Abstract
Contamination of meat products with food-borne pathogens usually results from the carcass coming in contact with the feces of an infected animal during processing. In the case of Salmonella, pigs can become colonized with the organism during transport and lairage from contaminated trailers and holding pens, resulting in increased pathogen shedding just prior to processing. Increased shedding, in turn, amplifies the likelihood of carcass contamination by magnifying the amount of bacteria that enters the processing facility. We conducted a series of experiments to test whether phage therapy could limit Salmonella infections at this crucial period. In a preliminary experiment done with small pigs (3 to 4 weeks old; 30 to 40 lb), administration of an anti-Salmonella phage cocktail at the time of inoculation with Salmonella enterica serovar Typhimurium reduced Salmonella colonization by 99.0 to 99.9% (2- to 3-log reduction) in the tonsils, ileum, and cecum. To test the efficacy of phage therapy in a production-like setting, we inoculated four market-weight pigs (in three replicates) with Salmonella enterica serovar Typhimurium and allowed the challenged pigs to contaminate a holding pen for 48 h. Sixteen naïve pigs were randomly split into two groups which received either the anti-Salmonella phage cocktail or a mock treatment. Both groups of pigs were comingled with the challenged pigs in the contaminated pen. Treatment with the anti-Salmonella phage cocktail significantly reduced cecal Salmonella concentrations (95%; P < 0.05) while also reducing (numerically) ileal Salmonella concentrations (90%; P = 0.06). Additional in vitro studies showed that the phage cocktail was also lytic against several non-Typhimurium serovars.
The U.S. Centers for Disease Control and Prevention report approximately 40,000 culture-confirmed cases of salmonellosis each year in the United States, which result in approximately 400 deaths (5). Many Salmonella outbreaks are associated with meat and poultry (20), with contamination usually resulting from the carcass coming into contact with the feces of a Salmonella-infected animal during processing (22).
There is an association between pork products and Salmonella, as swine are generally considered to be the second largest reservoir of the organism among food animals after poultry. Although infections in adult swine are normally asymptomatic, once colonized, pigs can shed the organism in the feces for weeks and sometimes months (7).
While a great deal of research has been done on developing on-farm anti-Salmonella intervention strategies, these methods are confounded by the fact that Salmonella prevalence in pigs often increases once the animals leave the farm as a result of (i) stress-induced reactivation of preexisting infections (14), (ii) new infections from contaminated transport trailers and processing facility holding pens (12, 15, 24, 31), or (iii) both. Consequently, animals with no history of previous Salmonella infection can begin shedding the organism just prior to processing, which is highly problematic in terms of food safety.
We hypothesized that phage therapy could be developed as an effective means to counteract transport- and lairage-associated increases in Salmonella colonization in swine. Phage therapy has the advantage of being natural, nontoxic, and relatively inexpensive and could be used just prior to slaughter, unlike many antibiotics (18, 28). Here we describe a series of experiments demonstrating that treating market-weight pigs with an anti-Salmonella phage cocktail prior to their comingling with Salmonella-infected pigs in a highly contaminated environment resulted in reductions in Salmonella colonization. We further show that the phage cocktail could be effectively microencapsulated, making feed or water delivery possible.
MATERIALS AND METHODS
Salmonella strain.
Salmonella enterica serovar Typhimurium γ4232 was used both for isolation of wild-type phages and in challenge experiments. This strain was originally isolated from diseased pigs and contains a nalidixic acid resistance selection marker (7, 11). The phage treatment consisted of 14 wild-type phages isolated specifically for this study, along with the Felix O1 phage (Université Laval, Quebec, Canada).
Wild-type phage isolation.
Wild-type phages were isolated using previously described protocols (19, 23) with some variations. Wastewater samples were collected from 14 different municipal wastewater treatment plants located throughout Indiana. Samples were frozen at −20°C until processed. Thawed samples (20 ml) were centrifuged (2,000 × g for 10 min), and the supernatants were filtered (0.2-μm filters) and treated with chloroform (1:100). Filtered samples were added to log-phase Salmonella enterica serovar Typhimurium in double-strength tryptic soy broth (TSB; Difco, Sparks, MD) containing 50 μg/ml nalidixic acid and incubated at 37°C overnight with shaking. Overnight cultures were centrifuged (1,000 × g for 10 min), filtered (0.2-μm filter), and treated with chloroform (1:100). Serial 10-fold dilutions of the filtered samples (100 μl) were combined with 100 μl of fresh log-phase S. Typhimurium, 67.5 μl of 1 M CaCl2, and 3 ml of nutrient agar (5%) overlay (containing 50 μg/ml nalidixic acid). The phage-overlay solution was then added to nutrient agar plates and incubated overnight at 37°C. One isolate from each wastewater treatment plant (n = 14) was plaque purified three times, suspended in SM buffer (0.1 M NaCl, 1 mM MgSO4, 0.2 M Tris [pH 7.5], 0.01% gelatin) containing 1% chloroform, and stored at −80°C until further use.
Characterization of phages.
Plaques produced by each individual virus (on nutrient plates) were examined for plaque morphology and measured (in mm). Individual viruses were then examined by transmission electron microscopy (TEM). For TEM, a mesh copper grid with carbon-coated Formvar film was floated on a droplet of the phage sample and then stained with 2% uranyl acetate. Images (15,500× and 28,500× magnifications) were collected using a Philips CM-100 TEM operated at 80 kV, spot 3, 200-μm condenser aperture, and 50-μm objective aperture.
