obtained by vortexing with glass beads as described previously (11). d-galacturonate dehydrogenase activity was measured in a reaction mixture containing 0.5 mM NAD, 4 mM d-galacturonate, 50 mM Tris-HCl (pH 7.5), and cell extract. The formation of NADH was monitored by measuring the absorption at 340 nm.
Biomass and intracellular galactarate determination.
Mycelia were collected by filtration through disposable cleaning cloth (X-tra) under vacuum and washed twice with an equal or greater volume of distilled H2O. Samples from flasks were collected by centrifugation and washed twice with one sample volume of distilled H2O. Mycelia were dried to a constant weight under vacuum at −80°C in a B. Braun Biotech International Christ Alpha 2-4 freeze-drier.
For analysis of intracellular galactarate concentrations, biomass which had been washed twice with an equal or greater volume of distilled water was frozen slowly at −20°C and then subjected to freeze-drying at −80°C. After weighing and partial disruption of the biomass by grinding, galactarate was extracted by the addition of 0.5 to 2 ml of NaOH (5 to 7.5 mM). Samples were extracted at 70°C for 0.5 to 1 h. Further physical disruption of the biomass (e.g., by vortexing with glass beads) was not necessary to remove intracellular galactarate from the biomass. To estimate the intracellular concentration, the volume of cytoplasm per g dry mass was assumed to be similar to that of Penicillium chrysogenum, which has been determined to be 2.86 ml per g dry mass (18).
Purification of galactaric acid from culture supernatant.
Galactaric aicd was precipitated from the culture supernatant by addition of 33 mM HCl to the supernatant of the culture at pH 4.0 and 63 mM HCl to the supernatant of the culture at pH 6.5. The rate of precipitation was enhanced by freezing and thawing the supernatant. Precipitate was collected by centrifugation and washed with 40 to 60% (vol/vol) ethanol in 0.2 M HCl. Ethanol was removed by evaporation at 45°C.
RESULTS
Deletion of the d-galacturonate reductase in the fungi H. jecorina and A. niger.
In H. jecorina QM6a, the gar1 gene, coding for the d-galacturonate reductase, was deleted by homologous recombination introducing a hygromycin B resistance gene at the location of gar1 and thereby removing the gene. To test whether the deletion had affected the ability to catabolize d-galacturonate, the deletion strain and QM6a were grown with or without d-galacturonate. QM6a produced ∼3 g of biomass liter−1 from 10 g of d-galacturonate liter−1 in 5 days. The gar1 deletion strains produced ∼0.5 g of biomass liter−1 from d-galacturonate in the same time. Both QM6a and the deletion strains produced ∼0.2 g of biomass liter−1 when no carbon source other than peptone was included in the medium. This demonstrated that gar1 is an essential gene in d-galacturonate catabolism.
The gaaA gene, encoding d-galacturonate reductase, was deleted in A. niger ATCC 1015 ΔpyrG. ATCC 1015 ΔpyrG (parental strain) and an ATCC 1015 ΔgaaA transformant were tested for growth on d-galacturonate as the main carbon source. The ΔgaaA transformant was unable to grow (Fig. 2).
FIG. 2.
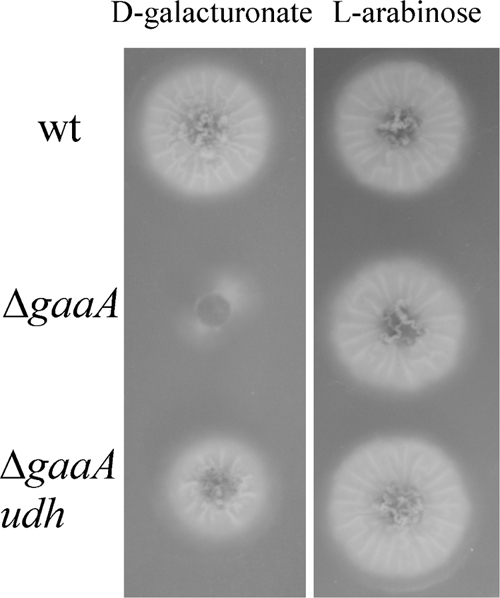
Growth of A. niger strains on d-galacturonate and l-arabinose on agar-solidified medium. In the top row is ATCC 1015 ΔpyrA (wt), in the middle row ATCC 1015 ΔgaaA (ΔgaaA), and in the lower row ATCC 1015 ΔgaaA udh (ΔgaaA udh), in which the endogenous gaaA is deleted and a heterologous udh, encoding d-galacturonate dehydrogenase, is expressed. Colonies were incubated at 28°C for 4 days and photographed from below.
