Abstract
Bioluminescence imaging (BLI) is emerging as a powerful tool for real-time monitoring of infections in living animals. However, since luciferases are oxygenases, it has been suggested that the requirement for oxygen may limit the use of BLI in anaerobic environments, such as the lumen of the gut. Strains of Escherichia coli harboring the genes for either the bacterial luciferase from Photorhabdus luminescens or the PpyRE-TS and PpyGR-TS firefly luciferase mutants of Photinus pyralis (red and green thermostable P. pyralis luciferase mutants, respectively) have been engineered and used to monitor intestinal colonization in the streptomycin-treated mouse model. There was excellent correlation between the bioluminescence signal measured in the feces (R2 = 0.98) or transcutaneously in the abdominal region of whole animals (R2 = 0.99) and the CFU counts in the feces of bacteria harboring the luxABCDE operon. Stability in vivo of the bioluminescence signal was achieved by constructing plasmid pAT881(pGB2ΩPamiluxABCDE), which allowed long-term monitoring of intestinal colonization without the need for antibiotic selection for plasmid maintenance. Levels of intestinal colonization by various strains of E. coli could be compared directly by simple recording of the bioluminescence signal in living animals. The difference in spectra of light emission of the PpyRE-TS and PpyGR-TS firefly luciferase mutants and dual bioluminescence detection allowed direct in vitro and in vivo quantification of two bacterial populations by measurement of red and green emitted signals and thus monitoring of the two populations simultaneously. This system offers a simple and direct method to study in vitro and in vivo competition between mutants and the parental strain. BLI is a useful tool to study intestinal colonization.
Among the wide variety of bacteria that colonize the gastrointestinal tracts of mammals, Escherichia coli is the most abundant facultative anaerobe of the human intestinal microflora. Aside from being part of the normal flora, E. coli is also a versatile organism capable of causing a variety of intestinal and extraintestinal diseases (18). The mechanisms that allow commensal E. coli to colonize the intestine and survive successfully in this niche remain poorly characterized. Conventional mice display natural resistance to colonization by commensal E. coli, but oral administration of streptomycin, which alters the intestinal microflora, allows colonization of the mouse large intestine by this species (25). The streptomycin-treated mouse model has been used extensively to study the factors of gram-negative bacteria implicated in the intestinal colonization process. However, this model is limited to the viable plate counts of bacteria in the feces and misses some critical information, such as the kinetics of colonization, the fate of the bacterial cells across the digestive tract, and the site of colonization. A better understanding of colonization would be facilitated by direct in vivo follow-up of this process.
Bioluminescence imaging (BLI) technology is emerging as a powerful tool for the study of a wide range of biological processes in live animals, including real-time monitoring of infections (16). Bioluminescence systems emit visible light due to the luciferase-mediated oxidation of a luciferin substrate. A variety of luciferin-luciferase systems with different peak emissions have been identified in nature from numerous species (14). The luciferase of the soil bacterium Photorhabdus luminescens has been expressed successfully in gram-negative and gram-positive bacteria. This system emits blue-green light, with an emission maximum of approximately 490 nm, and does not require the addition of an exogenous substrate since the luciferase operon contains the genes required for synthesis of the substrate. Therefore, this luciferase has been used extensively to monitor bacterial infections in the living mouse. One of the first investigations with Salmonella enterica serovar Typhimurium transformed with the lux operon of P. luminescens evaluated the tissue distribution and the virulence of various S. Typhimurium strains (9). Subsequent modification of the lux operon led to the generation of highly bioluminescent Staphylococcus aureus and allowed the monitoring of infections due to this species in living mice (11). The modified lux operon was engineered into a lux-kan transposon cassette for chromosomal integration in gram-positive bacteria, such as S. aureus, Streptococcus pneumoniae, group A Streptococcus, and Listeria monocytogenes (16). Replication of L. monocytogenes in the lumen of the gall bladder was demonstrated for the first time by BLI (13).
Bioluminescent E. coli was used in the neutropenic mouse thigh model of infection to evaluate the in vivo activity of antimicrobial agents (29). Bioluminescence was as indicative of therapeutic efficacy as CFU counts but, in addition, allowed real-time monitoring of the infection and of treatment efficacy in the same animal; however, only short-term monitoring (12 h) could be performed.
Because luciferases are oxygenases, it has been suggested that the requirement for oxygen may limit the use of BLI in anaerobic environments, such as the lumen of the gut. After oral administration of bioluminescent Salmonella to susceptible mice, the bioluminescent signal recorded in the abdominal region was greatly enhanced after air exposure (9). It was therefore assumed that direct bioluminescence imaging of intestine-colonizing microorganisms would not be optimal unless oxygen was provided exogenously or as the result of the close interaction between cells and the bacteria (9). However, the bacterial luciferase was used to trace in real time the colonization dynamics by Citrobacter rodentium of the gastrointestinal tracts of living animals, demonstrating that the gut represents a semianaerobic environment that allows the study of bacterial colonization by BLI (33).
Factors essential for colonization are best studied in cocolonization experiments (7, 17). There are several luciferases with distinct emission spectra (34) that could be used in competition experiments to trace simultaneously two bacterial populations in the same living animal. However, in order not to impose additional and different metabolic burdens on the bacteria under study, the exogenous luciferases ideally have to be similar to allow comparison between strains. The thermostable luciferase variants PpyRE-TS and PpyGR-TS, derived from wild-type luciferase from the North American firefly Photinus pyralis, emit red (612 nm) and green (552 nm) light, respectively, at 37°C and are encoded by single genes of 1,650 bp, differing by only 9 bp (4). Bioluminescence color is determined by the Ser284Thr (PpyRE-TS) and Val241Ile, Gly246Ala, and Phe250Ser (PpyGR-TS) amino acid changes (5, 34). By use of optical filters, the emission spectra are readily distinguishable (4, 5). Five additional mutations provide enhanced thermostability at 37°C (4), improving the compatibility of the enzymes with bacterial culture conditions and BLI in animal models.
While the luciferase mutants and all firefly luciferases use as substrates firefly luciferin and ATP to produce light, in vivo imaging is commonly performed with endogenous ATP and requires only exogenous administration of the luciferase substrate.