Phage cocktail preparation and microencapsulation.
Flasks containing 100 ml of nutrient broth were inoculated with 320 μl of log-phase Salmonella enterica serovar Typhimurium organisms and incubated for 4 h at 37°C with shaking. Individual flasks were then inoculated with 1 ml of phage stock (∼109 to 1010 PFU/ml) and incubated overnight at 37°C with shaking. Overnight cultures were centrifuged (1,000 × g for 10 min), filtered (0.2-μm filter), and treated with chloroform (1:100) as previously described. Each phage isolate was grown individually. The treatment cocktail was a combination of equal volumes of all 14 wild-type phages along with anti-Salmonella phage Felix O1, which was previously shown to have in vivo anti-Salmonella activity (16).
Phages were microencapsulated as a cocktail by using a previously described method with slight modifications (21). Briefly, 3.5 ml (∼1010 PFU/ml) of phage was suspended with 14 ml of 1.5% sodium alginate (Spectrum, Gardena, CA) solution, 1.58 ml Span-85 (Sigma, St. Louis, MO), and 70 ml of canola oil. The solution was mixed for 1 min at 5,500 rpm. While continuing mixing, 17.5 ml of a 0.5% CaCl2, 0.05% ZnCl2 solution was added dropwise. Microspheres were pelleted by centrifugation at 1,000 × g for 5 min, washed with 30 ml of sterile water, and pelleted once again. Microspheres were then washed with 10 ml of sterile water and 20 ml of 0.2% poly-l-lysine and pelleted. The pellet was resuspended in 3.5 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4 [pH 7.5]) and stored at 4°C overnight for use the next morning. Plaque assays were performed (as previously described) on the microencapsulated phages to determine the postprocessing viability of the phage cocktail.
Preliminary trial.
All protocols involving animals were conducted under the approval of the Purdue University Animal Care and Use Committee (PACUC). Sixteen small pigs (3 to 4 weeks old; 30 to 40 lb) were screened for Salmonella. Each pig was then concurrently inoculated (by oral gavage) with 5 × 108 CFU of Salmonella enterica serovar Typhimurium γ4232 and either the microencapsulated phage cocktail (5 ml by oral gavage; ∼109 PFU/ml) or a mock treatment (microencapsulation ingredients with no phage). Pigs were then housed in a group pen at the biosafety level 2 (BSL-2) facility of the USDA ARS Livestock Behavior Research Unit (West Lafayette, IN). Feed was removed from the pen, but all pigs had continual access to water. Pigs were readministered the phage or mock treatment every 2 hours for 6 hours (three doses in total for each pig).
Fecal samples were collected every 2 hours by fecal swab. At 6 hours postinoculation/treatment, a typical length of time that pigs are transported or held in lairage in the United States (25), all pigs were chemically euthanized and exsanguinated. Ileal and cecal contents, tonsil scrapings, mesenteric lymph nodes, and fecal samples were collected from all pigs to determine the concentration and prevalence of Salmonella colonization.
The challenge organism was isolated using previously published methods (7, 11). All bacteriologic media, unless otherwise noted, contained 50 μg/ml nalidixic acid for selective isolation of the challenge organism. Fecal swabs were first enriched in 10 ml tetrathionate broth (overnight at 37°C; Difco), followed by enrichment in Rappaport-Vassiliadis broth (1:10 dilution incubated overnight at 42°C; Difco). Enriched samples were then plated on XLT4 agar (Difco) plates and incubated overnight at 37°C. Mesenteric lymph nodes were homogenized in stomacher bags containing 50 ml tetrathionate broth and incubated at 37°C overnight followed by a second enrichment in Rappaport-Vassiliadis broth and plating on XLT4 agar plates. Tonsil, cecal, and ileal samples were not enriched but 10-fold serially diluted and directly plated on XLT4 agar plates for enumeration. Presumptive Salmonella colonies were chosen at random from each of the samples and confirmed biochemically by incubation in triple sugar iron agar (TSI; Difco) and lysine iron agar (LIA; Difco).
Main trial.
For the main trial, experiments were conducted in triplicate. In each replicate, four market-weight pigs (seeder pigs; ∼250 lb) were inoculated orally with 5 × 109 CFU of Salmonella enterica serovar Typhimurium γ4232. The four seeder pigs were housed in a common pen at the BSL-2 facility of the USDA ARS Livestock Behavior Research Unit (West Lafayette, IN) and fed ad libitum. Manure was allowed to accrue for 48 h, and pen floors were periodically sprayed with water to create a slurry like that often seen in commercial abattoir holding areas. Fecal samples were collected from each pig every 24 h to monitor Salmonella infection among the seeder pigs.
At 48 h postinoculation, eight naïve pigs were screened for Salmonella and administered the microencapsulated phage cocktail orally (15 ml; ∼109 PFU/ml). Eight additional naïve pigs were screened for Salmonella and administered a mock treatment (microencapsulation ingredients with no phage) as controls. Both sets of pigs were then comingled with the Salmonella-infected pigs (seeder pigs) in the Salmonella-contaminated holding pen. Feed was removed from the pen, but all pigs had continual access to water. Phage-treated pigs were readministered the phage cocktail every 2 hours for 6 hours (three doses in total per pig), while control pigs received the mock treatment every 2 hours for 6 hours. Fecal samples were collected from each pig every 2 hours for isolation of the challenge Salmonella.