Transcription analysis of the catabolic pathway in A. niger.
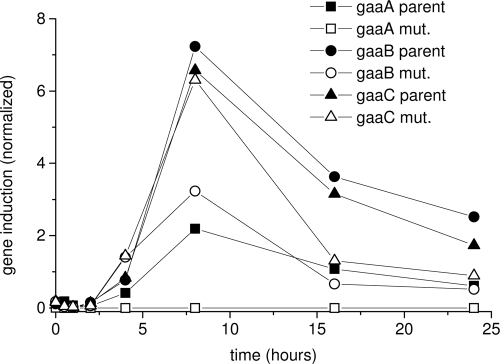
In A. niger the genes of the d-galacturonic acid catabolic pathway gaaA, gaaB, and gaaC, code for d-galacturonate reductase, l-galactonate dehydratase, and 3-deoxy-l-threo-hex-2-ulosonate aldolase, respectively, and are upregulated during growth on d-galacturonate or pectin (16, 17). In order to demonstrate the key regulatory function of d-galacturonate in transcriptional regulation, we analyzed expression of gaaA, gaaB, and gaaC and demonstrated that these genes were upregulated in the presence of d-galacturonate, regardless of whether it was further metabolized or not (Fig. 3). gaaB and gaaC were upregulated on d-galacturonate in both ATCC 1015 and in ATCC 1015 ΔgaaA (Fig. 3). gaaA was upregulated in the parent strain but was not detected in the ΔgaaA transformant, as expected since the gene was removed. Induction occurred within 4 h after transfer to d-galacturonate containing medium and continued for at least 24 h, although maximum induction was observed at ca. 8 h.
FIG. 3.
Transcription analysis of the catabolic d-galacturonate genes in A. niger after induction by d-galacturonate. The transcription of gaaA (squares), gaaB (circles), and gaaC (triangles) was analyzed in ATCC 1015 (solid symbols) and in the gaaA deletion strain (open symbols) after the transfer of mycelia to medium containing d-galacturonate. The mRNA was quantified by using qPCR and normalized to actin mRNA.
Expression of the d-galacturonate dehydrogenase.
When the A. tumefaciens udh gene was expressed in H. jecorina QM6a Δgar1, d-galacturonate dehydrogenase activity (1.3 to 1.5 nkat per mg of extracted protein) was detected in the cell extract of the transformant but not in QM6a Δgar1. Expression of the codon-optimized udh gene in A. niger ATCC 1015 ΔgaaA resulted in similar activity in the cell extract, whereas no activity was detected in ATCC 1015 ΔgaaA.
Expression of udh in A. niger ATCC 1015 ΔgaaA restored its ability to grow on d-galacturonate on agar-solidified medium (Fig. 2). However, growth was poor, and only 0.6 ± 0.1 g of biomass liter−1 were produced from d-galacturonate during 9 days incubation in liquid culture. Unlike in A. niger, expression of the udh gene in H. jecorina QM6a Δgar1 did not restore the ability to grow on d-galacturonate (data not shown).
Conversion of d-galacturonate to galactarate.