The aim of this study was to develop a dynamic mouse model using in vivo bioluminescence imaging systems to monitor bacterial colonization in situ and in real time in whole living animals. Various strains of E. coli harboring the genes for the bacterial luciferase from P. luminescens or the firefly luciferase mutants (PpyRE-TS and PpyGR-TS) from P. pyralis have been engineered and used to follow bacterial intestinal colonization in mice. BLI was found to be well adapted to compare the intestine-colonizing capacities of various E. coli strains and to monitor cocolonization in vivo by use of dual bioluminescence emission.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The properties of the strains and plasmids used are listed in Table 1. Spontaneous streptomycin-resistant mutants of human commensal E. coli K-12 MG1655 (24) (MG1655-Strr), diarrhea-associated enteroaggregative E. coli (EAEC) 55989 producing aggregative adhesion fimbriae type III (2) (55989-Strr), and E. coli BM2711, a RecA1 derivative of strain MM294 (12) (BM2711-Strr), were used. The strains were made bioluminescent by transformation with DNA of plasmids carrying the luxABCDE operon encoding the Photorhabdus luminescens bacterial luciferase or the lucG or lucR gene coding for the Photinus pyralis firefly luciferase green or red mutant, respectively (4).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant property(ies)a | Reference or source |
|---|---|---|
| E. coli strains | ||
| MG1655-Strr | Spontaneous Strr mutant of MG1655 (CGSC 7740); F− λ−ilvG rfb-50 rph-1, Strr | 24 |
| 55989 | Enteroaggregative strain producing AAF-IIIb | 2 |
| 55989-Strr | Spontaneous Strr mutant of 55989 | 22 |
| BM2711 | thi-1 endA1 hdR17(rK− mK−) glnV44 recA1 Δ(lac)X74 | 12 |
| BM2711-Strr | Spontaneous Strr mutant of BM2711 | This work |
| Plasmids | ||
| pSB2025 | Superlinker plasmid pSL1190ΩluxABCDE, Apr | 27 |
| pAL2 | pVA838ΩPamiluxABCDE, p15A and pVA749 ori, Emr | 1 |
| pGB2 | pSC101 derivative, Spr Smr | 8 |
| pAT881 | pGB2ΩPamiluxABCDE | This work |
| pGEX-6P-2ΩlucR | Source of lucR (Ser284Thr) | 4 |
| pGEX-6P-2ΩlucG | Source of lucG (Val241Ile, Gly246Ala, Phe250Ser) | 4 |
| pAT113 | pACYC184 derivative, Mob+, attTn1545, Emr Kmr | 32 |
| pAT882 | pAT113ΩPtetlucR | This work |
| pAT883 | pAT113ΩPtetlucG | This work |
| pTVC-erm | pACYC184 derivative, Mob+, ori pAMβ1, Emr Kmr | P. Trieu-Cuot, unpublished |
| pAT884 | pTVCΩPtetlucR | This work |
| pAT885 | pTVCΩPtetlucG | This work |
Ap, ampicillin; Em, erythromycin; Km, kanamycin; Sm, streptomycin (plasmid); Sp, spectinomycin; Str, streptomycin (chromosomal).
AAF-III, aggregative adhesion fimbriae type III.
The luxABCDE operon was carried by three vectors. Plasmid pSB2025 (27) carries the operon cloned into the superlinker plasmid pSL1190 (Amersham Pharmacia Biotech, Uppsala, Sweden). Plasmid pAL2 is a derivative of shuttle vector pVA838, in which the luxABCDE operon from pSB2025 has been cloned under the control of the Pami constitutive promoter of the Streptococcus pneumoniae aminopterin resistance operon (1). In the third construct, the luxABCDE operon fused to the Pami promoter was cloned in plasmid pGB2 (8) in three steps. First, the luxABCD BamHI fragment was cloned in the vector digested with the same enzyme, leading to pGB2ΩluxABCD. The XbaI-PstI fragment of pGB2ΩluxABCD containing the 3′ portion of luxD was then replaced by the XbaI-PstI fragment from pAL2 containing the 3′ portion of luxDE, generating pGB2ΩluxABCDE. Finally, an EcoRI fragment containing the Pami promoter from pAL2 was created by hybridization of two complementary primers and cloned in pGB2ΩluxABCDE digested with EcoRI to generate pAT881(pGB2ΩPamiluxABCDE).
Genes lucR and lucG for the red (PpyRE-TS) and green (PpyGR-TS) thermostable P. pyralis luciferase mutants (4) were cloned under the control of the strong Ptet promoter (21) in pCR-blunt (Invitrogen, Cergy-Pontoise, France). The PtetlucR and PtetlucG inserts were excised with PstI and BamHI and ligated to pAT113 (32) digested similarly, resulting in pAT882(pAT113ΩPtetlucR) and pAT883(pAT113ΩPtetlucG), respectively. Plasmid pAT113 is an integrative vector in gram-positive bacteria and a replicative vector in gram-negative bacteria. To obtain plasmids pAT884(pTVCΩPtetlucG) and pAT885(pTVCΩPtetlucR), PtetlucG and PtetlucR were, respectively, excised from pAT883(pAT113ΩPtetlucG) and pAT882(pAT113ΩPtetlucR) by digestion with PstI and BamHI and ligated to the pTVC-erm shuttle vector digested similarly. Plasmid DNA isolation, digestion with restriction endonucleases, amplification by PCR with Pfu DNA polymerase (Stratagene, La Jolla, CA), ligation with T4 DNA ligase (New England Biolabs, Beverly, MA), nucleotide sequencing, and transformation of E. coli were performed by standard methods (30). Strains were grown at 37°C in Luria-Bertani (LB) broth (Difco Laboratories, Detroit, MI) containing streptomycin (100 μg/ml) and erythromycin (150 μg/ml), kanamycin (50 μg/ml), spectinomycin (50 μg/ml), or ampicillin (100 μg/ml).
In vitro plasmid stability.
Stability of the various bioluminescent plasmids in E. coli was assessed by transferring cultures (1,000-fold diluted) to fresh LB broth every 24 h over 7 days. At every transfer, cells were plated on nonselective medium and the proportion of bioluminescent bacteria was determined by replica plating on the appropriate antibiotic followed by screening for bioluminescent colonies. When necessary, one ml of d-luciferin substrate (Caliper) at 150 μg/ml in phosphate-buffered saline (PBS) was added directly to the colonies just before bioluminescence measurement.
Mouse colonization.