At 6 hours postcomingling, a typical length of time that pigs are transported or held in lairage in the United States (25), all pigs were euthanized by captive bolt and exsanguination. Ileal and cecal contents, mesenteric lymph nodes, and fecal samples were collected to determine the level of Salmonella colonization and for phage reisolation. The challenge organism was isolated from each sample as previously described above. Anti-Salmonella phages were isolated by standard plaque assay as described above. All plaque assays were conducted under identical conditions in terms of bacterial concentrations, incubation times, etc., ensuring uniformity in phage enumeration across samples.
Phage specificity tests.
The specificity of the phage cocktail was measured by testing the ability of the treatment to lyse (in vitro) several non-Typhimurium Salmonella strains. Plaque assays were performed as described above for each of the following Salmonella serovars: Kentucky 1271-94, Tennessee 825-94, Litchfield 804-94, Schwarzengrund 507-94, Senftenberg 1402-94, Indiana 1110-94 (all kindly provided by Arun Bhunia, Department of Food Science, Purdue University), Dublin, and Enteriditis 7759 (kindly provided by John Patterson, Department of Animal Sciences, Purdue University).
Statistical analysis.
Bacterial counts were transformed to a logarithmic scale prior to statistical analysis. Bacterial concentrations (log10 CFU/g) were analyzed using the mixed models procedure of SAS (SAS Institute Inc., Cary, NC). The experimental unit was the individual pig. Statistical inferences were based on a P level of <0.05.
RESULTS
Phage isolation, characterization, and cocktail preparation.
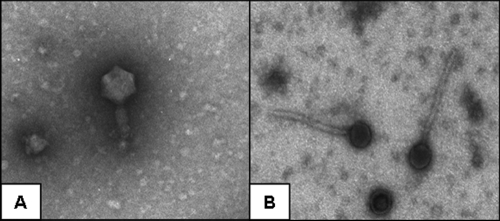
Wastewater samples were collected from 14 separate wastewater treatment plants throughout the state of Indiana. One phage isolate was selected from each facility and plaque purified three times, resulting in the isolation of 14 potentially distinct phage isolates: PEW 1 to 14. The 14 phage isolates were characterized first by plaque morphology. The majority of isolates created medium-sized (∼2-mm), round plaques. The exceptions were PEW 1, 5, 10, and 14, which generated larger, round plaques (∼4 to 5 mm) (data not shown). Isolates were further characterized by TEM. Two distinct types of phage were observed. Both types displayed icosahedral heads, but PEW 1, 2, 3, 8, and 10 were smaller with larger heads and thicker, shorter tails (Fig. 1A). Conversely, PEW 4, 5, 6, 7, 9, 11, 12, 13, and 14 were clearly larger, with smaller heads and longer, narrower tails (Fig. 1B).
FIG. 1.
Isolation and identification of anti-Salmonella phages. Fourteen wild-type anti-Salmonella phages were individually isolated from 14 different wastewater treatment plants. Images are of two phage isolates taken by transmission electron microscopy. Magnification, ×28,500. The isolated phages were separated into two types: smaller phages with larger heads and shorter, thicker tails (A) and larger phages with smaller heads and longer, narrower tails (B).
Prior to making the phage cocktail, titers of the individual phages (premicroencapsulation) were compared to that of the microencapsulated phage cocktail to ensure that the viruses remained viable throughout the microencapsulation process. Microencapsulation reduced the average phage titer from 4.6 × 1010 PFU/ml to 4.0 × 109 PFU/ml. For the treatment, the 15 phage isolates (14 wild-type phages and Felix O1) were grown individually on Salmonella enterica serovar Typhimurium, purified, combined into a cocktail based on volume, and microencapsulated together.
Preliminary trial.
All pigs tested negative for Salmonella prior to challenge. Ileal samples from pigs treated with the phage cocktail contained 99.0% fewer (2-log reduction) Salmonella organisms than ileal samples from pigs receiving the mock treatment (0.6 log10 CFU/ml versus 2.6 log10 CFU/ml, respectively) (Fig. 2). Five of six phage-treated pigs had no detectable Salmonella (below level of enumeration, <100 CFU/ml) in ileal samples (Fig. 2). Tonsil samples from phage-treated pigs contained 99.9% fewer (3-log reduction) Salmonella organisms than tonsil samples taken from pigs receiving the mock treatment (0.4 log10 CFU/ml versus 3.5 log10 CFU/ml, respectively) (Fig. 2). Similar to ileal samples, five of six phage-treated pigs had no detectable Salmonella (below level of enumeration, <100 CFU/ml) in tonsil samples (Fig. 2). Likewise, phage treatment reduced Salmonella counts in cecal samples 99.9% (3-log reduction) in phage-treated pigs compared to mock-treated pigs (0.4 log10 CFU/ml versus 3.6 log10 CFU/ml, respectively) (Fig. 2). Five of six phage-treated pigs had no detectable Salmonella (below level of enumeration, <100 CFU/ml) in cecal samples (Fig. 2).