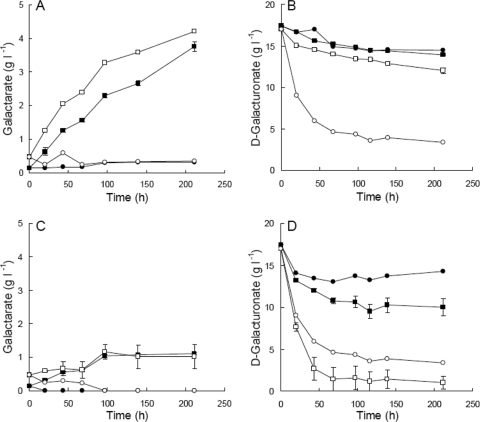
When incubated in 17.4 g of d-galacturonate liter−1 (initial pH 5) in flasks, H. jecorina Δgar1 udh produced 3.8 ± 0.1 g galactarate liter−1 in 211 h, at a yield of 1.08 ± 0.04 g galactarate (g of d-galacturonate)−1, since 3.5 ± 0.1 g of d-galacturonate was removed from the solution (Fig. 4). The initial production rate was 26 ± 2 mg of galactarate liter−1 h−1. The concentration of galactarate produced (4.2 ± 0.0 g liter−1) and initial production rate (36 ± 1 mg of galactarate liter−1 h−1) were increased (P > 0.05) when mycelia were incubated in d-glucose-containing medium (initial pH 5), rather than pure d-galacturonate (Fig. 4). However, 5.0 ± 0.3 g of d-galacturonate was removed from the medium, and the yield of galactarate on d-galacturonate was reduced to 0.86 ± 0.06 g g−1. Galactarate was extracted from the mycelia after 9 days. Mycelia incubated in d-galacturonate solution contained 11.8 ± 0.8 g of galactarate liter−1, which did not differ significantly (P > 0.05) from that obtained from mycelia incubated in medium (12.1 ± 0.1 g of galactarate liter−1).
FIG. 4.
Concentration of galactarate (A and C) and d-galacturonate (B and D) in flask cultures of H. jecorina QM6a Δgar1 udh (squares) and QM6a Δgar1 (circles) (A and B) or A. niger ATCC 1015 ΔgaaA udh (squares) and ΔgaaA (circles) (C and D) maintained at 200 rpm 30°C. Cultures were inoculated with 1.8 to 2.4 g of mycelia liter−1. Mycelia were incubated in 17 g of d-galacturonate liter−1 (pH 5.0; solid symbols) or 17 g of d-galacturonate liter−1 and 2 g of d-glucose liter−1 in modified Vogel's medium (pH 5.0; open symbols). Error bars represent ± the standard error of the mean for three replicate cultures of H. jecorina QM6a Δgar1 udh and three independent transformants of A. niger ATCC 1015 ΔgaaA udh. Single cultures were used for the control strains.
The H. jecorina Δgar1 strain did not produce galactarate (Fig. 4) in either d-galacturonate solution (pH 5) or medium containing d-galacturonate (pH 5) nor was galactaric acid extractable from the mycelia. Although the H. jecorina Δgar1 strain does not convert d-galacturonate to biomass, d-galacturonate was removed from the culture supernatant, and the pH of these cultures increased to 9.1. In contrast, the pH of H. jecorina Δgar1 udh cultures only increased to 6.8 and 5.2 in d-galacturonate solution and d-glucose medium, respectively.
A. niger ATCC 1015 ΔgaaA udh produced 1.0 ± 0.0 g of galactarate liter−1 in d-galacturonate solution (pH 5) in 4 days (Fig. 4), with a yield of 0.16 g of galactarate (g of d-galacturonate)−1. No further increase in galactarate production was observed during the following 5 days of incubation. The initial production rate was 11 ± 0 mg of galactarate liter−1 h−1. Mycelia harvested after 9 days incubation contained 0.9 ± 0.2 g of galactarate liter−1. The production of galactarate was more variable when A. niger ATCC 1015 ΔgaaA udh mycelia were incubated in d-glucose medium, rather than pure d-galacturonate solution, but was otherwise similar to that observed in the solution (1.1 ± 0.2 g of galactarate liter−1, rate 12 ± 2 mg of galactarate liter−1 h−1; Fig. 4). The mycelia contained 0.7 ± 0.1 g of galactarate liter−1 after 9 days of incubation in medium. The yield was reduced to 0.08 g of galactarate (g of d-galacturonate)−1 because of the greater (P < 0.05) consumption of d-galacturonate in medium which had contained glucose. Almost all d-galacturonate was consumed in A. niger cultures in the modified Vogel's medium with d-glucose, although this d-galacturonate was not converted to biomass.
