Abstract
This study investigated the potential utilization of lacto-N-biose I (LNB) by individual strains of bifidobacteria. LNB is a building block for the human milk oligosaccharides, which have been suggested to be a factor for selective growth of bifidobacteria. A total of 208 strains comprising 10 species and 4 subspecies were analyzed for the presence of the galacto-N-biose/lacto-N-biose I phosphorylase (GLNBP) gene (lnpA) and examined for growth when LNB was used as the sole carbohydrate source. While all strains of Bifidobacterium longum subsp. longum, B. longum subsp. infantis, B. breve, and B. bifidum were able to grow on LNB, none of the strains of B. adolescentis, B. catenulatum, B. dentium, B. angulatum, B. animalis subsp. lactis, and B. thermophilum showed any growth. In addition, some strains of B. pseudocatenulatum, B. animalis subsp. animalis, and B. pseudolongum exhibited the ability to utilize LNB. With the exception for B. pseudocatenulatum, the presence of lnpA coincided with LNB utilization in almost all strains. These results indicate that bifidobacterial species, which are the predominant species found in infant intestines, are potential utilizers of LNB. These findings support the hypothesis that GLNBP plays a key role in the colonization of bifidobacteria in the infant intestine.
Bifidobacteria are gram-positive anaerobic bacteria that naturally colonize the human intestinal tract and are believed to be beneficial to human health (21, 30). Breastfeeding has been shown to be associated with an infant fecal microbiota dominated by bifidobacteria, whereas the fecal microbiota of infants who are consuming alternative diets has been described as being mixed and adult-like (12, 21). It has been suggested that the selective growth of bifidobacteria observed in breast-fed newborns is related to the oligosaccharides and other factors that are contained in human milk (human milk oligosaccharides [HMOs]) (3, 4, 10, 11, 16, 17, 34). Kitaoka et al. (15) have recently found that bifidobacteria possess a unique metabolic pathway that is specific for lacto-N-biose I (LNB; Galβ1-3GlcNAc) and galacto-N-biose (GNB; Galβ1-3GalNAc). LNB is a building block for the type 1 HMOs [such as lacto-N-tetraose (Galβ1-3GlcNAcβ1-3Galβ1-4Glc), lacto-N-fucopentaose I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc), and lacto-N-difucohexaose I (Fucα1-2Galβ1-3[Fucα1-4]GlcNAcβ1-3Galβ1-4Glc)], and GNB is a core structure of the mucin sugar that is present in the human intestine and milk (18, 27). The GNB/LNB pathway, as previously illustrated by Wada et al. (33), involves proteins/enzymes that are required for the uptake and degradation of disaccharides such as the GNB/LNB transporter (29, 32), galacto-N-biose/lacto-N-biose I phosphorylase (GLNBP; LnpA) (15, 24) (renamed from lacto-N-biose phosphorylase after the finding of phosphorylases specific to GNB [23] and LNB [22]), N-acetylhexosamine 1-kinase (NahK) (25), UDP-glucose-hexose 1-phosphate uridylyltransferase (GalT), and UDP-galactose epimerase (GalE). Some bifidobacteria have been demonstrated to be enzymatically equipped to release LNB from HMOs that have a type 1 structure (lacto-N biosidase; LnbB) (33) or GNB from the core 1-type O-glycans in mucin glycoproteins (endo-α-N-acetylgalatosaminidase) (6, 13, 14). It has been suggested that the presence of the LnbB and GNB/LNB pathways in some bifidobacterial strains could provide a nutritional advantage for these organisms, thereby increasing their populations within the ecosystem of these breast-fed newborns (33).