FIG. 2.
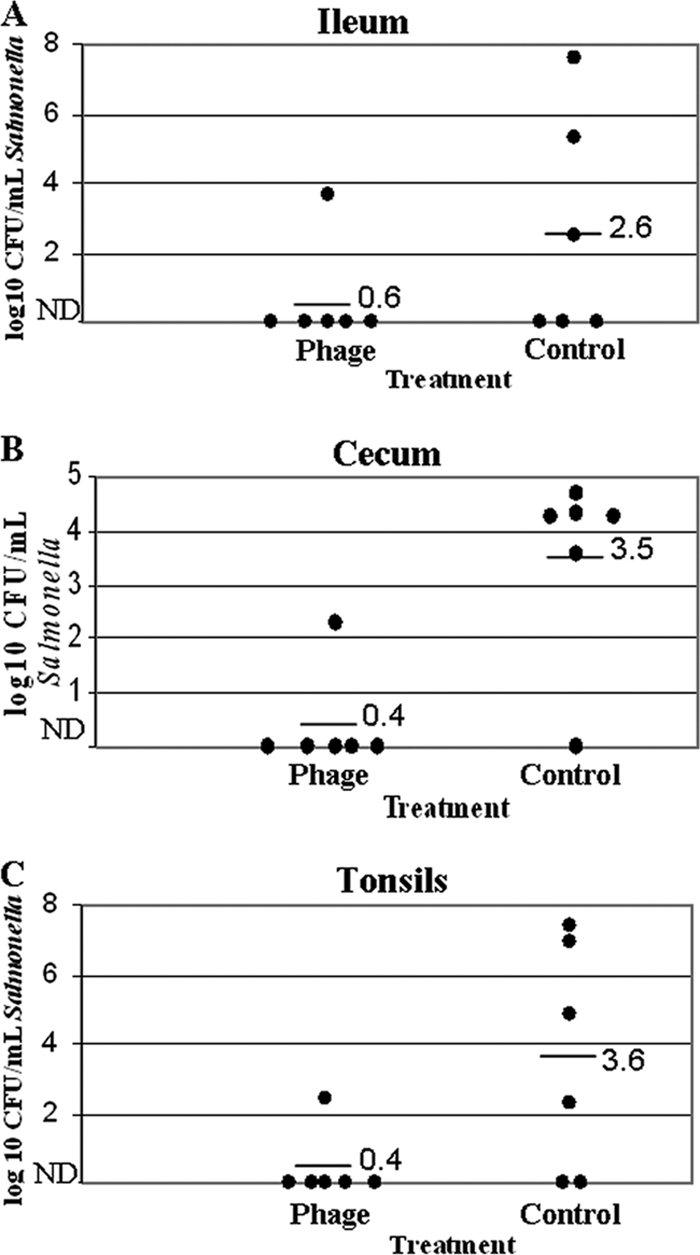
Phage therapy reduces Salmonella colonization in the ileum (A), cecum (B), and tonsils (C) of treated pigs. In a preliminary trial, small pigs (30 to 40 lb; n = 6 per treatment) were coadministered Salmonella enterica serovar Typhimurium and an anti-Salmonella phage cocktail. Black dots represent individual pigs. Horizontal bars and corresponding numbers represent averages within treatments. ND, not detectable/below level of enumeration (<100 CFU/ml).
In samples that were preenriched or enriched prior to plating, i.e., samples providing only qualitative (“yes/no” or “positive/negative”) data, there were no differences between phage-treated and mock-treated pigs. Three of six phage-treated pigs had Salmonella-positive lymph nodes, compared to four of six mock-treated pigs. Similarly, fecal samples from all pigs, both phage treated and mock treated, were positive for the challenge organism by 6 hours postinoculation (data not shown).
Main trial.
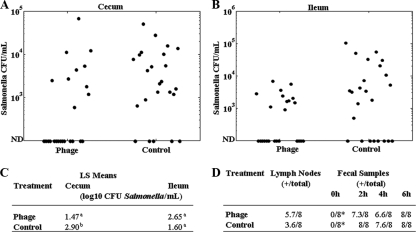
Samples collected from the seeder pigs and the pen (slurry) indicated that each seeder pig was successfully colonized, and the pen was consistently contaminated with the challenge Salmonella (Fig. 3). Cecal samples of phage-treated pigs contained significantly fewer Salmonella organisms (1.5 log10 CFU/ml) than those from mock-treated pigs (2.9 log10 CFU/ml), where phage treatment reduced cecal Salmonella counts by 95% (P < 0.05) (Fig. 4). Phage treatment numerically reduced (P = 0.06, not statistically significant) ileal Salmonella counts in phage-treated pigs (1.7 log10 CFU/ml) versus mock-treated pigs (2.7 log10 CFU/ml) as well, reducing Salmonella counts by 90.0% (Fig. 4).
FIG. 3.