The A. niger ATCC 1015 ΔgaaA strain did not convert d-galacturonate to biomass or galactarate (Fig. 4). Without the udh gene, less d-galacturonate was consumed than when the gene was present, in contrast with the H. jecorina strains where the strain lacking udh consumed more d-galacturonate than H. jecorina Δgar1 udh. No galactarate was extracted from the mycelia of the control strain. The pH of A. niger cultures increased to 7.9 ± 0.3 and 7.1 ± 0.1 in d-galacturonate solution and glucose medium, respectively. The pH of the control strain remained similar to that of the udh strains.
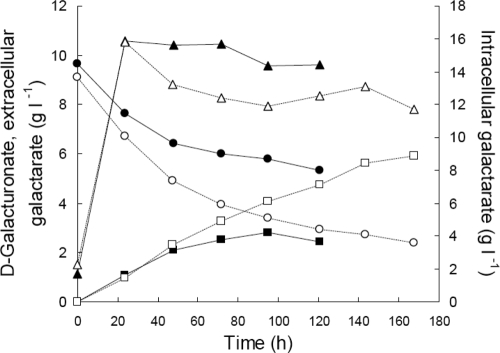
H. jecorina Δgar1 udh produced more galactarate at pH 6.5 (5.9 g of galactarate liter−1) than at pH 4.0 (2.6 g of galactarate liter−1) in pH-controlled bioreactor cultures (Fig. 5). Although the initial production rate (42 mg of galactarate liter−1 h−1) was the same at both pH values, this rate was sustained longer at pH 6.5 (∼95 h) than at pH 4.0 (∼48 h). The yield of galactarate on d-galacturonate was also higher at pH 6.5 [0.87 g of galactarate (g of d-galacturonate)−1] than at pH 4.0 [0.72 g of galactarate (g of d-galacturonate)−1].
FIG. 5.
Intracellular (triangles) and extracellular (squares) concentrations of galactarate produced by H. jecorina QM6a Δgar1 udh from d-galacturonate (circles) in pH-regulated bioreactors at pH 4.0 (solid symbols) and pH 6.5 (open symbols). Cultures were inoculated with 1.8 g of biomass liter−1 and maintained at 30°C and 500 rpm with 1 vvm aeration. In addition to d-galacturonate, the medium contained 4 g of glucose liter−1 and mineral salts as described in Materials and Methods.
Mycelia grown at both pH 4.0 and pH 6.5 contained 15.8 g of galactarate liter−1 after 24 h of incubation in d-galacturonate containing medium. The intracellular concentration in mycelia maintained at pH 4.0 remained above 14.4 g of galactarate liter−1 for the next 100 h, decreasing only gradually, but the intracellular concentration in hyphae maintained at pH 6.5 had decreased to 13.2 g of galactarate liter−1 by 48 h and was subsequently sustained at a concentration of 12.3 ± 0.3 g of galactarate liter−1.
Galactarate was removed from culture supernatants by simple acid precipitation. From supernatant at pH 4.0, 79% of the galactaric acid was recovered with ∼94% purity. The supernatant at pH 6.5 yielded 3.3 g of galactarate (97% recovery), which was 97.5% pure.
DISCUSSION
Through metabolic engineering we constructed fungal strains that converted d-galacturonate to galactarate. d-Galacturonate/d-galacturonic acid is the major component of pectin and is used by many fungi as a growth substrate. Such fungi are suitable candidates for engineering galactarate production since they are able to take up d-galacturonate. Proteins for d-galacturonate transport have not been described; however, Martens-Uzonova has identified several potential candidates, i.e., transporter proteins that are induced on d-galacturonate and pectin in A. niger (16). Some fungi, such as A. niger, have the added advantage of also producing pectinolytic enzymes to hydrolyze pectin to d-galacturonate.