The species that predominantly colonize the infant intestine are the bifidobacterial species B. breve, B. longum subsp. infantis, B. longum subsp. longum, and B. bifidum (21, 28). On the other hand, strains of B. adolescentis, B. catenulatum, B. pseudocatenulatum, and B. longum subsp. longum are frequently isolated from the adult intestine (19), and strains of B. animalis subsp. animalis, B. animalis subsp. lactis, B. thermophilum and B. pseudolongum have been shown to naturally colonize the guts of animals (1, 2, 7, 8). However, it is unclear whether there is a relationship between the differential colonization of the bifidobacterial species and the presence of the GNB/LNB pathway. In the present study, we investigated the ability of individual bifidobacterial strains in the in vitro fermentation of LNB and in addition, we also tried to determine whether or not the GLNBP gene (lnpA), which is a key enzyme of the GNB/LNB pathway, was present.
MATERIALS AND METHODS
Microorganisms.
Bifidobacterial strains were obtained from stock cultures maintained in the Morinaga Culture Collection (MCC; Morinaga Milk Industry Co., Ltd., Zama, Japan), the American Type Culture Collection (ATCC; Manassas, VA), Japan Collection of Microorganisms (JCM; Wako, Japan), and the German Collection of Microorganisms and Cell Cultures (DSMZ; Braunschweig, Germany). A total of 208 strains comprising 10 species and 4 subspecies originating from human, animals, or commercial products were studied (Table 1). The strains of the series IV (Table 1), which have also been deposited in other public collections, were originally a gift from Tomotari Mitsuoka (University of Tokyo, Tokyo, Japan). These microorganisms were cultured for 16 h at 37°C in ABCM broth (Eiken Chemical Co. Ltd., Tokyo, Japan). Microorganisms were collected by centrifugation, washed once with sterile saline, resuspended in a one-half volume of sterile saline, and used as seed cultures for fermentation studies.
TABLE 1.
Strains of bifidobacteria, fermentation on LNB, and detection of the GLNBP gene lnpA by PCR
| Speciesa | Strain(s) | Origin | PCR detection of lnpAb | Growth on LNBb | Growth pattern onc: |
|||
|---|---|---|---|---|---|---|---|---|
| LNB | Lactulose | Raffinose | Glucose | |||||
| B. longum subsp. longum | IV-51 (same as ATCC 15707T) | Feces of adult | + | 5 | 4 | 5 | 5 | |
| ATCC 15708 | Feces of infant | + | 2 | 2 | 4 | 4 | ||
| ATCC BAA-999 (BB536) | Feces of infant | + | 5 | 5 | 0 | 5 | ||
| MCC strains | Feces of adult or infant | 3/3 | 3/3 | 2-5 | 5 | 4-5 | 5 | |
| B. longum subsp. infantis | IV-8 (same as ATCC 15697T) | Feces of infant | + | 5 | 5 | 5 | 5 | |
| MCC strains | Feces of adult, infant, or from commercial products | 13/13 | 13/13 | 2-5 | 1-5 | 0-5 | 2-5 | |
| B. longum subsp. longum/infantis | MCC strains | Feces of adult, infant, or from commercial products | 7/7 | 7/7 | 2-5 | 5 | 5 | 2-5 |