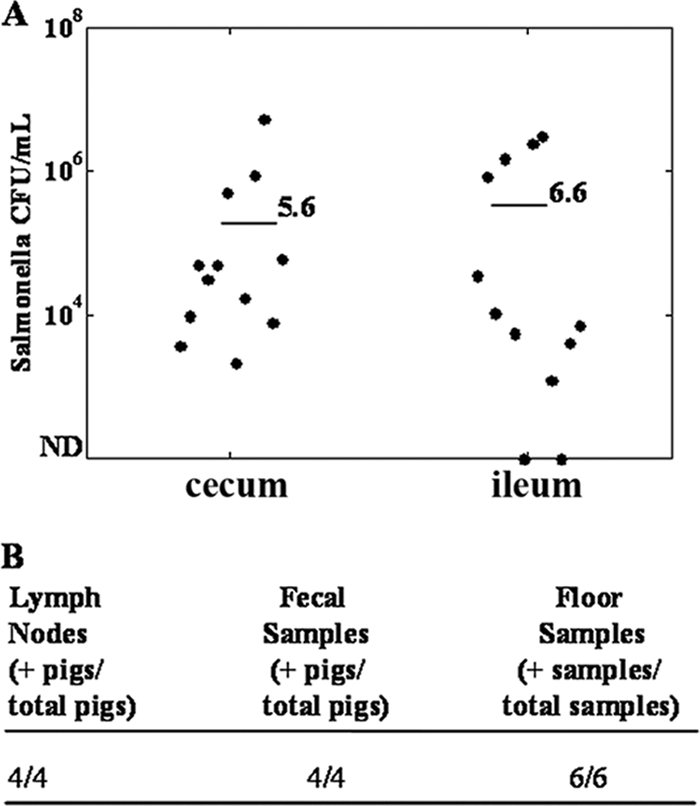
Colonization rates of seeder pigs. Seeder pigs were inoculated with 5 × 109 CFU of Salmonella enterica serovar Typhimurium (γ4232). (A) Colonization of the cecum and ileum. Black dots represent individual seeder pigs (four animals per three replicate experiments). Bars indicate average colonization rates (CFU/ml) over three replicates. (B) Colonization rates in lymph nodes, fecal samples, and floor samples. Data are averages over three replicates. ND, not detectable/below level of enumeration (<100 CFU/ml).
FIG. 4.
Phage therapy reduces Salmonella colonization under production-like settings. Four market-weight pigs (in three replicate trials) were inoculated with Salmonella enterica serovar Typhimurium and held in a common pen. At 48 h postinoculation, 16 naïve pigs were divided into two groups and received either an anti-Salmonella phage cocktail or a mock treatment (i.e., controls). All pigs were comingled in the Salmonella-contaminated pen for 6 hours. Upon necropsy, cecal and ileal samples were collected and Salmonella organisms were enumerated. (A and B) Black dots represent individual pigs. (C) Cecal and ileal colonization data, presented as least square means. Numbers with different subscripts are statistically different at P < 0.05. Comparisons are within sample (i.e., cecum or ileum). (D) Salmonella detection in lymph node and fecal samples averaged over three replicates. Data are qualitative (Salmonella positive versus negative). *, 0-h fecal samples were collected only in the first replicate (n = 8). ND, not detectable/below level of enumeration.
Similar to the trial done with smaller pigs, there were no differences in qualitative measurements (i.e., frequency of positive/negative samples). Phage-treated and mock-treated pigs had similar percentages of positive mesenteric lymph node samples (Fig. 4). Likewise, in each replicate, 100% of pigs in both the phage-treated and non-phage-treated groups tested positive for Salmonella by fecal swab sample collected at 6 h postcomingling (Fig. 4).
Reisolation of treatment phage.
Standard plaque assays were conducted on cecal and ileal samples from both phage-treated and mock-treated pigs to quantify the concentrations of anti-Salmonella phage in each sample. The concentrations of anti-Salmonella phage in ileal and cecal contents of phage-treated pigs were 1.6 × 106 and 9.0 × 106 PFU/ml, respectively (results are averages over three replicates, with eight pigs per treatment group in each replicate experiment). The concentrations of anti-Salmonella phage in ileal and cecal contents of mock-treated pigs were approximately 100-fold lower, at 9.4 × 104 and 6.6 × 104 PFU/ml, respectively.
Phage cocktail specificity.
It was of interest to determine the spectrum of the phage treatment and whether our cocktail was effective at lysing Salmonella serovars other than the challenge serovar (Typhimurium). The phage cocktail was effective in lysing four of eight Salmonella serovars tested. It was most effective against the Dublin serovar, producing a titer of over 109 PFU/ml in a standard plaque assay. Likewise, the phage cocktail effectively lysed the Enteriditis serovar, producing a titer of 5.8 × 108 PFU/ml. The phage cocktail also lysed both serovars Litchfield and Schwarzengrund, although to a much lesser extent (2 × 102 PFU/ml and 1.8 × 103 PFU/ml, respectively), but it was ineffective at lysing the Kentucky, Tennessee, Senftenberg, and Indiana serovars (Table 1).
TABLE 1.
Response to phage cocktail treatment of non-Typhimurium Salmonella serovarsa
| S. enterica serovar | Lysis by phage cocktail | Plaque assay titer (PFU/ml) |
|---|---|---|
| Dublin | Yes | >109 |
| Enteriditis | Yes | 5.8 × 108 |
| Indiana | No | |
| Kentucky | No | |
| Litchfield | Yes | 2.0 × 102 |
| Schwarzengrund | Yes | 1.8 × 103 |
| Senftenberg | No | |
| Tennessee | No |
The phage cocktail was prepared as described in Materials and Methods. Phages were purified and combined into a cocktail. Plaque assays were performed using the indicated Salmonella serovar as supporting bacteria.