Experiments were carried out with the streptomycin-treated mouse model that has been used extensively to study colonization of the large intestine by E. coli (25). Briefly, four 6-week-old BALB/c female mice (Janvier, Le Genest-Saint-Isle, France) were given drinking water containing 5 g/liter of streptomycin sulfate for 24 h and then starved for food overnight. Bioluminescent E. coli was grown in LB medium at 37°C with shaking to the exponential phase, washed twice, and resuspended at 108 CFU per ml in PBS. A portion (100 μl) of the bacterial suspension mixed with 100 μl of 20% sucrose plus 6% NaHCO3 was administered orogastrically with feeding needles, and food and streptomycin-water were returned to the mice. Fresh fecal samples collected at 24 h, 48 h, and 72 h and on every other day thereafter for 14 days were weighed, homogenized in PBS (0.1 g/ml), diluted, and plated on LB agar containing streptomycin and erythromycin, kanamycin, or spectinomycin to select, respectively, E. coli MG1655-Strr, 55989-Strr, or BM2711- Strr harboring bioluminescent plasmids pAL2(pVA838ΩPamiluxABCDE), pAT882(pAT113ΩPtetlucR), or pAT881(pGB2ΩPamiluxABCDE). At 72 h postfeeding, some mice colonized by E. coli MG1655-Strr/pAT881(pGB2ΩPamiluxABCDE) were sacrificed and the small and large intestines (cecum plus colon) were removed. The bioluminescence signal was monitored for intact or longitudinally opened organs. The small intestine and the large intestine were then homogenized mechanically in 3 ml and 5 ml of sterile PBS, respectively, and the number of viable bacteria per organ homogenate was determined by plating onto LB agar containing the appropriate antibiotics. The care and the use of experimental animals complied with local animal welfare laws and guidelines.
Bioluminescence quantification.
Bioluminescence imaging was performed using an IVIS 100 imaging system (100 series; Caliper Corp., Alameda, CA), which consists of a cooled charge-coupled-device camera mounted on a light-tight specimen chamber (dark box), a camera controller, a cryogenic refrigeration unit, and a Windows computer system. The bioluminescence of the bacterial luciferase was measured without filter (open). To separate spectral emissions of the green light- and red light-emitting firefly luciferase mutants, the imaging system was equipped with 2 band-pass filters that are 20 nm wide, with central wavelengths of 540 nm and 620 nm (4, 5).
The firefly luciferase mutants require the exogenous addition of d-luciferin substrate to produce light. d-Luciferin (chromaGlo reagent; Promega, Madison, WI) was used for in vitro experiments and d-luciferin potassium salt (Caliper) for in vivo experiments in which optimal signals were obtained 3 h after intragastric inoculation with feeding needles of 200 μl/mouse of d-luciferin at 15 mg/ml.
For feces analysis, 30 μl of the fecal samples homogenized in PBS (0.1 g/ml) was added to a 96-well black microplate. For living-animal imaging, mice were anesthetized in an oxygen-rich induction chamber with 2% isoflurane gas and anesthesia was maintained during imaging by using a nose cone isoflurane-oxygen delivery device in the specimen chamber.
The microplates or the mice in ventral position were placed in the warmed specimen chamber (37°C), and photon emission was measured, with exposure times ranging from 30 s to 1 min depending on the bioluminescence intensity signal.
For quantification of red and green bioluminescent signals in mixed cultures of red and green bioluminescent E. coli in the feces or in whole animals inoculated with various proportions of the two bioluminescent strains, images were acquired using the 540-nm filter to record the signal emitted by the bacterial population containing the green firefly luciferase mutant and the 620-nm filter for those producing the red firefly luciferase mutant. To determine the spectral overlaps of the luciferase mutants, the green and the red signals were also measured separately in similar mixtures of cells containing only one bioluminescent strain. The green overlap corresponds to the red emission measured with the green filter (540 nm) and the red overlap to the green emission measured with the red filter (620 nm). These independent measurements allowed subtraction of the red- and green-emission spectrum overlaps with the 620-nm and 540-nm filters, respectively, leading to individual quantification of the red and green emissions when they were in mixtures.
The data are presented as pseudocolor images indicating light intensity (red being the most intense and blue the least intense), which are superimposed over the grayscale reference photographs. The region of interest was selected manually and the signal expressed as the total number of photons emitted per second (photons/s) by using the IgorPro image analysis package.
RESULTS AND DISCUSSION
In vitro stability of the bioluminescent plasmids.
Bioluminescence imaging has been used previously to follow in vivo infectious processes or to measure drug efficacy, but mostly in short-term studies (less than 48 h). For long-term studies, stable maintenance of a bioluminescence plasmid usually requires concomitant antibiotic selection, which can interfere with the infectious process under study, particularly in the case of bacterial intestinal colonization. An alternative is to integrate the lux operon in the bacterial chromosome, but this requires engineering of every strain used.
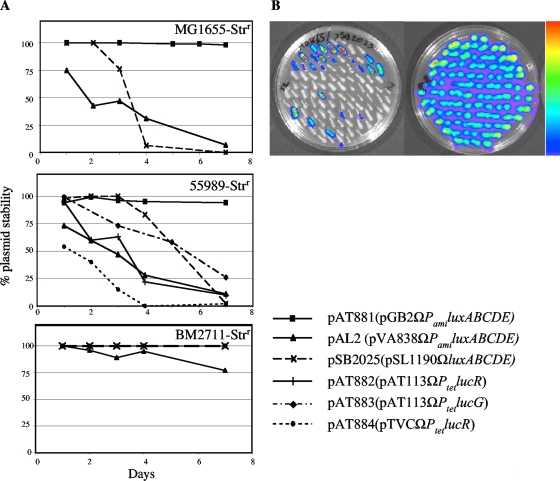
Stable bacterial maintenance of plasmids bearing the luciferase system in the absence of a selective agent is a prerequisite. The stability of three bioluminescent plasmids in three E. coli strains, MG1655-Strr, 55989-Strr, and BM2711-Strr (Table 1), was thus tested in vitro by subculture in LB broth over 7 days with replica plating every 24 h of an aliquot on medium containing the appropriate antibiotic; the bioluminescent signal of the colonies on the selective plates was also monitored in parallel (Fig. 1).
FIG. 1.
In vitro stability of bioluminescent plasmids. (A) The stability of the plasmids in E. coli MG1655-Strr, 55989-Strr, and BM2711-Strr was assessed by subculturing each strain in LB broth over 7 days and by replica plating on the appropriate antibiotic. The stability of a plasmid is expressed as the percentage of colonies on the selective plate relative to that on the nonselective plate. (B) The bioluminescent signal of every ampicillin- or erythromycin-resistant colony of E. coli MG1655-Strr/pSB2025 (left) and E. coli MG1655-Strr/pAL2 (right) after 2 days of culture was determined.