The first step in fungal catabolism of d-galacturonate is a reduction through an NAD(P)H requiring reductase. The corresponding genes have been described for H. jecorina and A. niger (12, 17). Deletion of gar1, encoding d-galacturonate reductase, in H. jecorina resulted in no growth on d-galacturonate, indicating that there are no alternative pathways of d-galacturonate catabolism. Similarly, deletion of gaaA in A. niger resulted in no growth on d-galacturonate. A. niger has another gene, gar2 (17), which has a high homology to gar1 of H. jecorina. That deletion of gaaA was sufficient to prevent growth on d-galacturonate (Fig. 2) suggests that gar2 is not functional in d-galacturonate catabolism. Removal of d-galacturonate from the culture medium by the d-galacturonate reductase deletion strains of both H. jecorina and A. niger suggests that uptake of d-galacturonate was not impaired by its deletion. d-Galacturonate was not observed in intracellular samples (data not shown) and was not used to produce biomass. d-Galacturonate was presumably converted into a nonmetabolizable compound via an unknown reaction, but this compound was not detected as an unknown in the HPLC profiles. Since the reaction can reduce the yield of galactarate from d-galacturonate, identification and disruption of this reaction would be advantageous. Futile metabolism of d-galacturonate has not previously been described.
d-Galacturonate or a metabolite derived from it has previously been shown to be responsible for the induction of pectinolytic and d-galacturonate catabolic genes in A. niger (7). To test whether d-galacturonic acid is itself the inducer, we analyzed the transcription of gaaB and gaaC in the A. niger strain lacking the gaaA gene, since d-galacturonate is not converted to l-galactonate in this strain. The induction of both gaaB and gaaC by d-galacturonate (Fig. 3) demonstrated that d-galacturonate is itself an inducer of the catabolic genes and that a metabolite derived from d-galacturonate is not required for induction. We observed, however, that the expression level of gaaB was lower in the gaaA deletion strain, whereas the expression levels of gaaC were similar.
Expression of the bacterial udh gene (d-galacturonate dehydrogenase) in H. jecorina and A. niger strains lacking d-galacturonate reductase activity was sufficient to generate strains which were able to convert d-galacturonate to galactarate. The reaction product of d-galacturonate dehydrogenase is galactaro-1,4-lactone (4) which can hydrolyze spontaneously to galactarate. The efficiency of this hydrolysis in fungi is not known. The relative efficiency of transport of the lactone and the linear form is also unknown. The lactone and linear forms of galactarate were not distinguished by HPLC.
H. jecorina Δgar1 udh was a better production host for galactarate than A. niger ΔgaaA udh (Fig. 4). Expression of udh in A. niger ΔgaaA restored the ability to grow on d-galacturonate, although the colony size increased more slowly than the parental strain (Fig. 2), and biomass production was not observed in liquid medium. This suggested that A. niger ΔgaaA udh can catabolize galactarate and that the poor accumulation of galactarate either intracellularly or extracellularly resulted from it subsequent catabolism. Growth of the parental strain ATCC 1015 on galactarate has been observed on agar-solidified medium but not in liquid (data not shown). Poor growth on galactarate may reflect poor galactarate uptake. In contrast, the similar intra- and extracellular concentrations of galactarate (∼1 g liter−1) suggested that export of galactarate was not a problem in A. niger. The relatively high extracellular pH (7.1 to 7.9), which resulted from the uptake of d-galacturonate from the culture supernatant, may have contributed to improved transport of galactarate and may also have reduced the amount of galactarate produced, since metabolism of many A. niger strains is reduced at pH values above 7. Since no measurable increase in biomass occurred, the metabolism of galactarate produced in the cytoplasm would not account for all of the removal of d-galacturonate from the medium. Both galactarate catabolism and d-galacturonate removal would contribute to the very low yield of galactarate from d-galacturonate in A. niger.