DISCUSSION
Several groups have shown marked increases in Salmonella prevalence in pigs just prior to processing. Hurd et al. (12) examined Salmonella isolation rates in pigs before and after transport (mean distance, 169 km) and lairage (2 to 3 h). Pigs necropsied at the abattoir had 7-fold-higher Salmonella isolation rates (39.9%) than pigs necropsied on the farm (5.3%). Likewise, Larsen and coworkers (15) reported increases in Salmonella prevalence (59%) in pigs after just 2 h of holding versus pigs slaughtered immediately upon entering the processing facility (44%).
It is clear that multiple intervention strategies are needed to effectively reduce the incidence of Salmonella contamination in processing plants (1, 3, 26). Toward this end, we hypothesized that phage therapy would prove very useful, as other groups have shown that phages can be used against difficult-to-treat infections, such as those caused by Pseudomonas and antibiotic-resistant tuberculosis (6, 10). Agricultural researchers have also shown that a properly selected phage can limit bacterial infections in food animals if administered at the appropriate time. Loc Carrillo et al. demonstrated that oral administration of phage to Campylobacter-infected chickens reduced the concentration of Campylobacter shed in the feces by as much as 5 log10 CFU/ml (17). Similar results were reported by Wagenaar et al., who also worked with Campylobacter in chickens (30), and by Fiorentin and coworkers, who worked with Salmonella (9). Using phage therapy in combination with competitive exclusion has also proved successful in limiting Salmonella infections in chickens (29), while phages have been used to treat different bacterial infections in ruminants as well (2, 4). There are fewer studies examining phage therapy in pork production. Lee and Harris infected small pigs with Salmonella followed by a single phage (Felix O1) in a lairage-type setting and found that phage treatment reduced Salmonella colonization in the cecum and tonsils but not the intestine (16). An older study by Smith and Huggins indicated that phage therapy could reduce both colonization of pathogenic Escherichia coli in the gut and associated fatalities in small pigs (27). To our knowledge, this is the first attempt at using phage therapy to combat lairage-associated Salmonella infections in market-weight pigs.
Several of these same earlier studies showed that there are limits to using phages as an antibacterial therapy (e.g., phage proliferation thresholds), but most of these limits become evident only when phages are suggested as a long-term or wholesale replacement for traditional antibiotics. If used under the right circumstances, phages still have excellent potential as antibacterial biotherapies. We predicted that employing phage therapy to combat reactivating or rapid Salmonella infections associated with transport/lairage would be an ideal use of phage therapy. First, as demonstrated here and by others (19, 23), Salmonella-specific virulent phages are readily isolated from various environments. Second, as pigs are not commonly held for over 12 hours during transport and holding, problems of phage resistance development are minimized which, while common in the laboratory, are actually rare under natural conditions (8, 13). Third, phages would be delivered in high initial concentrations and only be required to be effective for a very short period of time (∼6 to 12 h), thereby making phage proliferation threshold limits and rebounding growth of the target bacteria irrelevant.
The data presented here demonstrate that administering a phage cocktail to pigs prior to exposure to a Salmonella-contaminated environment can effectively reduce Salmonella colonization in naïve pigs. The preliminary studies were performed on small numbers of 3- to 4-week-old pigs to gauge the potential in vivo efficacy of the phage cocktail and to ascertain whether larger experiments employing more resources were justified. The results of these experiments, in which the pigs were coadministered Salmonella enterica serovar Typhimurium and the phage cocktail, were similar to those reported in poultry (9). Treatment with the phage cocktail reduced the extent of colonization by almost every parameter measured (e.g., ileal contents, cecal contents, tonsils). The only measurements not significantly lower in phage-treated pigs were lymph node samples and fecal samples. While contamination of pork products usually results from the carcass coming into contact with the feces of another infected animal, it is important to note that because of low initial Salmonella counts in both lymph node and fecal swab samples, these samples were enriched twice before plating on a solid medium and were not enumerated. Thus, lymph node and fecal samples provided only qualitative measurements (i.e., Salmonella positive versus Salmonella negative) and did not show the level/concentration of Salmonella colonization. As such, samples that were not enriched and the actual concentration of Salmonella directly determined (from ileal samples, cecal samples, and tonsils) probably provide more meaningful measurements of the effects of the phage cocktail on Salmonella colonization, and in each case these measurements were lower in phage-treated pigs.
When tested under much more realistic conditions (i.e., simulated lairage), the phage cocktail remained effective in reducing Salmonella infections in market-weight pigs as well. In these experiments, we were able to mimic a processing facility holding pen by letting the manure of infected pigs accrue for 48 h. Our infection model and its success mirrors that of Hurd et al. (11), who showed that Salmonella-negative pigs can become rapidly colonized when exposed to a Salmonella-contaminated environment. In their experiments, naïve pigs were exposed to pigs that had been challenged with Salmonella enterica serovar Typhimurium (γ4232, the same strain used in the present studies) and a Salmonella-contaminated slurry. The challenge organism was cultured from lymph nodes, cecal, ileal, or fecal samples of several pigs as early as 2 to 4 h postcomingling. By 6 hours postcomingling, all exposed pigs had at least one positive lymph node, cecal, ileal, or fecal sample. One shortcoming of our model, however, is that fecal samples were collected by rectal swabbing after the pigs had entered into a highly contaminated environment where manure was allowed to accrue for 48 h. By 2 h postcomingling, most pigs had a considerable amount of manure on their hides, which probably led to some contamination of fecal samples with manure from the environment and could account for a high percentage of Salmonella-positive fecal samples after only 6 hours. Again, samples taken at necropsy (lymph node, cecal, and ileal samples) probably offer a more accurate measurement of the extent of colonization and the effect of the phage treatment.