In E. coli MG1655-Strr, pSB2025 appeared stable during the first 2 days of culture, but thereafter rapid plasmid loss was observed (Fig. 1A). There was even more rapid loss of the bioluminescence signal, with only 24% bioluminescent colonies among the ampicillin-resistant colonies, after two transfers (Fig. 1B, left). Plasmid pAL2, which has been reported to be stable in vitro in S. pneumoniae (1), was also gradually lost from MG1655-Strr without additional loss of the luxABCDE operon; 100% of the erythromycin-resistant cells remained bioluminescent (Fig. 1B). The levels of stability of both plasmids were similar in E. coli 55989-Strr (Fig. 1A), and a loss of the bioluminescence signal was also observed for the plasmid pSB2025 during the successive transfers (data not shown). Surprisingly, in E. coli BM2711-Strr both pSB2025 and pAL2 were remarkably stable, with 100% and 77% of bioluminescent colonies after 7 days of subculture, respectively (Fig. 1A and B, right).
Low-copy-number plasmid pAT881(pGB2ΩPamiluxABCDE) was remarkably stable in E. coli MG1655-Strr, 55989-Strr, and BM2711-Strr, with, respectively, 98%, 94%, and 100% of bioluminescent and spectinomycin-resistant colonies after growth for 7 days (Fig. 1A).
For the firefly luciferase system, plasmid pAT884(pTVCΩPtetlucR) was very unstable in E. coli 55989-Strr, with almost a 50% loss after overnight culture (Fig. 1A), and was not used. After 3 days of growth, 63% and 73% of the bacteria still harbored plasmids pAT882 and pAT883, respectively, and 100% of the kanamycin-resistant cells remained bioluminescent, indicating no luciferase gene loss by the plasmid (data not shown).
These data confirm that plasmid stability depends on the plasmid, the genetic background of the host (15), and the presence of another plasmid in the strain (e.g., in 55989-Strr) (2). The new plasmid pAT881(pGB2ΩPamiluxABCDE) displayed good in vitro stability in the three E. coli strains studied and appears to be a good candidate for long-term in vivo studies.
Bioluminescence monitoring of mouse intestinal colonization.
Oral streptomycin treatment, which selectively removes facultative anaerobes of the intestinal tract while leaving the anaerobic microflora essentially intact, allows mouse intestinal colonization by E. coli K-12 (25). In this model, colonization ability is assessed by monitoring daily the number of CFU per gram of feces after intragastric inoculation. However, fecal counts are tedious and imply, in case of cocolonization, that strains have to be tagged with different selection markers. Moreover, this approach does not provide any direct indication about the anatomical localization of the colonization.
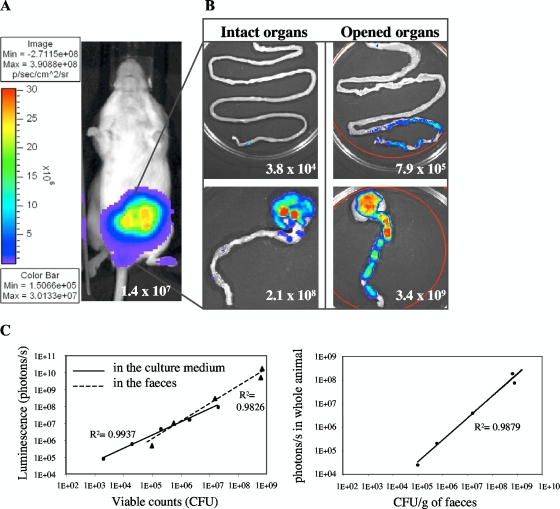
The very good correlation between the CFU counts and the bioluminescent signal in photons/s (R2 = 0.98 versus R2 = 0.99 for bioluminescent bacteria directly diluted in culture medium) (Fig. 2C) obtained with serial dilutions of feces from mice fed with E. coli MG1655-Strr/pAL2 indicated that photon emission levels accurately reflect bacterial numbers in the feces. The bioluminescence system allowed the detection of bacterial quantities as low as 103 CFU/ml in broth medium and ca. 105 CFU/g of feces in feces samples.
FIG. 2.
Monitoring of intestinal colonization by bioluminescence imaging in whole animals. (A) E. coli MG1655-Strr/pAL2(pVA838ΩPamiluxABCDE) (107 CFU) was inoculated intragastrically into four streptomycin-treated mice, and the bioluminescent signal was measured transcutaneously in whole animals 24 h postfeeding. The intensity of the transcutaneous photon emission is represented as a pseudocolor image. (B) The small (top) and large (bottom) intestines were dissected at 72 h, and the photon/s per organ was quantified on intact (left) or longitudinally opened (right) organs. (C) Linear correlation between the bacterial counts and the bioluminescent signal, determined by serial dilutions of bioluminescent bacteria in culture medium or in feces (left). Correlation between the transcutaneous photon emission measured in the whole animals and the number of bioluminescent bacteria in the feces during colonization (right).