Galactarate accumulated both intra- and extracellularly in H. jecorina (Fig. 4 and 5) and 5.9 g of galactarate liter−1 was produced at pH 6.5 without optimization of the system. The theoretical maximum yield of galactarate from d-galacturonate [1.08 g of galactarate (g of d-galacturonate)−1] was achieved when mycelia were incubated in pure d-galacturonate solution (pH 5.5), but the yield was reduced when d-glucose was provided as an energy source, even though galactarate was produced at a higher rate.
The high intracellular concentration of galactarate in H. jecorina Δgar1 udh suggests that export of galactarate may be limiting in H. jecorina. Intracellular concentrations were higher when mycelia were maintained at a lower pH (4.0) than at a higher pH (6.5), as would be expected since the higher concentration of undissociated acid in the culture supernatant would have resulted in more galactaric acid diffusing back into the cell and higher overall export costs (5). The cessation of galactarate production after approximately 48 h at pH 4.0 may reflect inhibition of the d-galacturonate dehydrogenase by the high intracellular concentration of galactarate, along with a need for an additional energy supply for active transport of the dissociated anion out of the cell. Interestingly, at pH 6.5 galactarate was still being exported after 168 h of incubation, even though all d-glucose had been consumed within less than 24 h. The rate of galactarate production at the end of this culture was similar to that observed in pure d-galacturonate solution in flasks, which may indicate that ATP generated from the reoxidation of NADH is sufficient for galactarate transport at pH values near neutral.
The low solubility of galactaric acid at low pH made it easy to purify from the culture supernatant. Interestingly, no precipitates were observed in the culture supernatant in cultures maintained at pH 4.0, since the extracellular concentration (2.6 g of galactarate liter−1) remained below that which is soluble at room temperature. Galactarate is considerably more soluble at neutral pH, so no precipitate was observed at pH 6.5, although more than twice as much galactarate was produced at pH 6.5 compared to pH 4.0. However, reducing the pH of the solution to 2.9 was sufficient to precipitate essentially all of the galactarate/galactaric acid.
This is the first report of engineering microbes for galactarate production. Conversion of d-galacturonate to galactarate was very efficient, achieving theoretical yields in H. jecorina, and the product was easy to purify. If the metabolism of galactarate in A. niger can be prevented, higher productivity from it can be expected and the direct conversion of pectin to galactarate would be made possible. Further enhancements are expected through optimization of cultural conditions and alternative energy supply.
Acknowledgments
This study was supported by the Academy of Finland through the following programs: Finnish Centre of Excellence in White Biotechnology-Green Chemistry, the Cadfiss program, and an Academy Research Fellowship for P.R.
We thank Tarja Laakso and Hanna Kuusinen for technical assistance.
Footnotes
Published ahead of print on 6 November 2009.
REFERENCES
- 1.Acree, S. F. 1931. Method of converting wood into mucic acid. U.S. patent 1,816,137.
- 2.Anonymous. 1922. Commercial production of mucic acid. Chem. Metallurg. Eng. 26:1118. [Google Scholar]
- 3.Bateman, D. F., T. Kosuge, and W. W. Kilgore. 1970. Purification and properties of uronate dehydrogenase from Pseudomonas syringae. Arch. Biochem. Biophys. 136:97-105. [DOI] [PubMed] [Google Scholar]
- 4.Boer, H., H. Maaheimo, A. Koivula, M. Penttilä, and P. Richard. 17 November 2009. Identification in Agrobacterium tumefaciens of the d-galacturonic acid dehydrogenase gene. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-009-2333-9. [DOI] [PubMed]
- 5.Casal, M., S. Paiva, O. Queirós, and I. Soares-Silva. 2008. Transport of carboxylic acids in yeasts. FEMS Microbiol. Rev. 32:974-994. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. F., and D. S. Feingold. 1969. Hexuronic acid dehydrogenase of Agrobacterium tumefaciens. J. Bacteriol. 99:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries, R. P., J. Jansen, G. Aguilar, L. Parenicova, V. Joosten, F. Wulfert, J. A. Benen, and J. Visser. 2002. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 530:41-47. [DOI] [PubMed] [Google Scholar]
- 8.Fauvarque, J. F., C. Guérin, S. Petit, and D. E. Baynast. 1994. Procédé de préparation de l'acide galactarique et cellule d'électrolyse utilisée à cet effet. French patent FR2699937(A1).