| B. breve | IV-14 (same as ATCC 15700T) | Feces of infant | + | 5 | 5 | 5 | 5 | |
| MCC strains | Feces of infant or from yogurt | 8/8 | 8/8 | 2-5 | 3-5 | 2-5 | 1-5 | |
| B. bifidum | IV-127 (same as ATCC 29521T) | Feces of infant | + | 1 | 3 | 0 | 3 | |
| ATCC 15696 | Intestine of infant | + | 5 | 5 | 0 | 4 | ||
| MCC strains | Feces of infant | 4/4 | 4/4 | 5 | 5 | 0 | 3-5 | |
| B. adolescentis | ATCC 15706 | Feces of adult | − | 0 | 1 | 2 | 2 | |
| ATCC 15705 | Feces of adult | − | 0 | 2 | 0 | 2 | ||
| ATCC 15704 | Feces of adult | − | 0 | 1 | 0 | 1 | ||
| MCC strains | Feces of adult or infant | 0/48 | 0/48 | 0 | 0-3 | 0-4 | 0-4 | |
| B. catenulatum | ATCC 27675 | Feces of human | − | 0 | 2 | 1 | 2 | |
| JCM7130 | Sewage | − | 0 | 4 | 4 | 4 | ||
| MCC strains | Feces of adult | 0/11 | 0/11 | 0 | 1-5 | 1-5 | 2-5 | |
| B. pseudocatenulatum | IV-152 (same as ATCC 27919T) | Feces of infant | − | 4 | 5 | 5 | 5 | |
| ATCC27676 | Sewage | − | 4 | 5 | 5 | 5 | ||
| ATCC27677 | Sewage | − | 4 | 4 | 5 | 4 | ||
| IV-40 (same as JCM7040) | Feces of adult | − | 5 | 5 | 5 | 5 | ||
| MCC strains | Feces of adult or infant | 0/57 | 29/57 | 0-4 | 1-5 | 3-5 | 2-5 | |
| B. dentium | ATCC 15424 | Pleural fluid from adult | − | 0 | 2 | 2 | 2 | |
| ATCC 27678 | Feces of human | − | 0 | 2 | 2 | 4 | ||
| ATCC 27679 | Human vagina | − | 0 | 1 | 2 | 2 | ||
| ATCC 27680 | Human dental caries | − | 0 | 4 | 4 | 5 | ||
| ATCC 15423 | Lung abscess in adult | − | 0 | 1 | 4 | 4 | ||
| JCM 7135 | Human dental caries | − | 0 | 4 | 4 | 4 | ||
| MCC strains | Feces of infant or adult | 0/10 | 0/10 | 0 | 1-4 | 2-5 | 2-5 | |
| B. angulatum | ATCC 27535T | Feces of human | − | 0-1 | 4 | 4 | 2 | |
| ATCC 27669 | Sewage | − | 0-1 | 1 | 2 | 1 | ||
| ATCC 27671 | Sewage | − | 0-1 | 2 | 4 | 2 | ||
| B. animalis subsp. animalis | IV-58 (same as ATCC 25527T) | Feces of rat | − | 0 | 4 | 5 | 4 | |
| IV-124 (same as ATCC 27672) | Feces of rat | + | 5 | 5 | 5 | 4 | ||
| MCC strains | Feces of rat | 2/2 | 2/2 | 4-5 | 3 | 4-5 | 3-4 | |
| B. animalis subsp. lactis | IV-113 (same as ATCC 27673) | Sewage | − | 0 | 5 | 5 | 3 | |
| IV-117 (same as ATCC 27674) | Feces of rabbit | − | 0 | 5 | 5 | 5 | ||
| IV-129 (same as JCM 1253) | Feces of chicken | − | 0 | 5 | 5 | 5 | ||
| DSM 10140 | Yogurt | − | 0 | 4 | 5 | 5 | ||
| MCC strains | Yogurt | 0/3 | 0/3 | 0 | 4-5 | 4-5 | 5 | |
| B. pseudolongum | IV-70 (same as ATCC 25526T) | Feces of swine | + | 5 | 4 | 4 | 4 | |
| MCC strains | Feces of animals | 4/5 | 4/5 | 0-5 | 3-4 | 3-4 | 3-4 | |
| B. thermophilum | IV-29 (same as ATCC 25525T) | Feces of swine | − | 0 | 4 | 5 | 3 | |
| MCC strains | Feces of animals | 0/4 | 0/4 | 0 | 0-5 | 5 | 4-5 | |
These microorganisms were identified by cellular morphology, patterns of sugar fermentation, DNA homology, and species- or subspecies-specific PCR.
Number of positive strains (growth pattern category ≥ 1)/number of total strains tested.