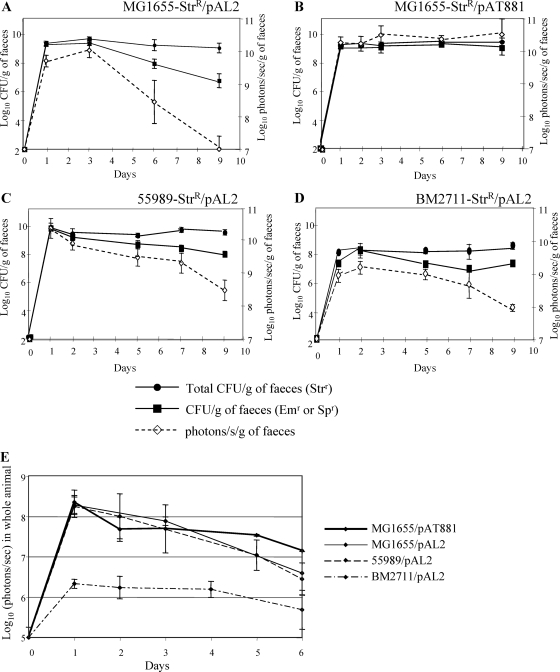
E. coli MG1655-Strr/pAL2(pVA838ΩPamiluxABCDE) was used to monitor intestinal colonization of streptomycin-treated mice by bioluminescence. Sets of four mice were intragastrically inoculated with 107 CFU in 200 μl PBS with 10% sucrose plus 3% NaHCO3, and the bioluminescent signal was monitored, both transcutaneously on whole animals and in the feces (Fig. 2). As early as 24 h postfeeding, a strong bioluminescent signal was observed transcutaneously in the abdominal region (Fig. 2A) and in the feces (Fig. 3A). Counts on streptomycin-containing plates, which represent all E. coli CFU, showed that MG1655-Strr/pAL2 had the classical colonization profile of E. coli K-12, with an initiation stage (0 to 3 days postfeeding) followed by a maintenance stage (3 days postfeeding and beyond), at 109 CFU/g of feces (7), indicating that the presence of pAL2 in the strain did not interfere with the colonization ability. The signal in the feces remained at a plateau of 1010 photons/s/g of feces until day 3 and then slowly decreased to 107 photons/s/g of feces at day 9 (Fig. 3A); the similar decrease, from 109 to 106 CFU/g of feces, of colonies on erythromycin-supplemented plates indicated a loss of plasmid pAL2 by the host. Similarly, the signal measured in whole animals decreased slowly, from 108 photons/s at day 3 postinoculation to 105 photons/s at day 9 of colonization (Fig. 3E). There was very good correlation (R2 = 0.98) between the bioluminescent signal in the abdominal region measured transcutaneously in whole animals and the CFU counts of bioluminescent bacteria in the feces (Fig. 2C). These results indicated that intestinal colonization can be detected directly by simple recording of the bioluminescent signal in living animals. However, because of its instability in E. coli MG1655-Strr, the bioluminescent reporter plasmid pAL2 allowed real-time monitoring only during the initial steps of intestinal colonization.
FIG. 3.
Comparison of the colonizing abilities of various E. coli strains by bioluminescence. Sets of four mice were fed with 107 CFU of MG1655-Strr/pAL2(pVA838ΩPamiluxABCDE) (A), MG1655-Strr/pAT881(pGB2ΩPamiluxABCDE) (B), 55989-Strr/pAL2(pVA838ΩPamiluxABCDE) (C), or BM2711-Strr/pAL2(pVA838ΩPamiluxABCDE) (D). At the indicated times, colonization was monitored by enumeration of bacteria in the feces plated on streptomycin (100 μg/ml), erythromycin (Em) (150 μg/ml), or spectinomycin (Sp) (50 μg/ml) and by measuring the bioluminescent signal in feces or (E) transcutaneously in whole animals. At every time point, log10 means of the CFU per gram of feces, photons/s per gram of feces, and photons/s in whole animals for each set of four mice in a given experiment are plotted, with standard errors shown.
A long-term study of intestinal colonization was performed with E. coli MG1655-Strr harboring pAT881(pGB2ΩPamiluxABCDE), which was remarkably stable in vitro (Fig. 1A). The level of colonization by this strain was similar to that obtained with MG1655-Strr (Fig. 3B), and the bioluminescent signals obtained in the feces (Fig. 3B) and transcutaneously (Fig. 3E) remained stable until at least day 13 (data not shown). Long-term colonization can thus reliably be assessed by bioluminescence monitoring of MG1655-Strr/pAT881.
Since bioluminescence emission is optimal in an aerobic environment (14), the signal recorded transcutaneously may underestimate the actual colonization due to the limitation in oxygen availability in the intestinal cavity. The small and large intestines (cecum and colon) of mice colonized with E. coli MG1655-Strr/pAL2 were dissected at 72 h, and the bioluminescent signal was recorded on intact or longitudinally opened organs (Fig. 2B). A ca. 10-fold-higher bioluminescent signal was observed for the cecum and large intestine when these organs were opened. No significant bioluminescent signal was recorded from the intact small intestine, but a slight signal was detected, albeit inconsistently, in the distal part of the small intestine when it was longitudinally opened (Fig. 2B). The CFU counts confirmed that the large intestine was the major site of E. coli colonization (109 CFU per organ), whereas only ca. 106 CFU were grown from the small intestine. These results are consistent with previous investigations that identified the mucus overlying the epithelial cells of the large intestine as the principal site of colonization by various E. coli strains, such as MG1655 (25, 26). Taken together, these data indicate that transcutaneous BLI therefore appears to be a good indicator of intestinal colonization. These observations support the recently established notion that aerobic respiration, required for optimal colonization by commensal and pathogenic E. coli, can take place in the intestine in mice (17).
Comparison of intestinal colonization by various E. coli strains by bioluminescence.
Strain 55989-Strr is a diarrhea-associated enteroaggregative E. coli strain with particular adhesion properties (fimbriae type III) and constitutes an emerging pathotype responsible for enteric illness (2). The biochemical characteristics and virulence factors implicated in host infectivity and in intestinal colonization ability of this pathogenic strain have recently been studied (3, 22). The levels of colonization by 55989-Strr/pAL2, between 108 and 109 CFU/g of feces, and bioluminescence, from 109 to 1010 photons/s/g of feces, were similar to those obtained with MG1655-Strr/pAL2 (Fig. 3C). E. coli BM2711-Strr is a RecA1 derivative of K-12 MM294 (Table 1). The level of intestinal colonization by E. coli BM2711-Strr/pAL2, from 107 to 108 CFU/g of feces and 109 photons/s/g of feces (Fig. 3D), was lower than those of 55989-Strr/pAL2 and MG1655-Strr/pAL2. This difference is within the range observed when strains with colonizing defects, related for example to differences in sugar consumption, are studied (10). The difference in colonization ability among MG1655-Strr, 55989-Strr, and BM2711-Strr was easily detected by bioluminescence measure of whole animals; the transcutaneous photon emission of MG1655-Strr/pAL2 and 55989-Strr/pAL2 was ca. 108 photons/s, whereas BM2711-Strr/pAL2 colonization resulted in a signal of only ca. 106 photons/s (Fig. 3E). Transcutaneous BLI can therefore be used for rapid screening of the colonizing ability of mutant strains or of enteropathogenic E. coli.
Streptomycin treatment regimen for intestinal colonization.