While outside the scope of these studies, it will be of interest in the future to determine where the phages and bacteria intersect (e.g., upper versus lower intestine). Although we are unsure whether the phages reach the lower gastrointestinal tract, we did isolate higher concentrations of anti-Salmonella phages in the ileum and cecum of treated pigs, indicating that the phages did reach several potential sites of infection.
It should be noted that the differences in our main trial, while significant, were not as large as those seen in the preliminary trial. The reduced efficacy could be due to effects of the differences in the microflora of the pigs between replications, differences in clearance rates, or variability due to endogenous phages, as anti-Salmonella phages were isolated from nontreated pigs after processing. While pigs were screened for Salmonella prior to challenge, they were not screened for anti-Salmonella phages. We are currently exploring whether these issues could impact our results.
In these experiments, treated pigs were administered the phage cocktail by gavage. This method was chosen to ensure that each pig received the treatment. Current experiments are focused on administering the phages through water systems or in the feed, making the treatment more practical from a commercial standpoint. As we are able to effectively microencapsulate the phages, both water- and feed-based systems are feasible. In addition, the phages in our cocktail were isolated based upon their ability to lyse the challenge strain (Salmonella enterica serovar Typhimurium). While the cocktail has some extended activity, as shown by its ability to lyse some non-Typhimurium serovars, the spectrum of the cocktail would have to be expanded to include the Salmonella serovars most common to both swine and processing facilities to be most effective. This would, of course, require a much larger phage library, but as we have shown that phages are easily isolated from several different common sources, building such a library is entirely possible. It will be of interest to ascertain whether the treatment is effective at decreasing Salmonella colonization outside of the pig, such as on the skin, as the hide can also serve as a source of contamination during processing (e.g., during dehairing). Likewise, future studies will have to comprehensively examine the possible emergence of resistant mutants to ensure that the treatment remains effective at controlling pathogens over much longer periods of time.
Acknowledgments
This work was supported by NPB grant 06-167 from the National Pork Board.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Alban, L., and K. D. C. Stark. 2005. Where should the effort be put to reduce the Salmonella prevalence in the slaughtered swine carcass effectively. Prev. Vet. Med. 68:63-79. [DOI] [PubMed] [Google Scholar]
- 2.Barrow, P., M. A. Lovell, and A. Berchieri, Jr. 1998. Use of lyticbacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloeil, P. A., C. Chauvin, K. Proux, F. Madec, P. Fravalo, and A. Alioum. 2004. Impact of the Salmonella status of market-age pigs and the pre-slaughter process on Salmonella caecal contamination at slaughter. Vet. Res. 35:513-530. [DOI] [PubMed] [Google Scholar]
- 4.Callaway, T. R., T. S. Edrington, A. D. Brabban, R. C. Anderson, M. L. Rossman, M. J. Engler, M. A. Carr, K. J. Genovese, J. E. Keen, M. L. Looer, E. M. Kutter, and D. J. Nisbet. 2008. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157:H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog. Dis. 5:183-191. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2008. Salmonellosis. http://www.cdc.gov/ncidod/diseases/submenus/sub_salmonella.htm.