The growth pattern was categorized from 0 to 5 based on OD660 measured at 24 h (OD24 h) and 144 h (OD144 h). 0, no growth (OD144 h < 0.1); 1, slow and mild growth (OD24 h < 0.1; OD144 h = 0.1 to 0.5); 2, slow but strong growth (OD24 h < 0.1; OD144 h > 0.5); 3, intermediate and mild growth (OD24 h = 0.1 to 0.5; OD144 h = 0.1 to 0.5); 4, intermediate and strong growth (OD24 h = 0.1 to 0.5; OD144 h > 0.5); 5, quick and strong growth (OD24 h > 0.5).
Substrate fermentation.
A medium previously developed for carbohydrate substrate fermentation studies (20) was used, with some modifications. The medium contained the following (per liter): Difco Bacto liver (Becton Dickinson [BD], Sparks, MD) extract, 250 ml; Difco proteose peptone no. 3 (BD), 10 g; BBL Trypticase peptone (BD), 5 g; Difco yeast extract (BD), 3 g; Tween 80, 1 g; l-cysteine·HCl·H2O, 0.2 g; MgSO4·7H2O, 0.2 g; FeSO4·7H2O, 0.01 g; NaCl, 0.01 g; MnSO4·4H2O, 0.0068 g (with the pH adjusted to 7.2 and autoclaved for 20 min at 115°C). For the preparation of Bacto liver extract, 11 g powder was added to 500 ml distilled water and extracted at 50 to 60°C for 1 h with mixing. LNB, which was used as the sole carbohydrate source, was prepared as previously described (26). After being filter sterilized (0.22 μm), it was added to a sterile medium in order to attain a final concentration of 10 g liter−1. The same concentrations (10 g liter−1) of glucose, lactulose, and raffinose were used as the positive controls, while the medium without sugar supplementation was used as the negative control. The media were inoculated with 1.5% of the seed cultures and incubated under anaerobic conditions at 37°C for up to 144 h. Bacterial growth was measured for absorption at 660 nm using a spectrophotometer (Multiskan Spectrum; Thermo LabSystems, Vantaa, Finland) at 24 and 144 h. Growth on each substrate was evaluated on a scale from 0 to 5 based on values of the optical density at 660 nm (OD660) measured at 24 h (OD24 h) and 144 h (OD144 h) after subtracting the values for the negative control, as follows: 0, no growth (OD24 h and OD144 h, <0.1); 1, slow and mild growth (OD24 h < 0.1; OD144 h = 0.1 to 0.5); 2, slow but sound growth (OD24 h < 0.1; OD144 h > 0.5); 3, intermediate and mild growth (OD24 h = 0.1 to 0.5; OD144 h = 0.1 to 0.5); 4, intermediate and sound growth (OD24 h = 0.1 to 0.5; OD144 h > 0.5); and 5, fast and sound growth (OD24 h and OD144 h, >0.5). A growth scale of >1 was considered to be positive for utilizing each substrate.
PCR detection of the GLNBP gene (lnpA).
The presence of lnpA was detected by PCR analysis that used the primer pair DPVE-f (5′-GACCGGTCGAGATGTACAA-3′) and KFPK-r (5′-TCCACGAACTTCGGGAACTT-3′). The primer, which was designed from sequences previously published or from deduced GLNBP genes of bifidobacteria (15, 24; http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) and from three unpublished bifidobacterial genomic sequences of B. longum subsp. longum, B. breve, and B. longum subsp. infantis existing in the Food Science and Technology Institute, Morinaga Milk Industry Co., Ltd., Kanagawa, Japan, was expected to result in a product of approximately 760 bp. The temperature program used was as follows: initial denaturation (94°, 5 min); 30 cycles of denaturation (94°C, 20 s), annealing (56°C, 30 s), and extension (72°C, 30 s); and final extension (72°C, 10 min). Electrophoresis on a 1% agarose gel stained with ethidium bromide was used for the visualization of the PCR products.
Detection of enzymatic activity of GLNBP.