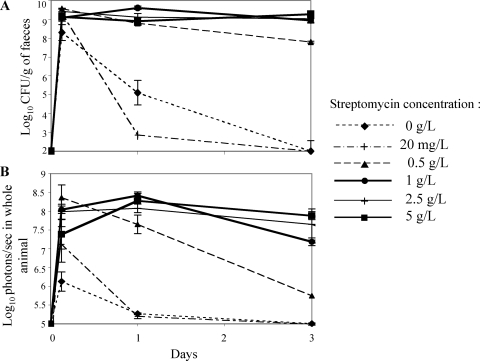
The standard protocol for intestinal colonization of mice has remained as originally described in 1982 by Myhal et al. (25), with mice fed orally with 5 g/liter of streptomycin in the drinking water. To determine the minimal dose of streptomycin required to eliminate the resident facultative anaerobic microflora to allow colonization by E. coli, mice were given, prior to colonization, water containing various concentrations of streptomycin (20 mg/liter, 0.5 g/liter, 1 g/liter, 2.5 g/liter, and 5 g/liter). Colonization by MG1655-Strr/pAL2 was monitored by feces counts (Fig. 4A) and bioluminescence (Fig. 4B). There was no colonization in the absence or in the presence of 20 mg/liter of streptomycin. However, at a streptomycin concentration as low as 1 g/liter, colonization reached levels similar to those obtained with the standard concentration of 5 g/liter. When mice were treated with 0.5 g/liter, colonization reached the same high level but decreased slowly from 10 to 8 log10 CFU/g of feces and from 8.5 to 5.5 log10 photons/s/g of feces between days 1 and 3, suggesting that this concentration was too low to maintain colonization in the digestive tracts of mice. Bioluminescence is therefore a useful tool to directly assess the host parameters involved in intestinal colonization, such as the role of the innate immune system.
FIG. 4.
Intestinal colonization by E. coli MG1655-Strr/pAL2 of mice treated with various concentrations of streptomycin. The log10 means of CFU/g of feces (A) and photons/s in whole animal (B) for each set of two mice are presented for every time point, with bars indicating standard errors.
Firefly luciferase can be used to trace intestinal colonization by E. coli.
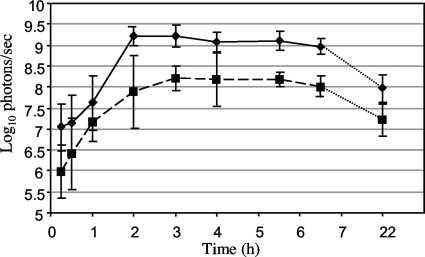
The 6-kb lux operon from P. luminescens encodes both the luciferase and the luciferin substrate required for the bioluminescence reaction (23). Mutants PpyRE-TS and PpyGR-TS, derived from wild-type firefly luciferase, are encoded by single genes of 1,650 bp that are 99.4% identical (4). Bacteria expressing the firefly luciferases do not produce firefly luciferin, and the substrates, identical for the two variants, have to be added exogenously whereas the ATP is available endogenously. d-Luciferin is usually injected via the intraperitoneal route and distributes rapidly throughout the mice (31). After intraperitoneal injection of 200 μl/mouse of the substrate at 15 mg/ml in PBS to mice colonized by E. coli 55989-Strr/pAT882(pAT113ΩPtetlucR) or E. coli 55989-Strr/pAT882(pAT113ΩPtetlucG), the bioluminescent signal peaked 15 to 20 min postinoculation (6.6 log10 photons/s) and decreased rapidly (data not shown). However, after administration of the same amount of substrate via the intragastric route, the transcutaneous red and green bioluminescent signals increased regularly during the first 2 h (approximately 2 log10), reaching a plateau that lasted at least 4 h (7.9 log10 photons/s for PpyGR-TS and 9.2 log10 photons/s for PpyRE-TS), and then started to decrease slowly 6 h postinoculation (Fig. 5). The transcutaneous signals obtained with PpyRE-TS and the bacterial luciferase were comparable (10 log10 photons/s). In all subsequent experiments, the bioluminescent signal was therefore measured at an early stage of the plateau, i.e., 3 h after intragastric inoculation of the substrate. Of note, the red signal was always approximately 1 log10 higher than the green one (Fig. 5), consistent with the notion that blue and green emission spectra are more absorbed by tissue hemoglobin than the red emission spectrum (35).
FIG. 5.
Transcutaneous bioluminescence kinetic profile after intragastric administration of d-luciferin. d-Luciferin was administered intragastrically 24 h after bacterial inoculation of mice colonized with E. coli 55989-Strr harboring pAT882(pAT113ΩPtetlucR) or pAT883(pAT113ΩPtetlucG), and the red (♦) or green (▪) bioluminescent signal was monitored for 22 h.
Intestinal colonization by E. coli 55989-Strr was monitored with the firefly PpyRE-TS luciferase reporter gene using plasmid pAT882(pAT113ΩPtetlucR) (Table 1). In two independent experiments, E. coli 55989-Strr/pAT882 gave colonization levels similar to those of E. coli 55989-Strr/pAL2(pVA838ΩPamiluxABCDE) by CFU counts and bioluminescence detection (7.0 ± 0.3 and 6.9 ± 0.4 log10 CFU/g of feces plated on medium containing erythromycin and 8.6 ± 0.2 and 8.3 ± 0.4 log10 photons/s/g of feces, respectively), suggesting that plasmid pAT882 did not interfere with the colonization capacity of the host strain and that the two luciferases have similar levels of luminescence emission in vivo in the microaerobic environment of the mouse intestine.
One potential way to improve luc gene efficiency is to optimize codon usage for bacteria and even more so to the specific host strain. We recently found that human codon optimization of a related red light-emitting luciferase led to a 10-fold improvement in luciferase detectability in the HEK293 cell line (6). The stability of the bioluminescence signal could be improved by insertion of the luciferase reporter gene in the chromosome (20). Preliminary colonization studies have been undertaken with bioluminescent engineered strains of Enterococcus faecalis that contain the red firefly luciferase gene mutant stably inserted in the chromosome at a single copy. Luminescent bacteria could easily be detected transcutaneously and in the feces of gnotobiotic mice during long-term colonization without signal loss (unpublished data). Thus, bioluminescence technology can be applied to other intestine colonizers, such as Enterococcus, for which specific factors implicated in the colonization process are being characterized (28).