- 6.Danelishvili, L., L. S. Young, and L. E. Bermudez. 2006. In vivo efficacy of phage therapy for Mycobacterium avium infection as delivered by a nonvirulent mycobacterium. Microb. Drug Resist. 12:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Ebner, P. D., and A. G. Mathew. 2000. Effects of antibiotic regimens on the fecal shedding patterns of pigs infected with Salmonella typhimurium. J. Food Prot. 63:709-714. [DOI] [PubMed] [Google Scholar]
- 8.El-Shibiny, A., A. Scott, A. Timms, Y. Metawea, P. Connerton, and I. Connerton. 2009. Application of group II Campylobacter bacteriophage to reduce strains of Campylobacter jejuni and Campylobacter coli colonizing broiler chickens. J. Food Prot. 72:733-740. [DOI] [PubMed] [Google Scholar]
- 9.Fiorentin, L., N. D. Vieira, and W. Barioni. 2005. Oral treatment with bacteriophages reduces the concentration of Salmonella enteritidis PT4 in caecal contents of broilers. Avian Pathol. 34:258-263. [DOI] [PubMed] [Google Scholar]
- 10.Hagens, S., A. Habel, U. von Ahsen, A. von Gabain, and U. Blasi. 2004. Therapy of experimental Pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 48:3817-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurd, H. S., J. K. Gailey, J. D. McKean, and M. H. Rostagno. 2001. Rapid infection in market-weight swine following exposure to a Salmonella typhimurium-contaminated environment. Am. J. Vet. Res. 62:1194-1197. [DOI] [PubMed] [Google Scholar]
- 12.Hurd, H. S., J. D. McKean, R. W. Griffith, I. V. Wesley, and M. H. Rostagno. 2002. Salmonella enterica infections in market swine with and without transport and holding. Appl. Environ. Microbiol. 68:2376-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurley, A., J. J. Maurer, and M. D. Lee. 2008. Using bacteriophages to modulate Salmonella colonization of the chicken's gastrointestinal tract: lessons learned from in silico and in vivo modeling. Avian Dis. 52:599-607. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson, R. E., L. D. Firkins, R. M. Weigel, F. A. Zuckerman, and J. A. DiPietro. 1999. Effect of transportation and feed withdrawal on shedding of Salmonella typhimurium among experimentally infected pigs. Am. J. Vet. Res. 60:1155-1158. [PubMed] [Google Scholar]
- 15.Larsen, S. T., H. S. Hurd, J. D. McKean, R. W. Griffith, and I. V. Wesle. 2004. Effect of short-term lairage on the prevalence of Salmonella enterica in cull sows. J. Food Prot. 67:1489-1493. [DOI] [PubMed] [Google Scholar]
- 16.Lee, N., and D. L. Harris. 2001. Effect of bacteriophage treatment as a preharvest intervention strategy to reduce the rapid dissemination of Salmonella typhimurium in pigs, p. 555-557. Proc. Am. Assoc. Swine Vet. American Association of Swine Veterinarians, Perry, IA.
- 17.Loc Carrillo, C., R. J. Atterbury, A. El-Shibiny, P. L. Connerton, E. Dillon, A. Scott, and I. F. Connerton. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71:6554-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew, A. G., and P. D. Ebner. 2004. Issues of drug use and antibiotic resistance in pig production. Pig News Inform. 25:133-147. [Google Scholar]
- 19.McLaughlin, M. R., M. F. Balaa, J. Sims, and R. King. 2006. Isolation of Salmonella bacteriophages from swine effluent lagoons. J. Environ. Qual. 35:522-528. [DOI] [PubMed] [Google Scholar]
- 20.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittal, S. K., N. Aggarwal, G. Sailaja, A. van Olphen, H. HogenEsch, A. North, J. Hays, and S. Moffatt. 2001. Immunization with DNA, adenovirus or both in biodegradable alginate microspheres: effect of route of inoculation on immune response. Vaccine 19:253-263. [DOI] [PubMed] [Google Scholar]
- 22.Morgan, I. R., F. L. Krautil, and J. A. Craven. 1987. Effect of time in lairage on caecal and carcass Salmonella contamination of slaughter pigs. Epidemiol. Infect. 98:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Flynn, G., A. Coffey, G. F. Fitzgerald, and R. P. Ross. 2006. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J. Appl. Microbiol. 101:251-259. [DOI] [PubMed] [Google Scholar]
- 24.Rostagno, M. H., H. S. Hurd, J. D. McKean, C. J. Ziemer, J. K. Gailey, and R. C. Leite. 2003. Preslaughter holding environment in pork plants is highly contaminated with Salmonella enterica. Appl. Environ. Microbiol. 69:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanga, J. A., F. K. McKeith, J. W. Savell, K. E. Belk, D. B. Griffin, L. I. Wright, A. J. Stetzer, R. C. Person, S. M. Lonergan, T. H. Powell, D. J. Meisinger, and G. C. Smith. 2003. Benchmarking value in the pork supply chain: quantitative strategies and opportunities to improve quality. Final report to the National Pork Board by Colorado State University, University of Illinois at Urbana, Texas A&M University, and Iowa State University to the American Meat Science Association, Savoy, IL.
- 26.Schmidt, P. L., A. M. O'Connor, J. D. McKean, and H. S. Hurd. 2004. The association between cleaning and disinfection of lairage pens and the prevalence of Salmonella enterica in swine at harvest. J. Food Prot. 67:1384-1388. [DOI] [PubMed] [Google Scholar]
- 27.Smith, H. W., and M. B. Huggins. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129:2659-2675. [DOI] [PubMed] [Google Scholar]
- 28.Sulakvelidze, A., and P. Barrow. 2005. Phage therapy in animals and agribusiness, p. 333-376. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL.
- 29.Toro, H., S. B. Price, A. S. McKee, F. J. Hoerr, J. Krehling, M. Perdue, and L. Bauermeister. 2005. Use of bacteriophages in combination with competitive exclusion to reduce Salmonella from infected chickens. Avian Dis. 49:118-124. [DOI] [PubMed] [Google Scholar]
- 30.Wagenaar, J. A., M. A. P. Van Bergen, M. A. Mueller, T. M. Wassenaar, and R. M. Carlton. 2005. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 109:275-283. [DOI] [PubMed] [Google Scholar]
- 31.Wondwossen, A. G., P. R. Davies, P. Turkson, W. E. Morgan Morrow, J. Funk, C. Altier, and S. Thakur. 2004. Characterization of antimicrobial-resistant phenotypes and genotypes among Salmonella enterica recovered from pigs on farms, from transport truck, and pigs after slaughter. J. Food Prot. 67:698-705. [DOI] [PubMed] [Google Scholar]