The enzymatic activity of GLNBP was examined for some strains that with or without GLNBP exhibited LNB utilization. After being cultured for 24 h on glucose or LNB, as described above, cells from 50 ml of each culture broth were collected by centrifugation (8,000 × g, 10 min) and then suspended in 1 ml MOPS (morpholinepropanesulfonic acid) buffer (20 mM, pH 7.5). The suspension was treated with ultrasonic oscillation (output control 4, constant) for 10 min in ice via the use of a sonifier (Branson Sonifier 250; Branson Ultrasonic Corporation, Danbury, CT). After the homogenate was centrifuged at 8,000 × g for 10 min, the supernatant was then collected as a cell extract.
To detect the phosphorolytic activity, 80 μl of a mixture containing 25 mM LNB and 25 mM phosphate in 100 mM MOPS buffer (pH 7.5) was mixed with 20 μl of the cell extract, and the reaction was allowed to proceed at 37°C for up to 18 h. Heat-treated cell extract (boiled for 10 min) served as the negative control. Subsequently, 1 μl of the reaction mixture was spotted on a silica gel thin-layer chromatography (TLC) plate, with the plate being developed using 75% acetonitrile. To visualize the carbohydrates on the plate, each plate was dipped in 5% sulfuric acid in methanol and then heated in a radiation oven.
To detect the synthetic activity of LNB, 80 μl of a mixture containing 25 mM GlcNAc and 25 mM Gal1P in 100 mM MOPS buffer (pH 7.5) was mixed with 20 μl of the cell extract. After the reaction mixture was incubated at 37°C for up to 66 h, formation of LNB was detected by TLC, as described above. Some samples were applied to high-performance liquid chromatography with a Corona charged aerosol detector (ESA, Inc., Chelmsford, MA), a Shodex Asahipak NH2P-50 4E column (4.6 mm by 250 mm; Showa Denko K.K., Tokyo, Japan), and a mobile phase of acetonitrile-H2O (75:25 dilution by volume) at a flow rate of 1 ml/min for the determination of LNB concentration.
RESULTS
Substrate fermentation.
Table 1 summarizes the results of fermentation of LNB, lactulose, raffinose, and glucose by the bifidobacterial strains. While all strains of B. longum subsp. longum, B. longum subsp. infantis, B. breve, and B. bifidum were able to grow on LNB, no growth was seen for B. adolescentis, B. catenulatum, B. dentium, B. angulatum, B. animalis subsp. lactis, and B. thermophilum. Out of the 61 strains of B. pseudocatenulatum, there were 33 that were able to grow on LNB. There also was utilization of LNB by 3 out of the 4 strains of B. animalis subsp. animalis and by 5 out of the 6 strains of B. pseudolongum. Fermentation ability of each carbohydrate source by the 208 individual strains is shown in Table S1 in the supplemental material.
PCR detection of lnpA.
Like the results for LNB growth, PCR product for lnpA was found for all strains of B. longum subsp. longum, B. longum subsp. infantis, B. breve, and B. bifidum, while no product was found for B. adolescentis, B. catenulatum, B. dentium, B. angulatum, B. animalis subsp. lactis, and B. thermophilum (Table 1; see also Table S1 in the supplemental material). PCR products were also detected for those strains of B. animalis subsp. animalis and B. pseudolongum that displayed growth on LNB (Table 1). However, no product was found for the strains of B. pseudocatenulatum, even though some of the strains had exhibited LNB growth (Table 1).
Detection of enzymatic activity for GLNBP.
Strains with or without LNB utilization were examined for the enzymatic activity of GLNBP. Figure 1 shows TLC analysis of LNB-degrading activity for B. animalis subsp. animalis ATCC 27672 (lnpA+) and for B. pseudocatenulatum ATCC 27677 (lacking lnpA). GlcNAc was detected in both lanes, indicating that both strains were able to degrade LNB. Gal1P, an indicator of LNB-phosphorolytic activity, was detected only with B. animalis. Table 2 summarized the results of GLNBP activity for bifidobacterial strains with or without LNB utilization/PCR detection of lnpA. LNB-phosphorolyzing activity of GLNBP, as shown by the detection of Gal1P, was observed on extracts from cells cultivated on substrates of LNB for those strains harboring the lnpA gene. LNB-phosphorolyzing activity was also observed for strains B. breve ATCC 15700T and B. bifidum ATCC 29521T, with glucose as the carbohydrate substrate. Mild activity was observed for B. bifidum ATCC 29521T, which displayed mild growth on LNB. No enzyme activity of GLNBP was found for those strains of B. pseudocatenulatum, regardless of their LNB fermentation ability.