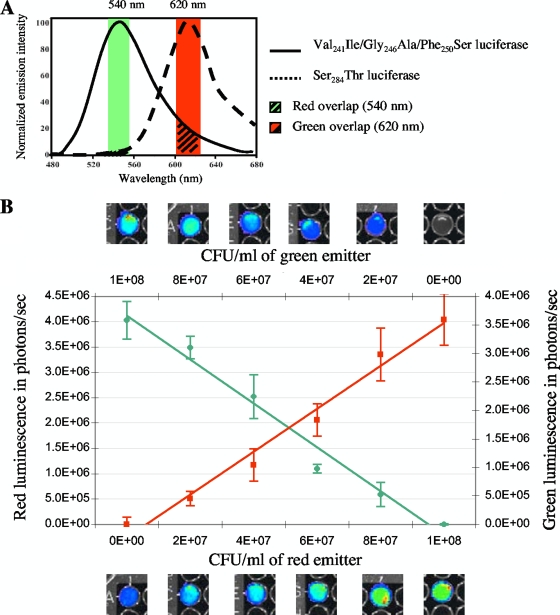
Quantification in vitro and in vivo of two simultaneous bioluminescent signals.
Cocolonization by two strains is frequently performed to study growth rates or nutrient requirements during intestinal colonization or to compare commensal or pathogenic E. coli strains (10). Bioluminescence could be used to distinguish the strains without the need for introducing distinct selectable markers that could alter the fitness of the host. The green and red luciferase mutants have emission spectra at 546 and 610 nm at 25°C, respectively, which can be separated by narrow band-pass filters to minimize spectral overlap (Fig. 6). The green overlap with the red filter (620 nm) is more important than the negligible red overlap with the green filter (540 nm). To determine the consequences of spectral overlap on the ability to measure accurately the individual red and green bioluminescence emissions, the signals of E. coli 55989-Strr suspensions composed of various proportions of bacteria harboring pAT882(pAT113ΩPtetlucR) or pAT883(pAT113ΩPtetlucG) were measured in vitro (Fig. 6). The results were then compared with the bioluminescent signals obtained from green light- or red light-emitting bacteria mixed in the same proportion with nonbioluminescent 55989-Strr. The green and red signals of the bacterial mixtures measured simultaneously were, after correction for overlap, proportional to the corresponding bacterial numbers in the suspension (R2 = 0.98 for both the red and the green signal) (Fig. 6). These results indicate that the dual bioluminescence detection system allows direct in vitro quantification of two bacterial populations by simultaneous measurement of red and green emitted signals, thus avoiding bacterial enumeration.
FIG. 6.
In vitro simultaneous bioluminescence monitoring of various bacterial mixtures of green light- and red light-emitting E. coli 55989-Strr. (A) Spectral emissions of the green light-emitting (Val241Ile/Gly246Ala/Phe250Ser) and red light-emitting (Ser284Thr) luciferase mutants. (B) Suspensions of ca. 108 CFU/ml of mixed green light- and red light-emitting bacteria at ratios of 0:100, 20:80, 40:60, 60:40, 80:20, and 100:0 were analyzed. Similar ratios of green light- or red light-emitting bacteria with nonbioluminescent E. coli 55989-Strr were analyzed. The enzymatic reaction was initiated by adding chromaGlo reagent just before imaging. Mean values are plotted for each dilution, and standard errors are indicated by the bars.
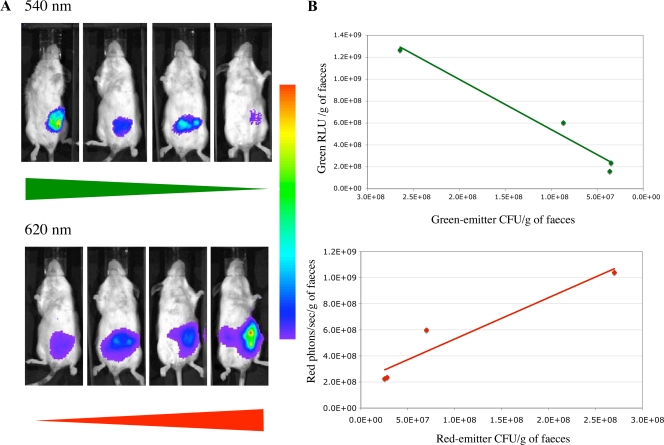
Various proportions (20:80, 50:50, 80:20, and 100:0) of red light- and green light-emitting E. coli 55989-Strr were inoculated orogastrically into streptomycin-treated mice, and the bioluminescent signal was measured transcutaneously and in the feces 3 h after inoculation of the substrate (Fig. 7). The mice were assigned to three groups: the G group was inoculated with various proportions of E. coli 55989-Strr with or without pAT883(pAT113ΩPtetlucG), the R group was inoculated with mixtures of E. coli 55989-Strr with or without pAT882(pAT113ΩPtetlucR), and the M group was inoculated with various mixtures of E. coli 55989-Strr containing pAT882 or pAT883. The groups inoculated with bacteria emitting a single bioluminescent signal allowed determination of the spectral overlaps of the 540- and 620-nm filters. Since the red light- and green light-emitting bacteria in a mouse were indistinguishable on the basis of antibiotic resistance, the CFU counts of the two types of bacteria in the mixture were deduced from the counts of the bioluminescent bacteria in the R and G groups. Twenty-four hours postinoculation, there was very good correlation between the bioluminescent red and green signals and the red light- and green light-emitting CFU counts in the feces (Fig. 7B).
FIG. 7.
In vivo BLI of mice colonized with mixtures of red light- and green light-emitting E. coli. Various proportions (20:80, 50:50, 80:20, and 100:0) of red light- and green light-emitting 55989-Strr were inoculated orogastrically into streptomycin-treated mice. The bioluminescence signal was measured transcutaneously (A) and in fecal samples (B) 24 h after inoculation of mice, using optical filters. Values are means from two mice.
Similar results were observed when the bioluminescent signals were measured transcutaneously in whole animals, but, as already mentioned, the red signal was stronger due to greater green absorption by animal tissues (Fig. 7A). However, the transcutaneous signal was more variable than that in the feces (data not shown).
These results indicate that monitoring of two bacterial populations in the gut of a single animal can be achieved by quantification of bioluminescence signals, either in the feces or transcutaneously in whole animals. This system, which allows direct monitoring of two bacterial populations simultaneously, offers a simple and direct method to study in vitro and in vivo competition between mutants and the parental strain.