- 9.Hilditch, S., S. Berghäll, N. Kalkkinen, M. Penttilä, and P. Richard. 2007. The missing link in the fungal d-galacturonate pathway: identification of the l-threo-3-deoxy-hexulosonate aldolase. J. Biol. Chem. 282:26195-26201. [DOI] [PubMed] [Google Scholar]
- 10.Kiely, D. E., L. Chen, and T.-H. Lin. 2000. Synthetic polyhydroxypolyamides from galactaric, xylaric, d-glucaric, and d-mannaric acids and alkylenediamine monomers—some comparisons. J. Polymer Sci. A Polymer Chem. 38:594-603. [Google Scholar]
- 11.Kuorelahti, S., P. Jouhten, H. Maaheimo, M. Penttilä, and P. Richard. 2006. l-Galactonate dehydratase is part of the fungal path for d-galacturonic acid catabolism. Mol. Microbiol. 61:1060-1068. [DOI] [PubMed] [Google Scholar]
- 12.Kuorelahti, S., N. Kalkkinen, M. Penttilä, J. Londesborough, and P. Richard. 2005. Identification in the mold Hypocrea jecorina of the first fungal d-galacturonic acid reductase. Biochemistry 44:11234-11240. [DOI] [PubMed] [Google Scholar]
- 13.Lewkowski, J. 2001. Synthesis, chemistry and applications of 5-hydroxymethyl-furfural and its derivatives. ARKIVOC 2001:17-54. [Google Scholar]
- 14.Liepins, J., S. Kuorelahti, M. Penttilä, and P. Richard. 2006. Enzymes for the NADPH-dependent reduction of dihydroxyacetone and d-glyceraldehyde and l-glyceraldehyde in the mould Hypocrea jecorina. FEBS J. 273:4229-4235. [DOI] [PubMed] [Google Scholar]
- 15.Mandels, M., and J. Weber. 1969. The production of cellulases, p. 391-414. In G. J. Hajny and E. T. Reese (ed.), Cellulases and their applications, vol. 95. American Chemical Society, Washington, DC. [Google Scholar]
- 16.Martens-Uzonova, E. 2008. Assessment of the pectinolytic network of Aspergillus niger by functional genomics: insight from the transcriptome. Ph.D. thesis. University of Wageningen, Wageningen, the Netherlands.
- 17.Martens-Uzunova, E. S., and P. J. Schaap. 2008. An evolutionary conserved d-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet. Biol. 45:1449-1457. [DOI] [PubMed] [Google Scholar]
- 18.Nestaas, E., and D. I. C. Wang. 1981. A new sensor, the “filtration probe,” for quantitative characterization of the penicillin fermentation. I. Mycelial morphology and culture activity. Biotechnol. Bioeng. 23:2803-2813. [DOI] [PubMed] [Google Scholar]
- 19.Richard, P., and S. Hilditch. 2009. d-Galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl. Microbiol. Biotechnol. 82:597-604. [DOI] [PubMed] [Google Scholar]
- 20.Sharma, B. S., and H. J. Blumenthal. 1973. Catabolism of d-gluaric acid to alpha-ketoglutarate in Bacillus megaterium. J. Bacteriol. 116:1346-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36:79-81. [Google Scholar]
- 22.Wagner, G., and S. Hollmann. 1976. Uronic acid dehydrogenase from Pseudomonas syringae: purification and properties. Eur. J. Biochem. 61:589-596. [DOI] [PubMed] [Google Scholar]
- 23.Vogel, H. J. 1956. Convenient growth medium for Neurospora (medium N). Microb. Genet. Bull. 243:112-119. [Google Scholar]
- 24.Yoon, S. H., T. S. Moon, P. Iranpour, A. M. Lanza, and K. J. Prather. 2008. Cloning and characterization of uronate dehydrogenases from two pseudomonads and Agrobacterium tumefaciens strain C58. J. Bacteriol. 191:1565-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]