FIG. 1.
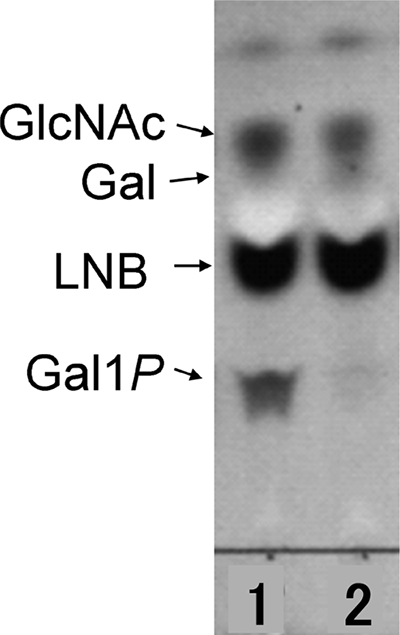
TLC analysis of products from a LNB-phosphorolyzing reaction by cell extracts of bifidobacteria in the presence of phosphate. Lane 1, B. animalis subsp. animalis ATCC 27672; lane 2, B. pseudocatenulatum ATCC 27677.
TABLE 2.
Detection of enzymatic activity of LNB phosphorylase in bifidobacterial strains
| Species | Strain(s) | PCR detection of lnpAe | Growth ond: |
Type of activityc,e |
||||
|---|---|---|---|---|---|---|---|---|
| LNB phosphorolyzinga |
LNB formingb |
|||||||
| Glucose | LNB | Glucose | LNB | Glucose | LNB | |||
| B. pseudocatenulatum | IV-152 (same as ATCC 27919T) | − | 5 | 5 | − | − | − | − |
| ATCC 27677 | − | 5 | 4 | − | − | − | − | |
| ATCC 27676 | − | 4 | 4 | − | − | − | − | |
| MCC 0310 | − | 5 | 0 | − | ND | − | ND | |
| MCC 0337 | − | 5 | 0 | − | ND | − | ND | |
| MCC 0376 | − | 5 | 0 | − | ND | − | ND | |
| B. animalis subsp. | IV-58 (same as ATCC 25527T) | − | 4 | 0 | − | ND | − | ND |
| animalis | IV-124 (same as ATCC 27672) | + | 4 | 5 | − | + | − | + |
| B. longum subsp. longum | IV-51 (same as ATCC 15707T) | + | 5 | 5 | − | + | − | + |
| B. longum subsp. infantis | IV-8 (same as ATCC 15697T) | + | 5 | 5 | − | + | − | + |
| B. breve | IV-14 (same as ATCC 15700T) | + | 5 | 5 | + | + | + | + |
| B. bifidum | IV-127 (same as ATCC 29521T) | + | 3 | 1 | + | ± | + | ± |
| B. pseudolongum | IV-70 (same as ATCC 25526T) | + | 4 | 5 | − | + | − | + |
LNB-phosphorolyzing activity was indicated by the detection of Gal1P degraded from LNB in the presence of phosphate by TLC after incubation for 5 to 18 h.
LNB-forming activity was indicated by the detection of LNB formation from Gal1P and GlcNAc by TLC after incubation for 24 to 66 h.
LNB and glucose were the carbohydrate sources used in cultivating each strain for the preparation of crude cell extract.
Growth patterns as described in Table 1.