In conclusion, we have demonstrated that intestinal colonization can be monitored in whole living animals with genetically engineered E. coli strains that produce either the bacterial luciferase from P. luminescens or the firefly luciferase mutants PpyRE-TS and PpyGR-TS from P. pyralis and that two bioluminescent signals can simultaneously be monitored and quantified in the same living mouse. BLI is therefore a very useful tool to compare the relative colonization efficiencies of commensal and pathogenic E. coli strains and further elucidate the mechanism of colonization resistance (19). The stability of the bioluminescence signal in vivo has been improved by constructing plasmid pAT881(pGB2ΩPamiluxABCDE), which allows long-term monitoring of intestinal colonization without the need for antibiotic selection for plasmid maintenance. This reporter system has numerous potential applications for monitoring the evolution of infectious diseases in animal models in the absence of antibiotic selective pressure and without killing the animals.
Acknowledgments
We thank P. S. Cohen for providing E. coli MG1655-Strr, C. Le Bouguénec for providing E. coli 55989-Strr and for helpful discussions, M.-A. Nicolas for help with bioluminescence experiments, and P. E. Reynolds for critical reading of the manuscript.
This work was supported by grants from the European Commission; grant no. LSHM CT 2005 518152-EAR, which included a fellowship in support of M.-L.F.; and grants from the Air Force Office of Scientific Research (FA9550-07-1-0043), the National Science Foundation (MCB0842831), and the Hans & Ella McCollum 21 Vahlteich Endowment.
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Beard, S. J., V. Salisbury, R. J. Lewis, J. A. Sharpe, and A. P. MacGowan. 2002. Expression of lux genes in a clinical isolate of Streptococcus pneumoniae: using bioluminescence to monitor gemifloxacin activity. Antimicrob. Agents Chemother. 46:538-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernier, C., P. Gounon, and C. Le Bouguenec. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70:4302-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier-Fébreau, C., L. du Merle, E. Turlin, V. Labas, J. Ordonez, A. M. Gilles, and C. Le Bouguenec. 2004. Use of deoxyribose by intestinal and extraintestinal pathogenic Escherichia coli strains: a metabolic adaptation involved in competitiveness. Infect. Immun. 72:6151-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branchini, B. R., D. M. Ablamsky, M. H. Murtiashaw, L. Uzasci, H. Fraga, and T. L. Southworth. 2007. Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal. Biochem. 361:253-262. [DOI] [PubMed] [Google Scholar]
- 5.Branchini, B. R., T. L. Southworth, N. F. Khattak, E. Michelini, and A. Roda. 2005. Red- and green-emitting firefly luciferase mutants for bioluminescent reporter applications. Anal. Biochem. 345:140-148. [DOI] [PubMed] [Google Scholar]
- 6.Caysa, H., R. Jacob, N. Muther, B. Branchini, M. Messerle, and A. Soling. 2009. A red shifted codon-optimized firefly luciferase is a sensitive reporter for bioluminescence imaging. Photochem. Photobiol. Sci. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 9.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 10.Fabich, A. J., S. A. Jones, F. Z. Chowdhury, A. Cernosek, A. Anderson, D. Smalley, J. W. McHargue, G. A. Hightower, J. T. Smith, S. M. Autieri, M. P. Leatham, J. J. Lins, R. L. Allen, D. C. Laux, P. S. Cohen, and T. Conway. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grillot-Courvalin, C., S. Goussard, F. Huetz, D. M. Ojcius, and P. Courvalin. 1998. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 16:862-866. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 14.Hastings, J. W. 1996. Chemistries and colors of bioluminescent reactions: a review. Gene 173:5-11. [DOI] [PubMed] [Google Scholar]
- 15.Helinski, D. R. 2004. Introduction to plasmids: a selective view of their history, p. 1-21. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. American Society for Microbiology, Washington, DC.
- 16.Hutchens, M., and G. D. Luker. 2007. Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 9:2315-2322. [DOI] [PubMed] [Google Scholar]
- 17.Jones, S. A., F. Z. Chowdhury, A. J. Fabich, A. Anderson, D. M. Schreiner, A. L. House, S. M. Autieri, M. P. Leatham, J. J. Lins, M. Jorgensen, P. S. Cohen, and T. Conway. 2007. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75:4891-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 19.Leatham, M. P., S. Banerjee, S. M. Autieri, R. Mercado-Lubo, T. Conway, and P. S. Cohen. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect. Immun. 77:2876-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, P., P. Trieu-Cuot, and P. Courvalin. 1986. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 14:7047-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Jéhanne, V., L. du Merle, C. Bernier-Fébreau, C. Usein, A. Gassama-Sow, A.-A. Wane, M. Gouali, M. Damian, A. Aïdara-Kane, Y. Germani, A. Fontanet, B. Coddeville, Y. Guérardel, and C. Le Bouguénec. 2009. Role of deoxyribose catabolism in colonization of the murine intestine by pathogenic Escherichia coli strains. Infect. Immun. 77:1442-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meighen, E. A. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7:1016-1022. [DOI] [PubMed] [Google Scholar]
- 24.Møller, A. K., M. P. Leatham, T. Conway, P. J. Nuijten, L. A. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myhal, M. L., D. C. Laux, and P. S. Cohen. 1982. Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur. J. Clin. Microbiol. 1:186-192. [DOI] [PubMed] [Google Scholar]
- 26.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, L. B., V. Lakticova, L. L. Carias, S. Rudin, R. Hutton, and S. H. Marshall. 2009. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J. Infect. Dis. 199:342-349. [DOI] [PubMed] [Google Scholar]
- 29.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iversen, D. L. Le Tourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr, Jr. 2001. Validation of a noninvasive, real-time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thigh model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shinde, R., J. Perkins, and C. H. Contag. 2006. Luciferin derivatives for enhanced in vitro and in vivo bioluminescence assays. Biochemistry 45:11103-11112. [DOI] [PubMed] [Google Scholar]
- 32.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene 106:21-27. [DOI] [PubMed] [Google Scholar]
- 33.Wiles, S., K. M. Pickard, K. Peng, T. T. MacDonald, and G. Frankel. 2006. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 74:5391-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood, K. V., Y. A. Lam, H. H. Seliger, and W. D. McElroy. 1989. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science 244:700-702. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, H., T. C. Doyle, O. Coquoz, F. Kalish, B. W. Rice, and C. H. Contag. 2005. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J. Biomed. Opt. 10:41210. [DOI] [PubMed] [Google Scholar]