+, presence of lnpA or enzymatic activity; −, absence of lnpA or enzymatic activity; ±, weak activity; ND, not examined. Crude cell extract boiled for 10 min showed no activity (data not shown).
For confirmation, these strains were examined for LNB-forming activity from Gal1P and GlcNAc. LNB-forming activity was detected on extracts from those strains exhibiting LNB-phosphorolyzing activity (Table 2), which further supports the notion that B. pseudocatenulatum does not have any GLNBP activity.
DISCUSSION
LNB is a building block of the type 1 HMOs that are known to be rich in human milk. In addition, it has also been suggested that the HMOs might be a factor for selective growth of bifidobacteria (15). Interestingly, the present results indicated that all strains of the species that naturally colonize an infant's intestine, such as B. longum subsp. longum, B. longum subsp. infantis, B. breve, and B. bifidum (21, 28), were able to grow on LNB and that they all possessed the GLNBP gene. In contrast, strains of species that are frequently found in the adult intestine or in human dental caries, such as B. adolescentis, B. catenulatum, and B. dentium, or strains of species that are found in the guts of domestic animals, such as B. animalis subsp. lactis and B. thermophilum, are not able to utilize LNB and appear to have no GLNBP gene. In support of these findings is a survey that examined 18 bifidobacterial strains in the Genomic BLAST (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), which confirmed the presence of highly homologous sequences that have been previously described for GLNBP (15, 24) in the strains of B. longum subsp. longum (2 strains), B. longum subsp. infantis (3 strains), B. bifidum (1 strain), and B. breve (1 strain) but not in those of B. adolescentis (2 strains), B. angulatum (1 strain), B. animalis subsp. lactis (4 strains), B. catenulatum (1 strain), B. dentium (1 strain), B. gallicum (1 strain), and B. pseudocatenulatum (1 strain).
Furthermore, among the strains to be tested, enzymatic activity of GLNBP, as indicated by LNB-degrading (release of Gal1P) and -forming activities, was recognized among strains of bifidobacterial species harboring the GLNBP genes when LNB was used as the substrate. Mild activity was observed for B. bifidum ATCC 29521T that displayed mild growth on LNB. GLNBP activity was also observed for strains B. breve ATCC 15700T and B. bifidum ATCC 29521T, with glucose as the carbohydrate substrate. These results suggested that GLNBP is constitutive for the type strains of B. breve and B. bifidum. Kitaoka et al. previously reported that GLNBP activity was detected for B. bifidum JCM1254 but not for B. longum JCM1217 (which is ATCC 15707T) when cultivated in medium containing glucose as the sole carbohydrate source (15). Two genes (lnpA1 and lnpA2) existed in the genome of B. bifidum JCM1254, but only lnpA2 was expressed constitutively (24). The lnpA gene of B. longum JCM1217 was found to be located at loci similar to those of the lnpA1 gene of B. bifidum JCM1254, which was suggested to be LNB inducible (24).
Some strains of B. animalis subsp. animalis and B. pseudolongum, which are known to be able to colonize the guts of domestic animals, were also found to be able to utilize LNB. Both the lnpA gene and GLNBP activity were detected in some strains of B. animalis subsp. animalis and B. pseudolongum (Table 2). Since LNB is supposed to be a building unit specific for HMOs (31), the significance of the presence of the GLNBP gene in these strains is unclear at the current time. However, the GNB/LNB pathway is also related to the use of GNB, which is the core structure of the sugar of mucin. In addition, it is also been proposed to be associated with intestinal colonization (5). Besides, different substrate affinity has been described for GLNBP in different microorganisms; the enzyme from Clostridium perfringens ATCC 13124 showed a strong preference for GNB over LNB, while LnpA from B. longum subsp. longum JCM1217 showed identical affinity for the two disaccharides (23). Further studies of the structures and substrate affinity of these enzymes may show some insight into the significance of these microorganisms. The lnpA gene and GLNBP activity were not found for B. pseudocatenulatum, which was the only species that was able to grow on LNB while being negative for GLNBP. These particular strains seem to have the ability to utilize LNB via the use of a hydrolytic enzyme. LNB-hydrolyzing activity is interesting because all four of the β-galactosidases of B. bifidum have been reported to be unable to hydrolyze LNB to any degree (9). In order to clarify and completely understand the LNB utilization mechanism for these B. pseudocatenulatum strains, further studies will need to be undertaken.
Recently, Wada et al. cloned LnbB, which is a critical enzyme for the degradation of LNB from HMOs with a type 1 structure (33). Although the number of strains tested was limited, LnbB activity was found only in the strains of B. longum and B. bifidum and not in any of the other bifidobacteria, including B. breve, or in any of the strains belonging to other genera, such as Bacteroides, Clostridium, and Lactobacillus (33). Considering the fact that B. breve is generally the dominant species found in an infant's microbiota and that it has been determined to be capable of potentially utilizing LNB, but LNB does not exist in free style in the milk, the possibility exists that some of the LnbB-negative candidates might be able to utilize LNB released by the positive candidates in the intestine of humans. Another possibility is that bifidobacterium factors other than LNB in breast milk are involved in the growth of B. breve and other bifidobacteria.
Breastfeeding has long been associated with a microbiota in which bifidobacteria are the predominant bacteria. The current results demonstrate that the bifidobacterial species that are the predominant species in infant intestines can potentially utilize LNB. These findings support the hypothesis that by utilizing LNB released from HMOs, GLNBP may play a key role in the rapid and selective colonization of individual bifidobacterial species that is seen in the intestines of breast-fed infants.
Supplementary Material
Acknowledgments
This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) of Japan.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Benno, Y., and T. Mitsuoka. 1986. The development of gastrointestinal micro-flora in humans and animals. Bifidobacteria Microflora 5:13-25. [Google Scholar]
- 2.Benno, Y., K. Endo, T. Mizutani, Y. Namba, T. Komori, and T. Mitsuoka. 1989. Comparison of fecal microflora of elderly persons in rural and urban areas of Japan. Appl. Environ. Microbiol. 55:1100-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bode, L. 2006. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J. Nutr. 136:2127-2130. [DOI] [PubMed] [Google Scholar]
- 4.Coppa, G. V., S. Bruni, L. Morelli, S. Soldi, and O. Gabrielli. 2004. The first prebiotics in humans: human milk oligosaccharides. J. Clin. Gastroenterol. 38:S80-S83. [DOI] [PubMed] [Google Scholar]
- 5.Derensy-Dron, D., F. Krzewinski, C. Brassart, and S. Bouquelet. 1999. β-1,3-Galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol. Appl. Biochem. 29:3-10. [PubMed] [Google Scholar]
- 6.Fujita, K., F. Oura, N. Nagamine, T. Katayama, J. Hiratake, K. Sakata, H. Kumagai, and K. Yamamoto. 2005. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 280:37415-37422. [DOI] [PubMed] [Google Scholar]
- 7.Gavini, F., A. M. Pourcher, C. Neut, D. Monget, C. Romond, C. Oger, and D. Izard. 1991. Phenotypic differentiation of bifidobacteria of human and animal origins. Int. J. Syst. Bacteriol. 41:548-557. [DOI] [PubMed] [Google Scholar]
- 8.Gavini, F., V. Delcenserie, K. Kopeinig, S. Pollinger, H. Beerens, C. Bonaparte, and M. Upmann. 2006. Bifidobacterium species isolated from animal feces and from beef and pork meat. J. Food Prot. 69:871-887. [DOI] [PubMed] [Google Scholar]
- 9.Goulas, T., A. Goulas, G. Tzortzis, and G. R. Gibson. 2009. Comparative analysis of four β-galactosidases from Bifidobacterium bifidum NCIMB41171: purification and biochemical characterization. Appl. Microbiol. Biotechnol. 82:1079-1088. [DOI] [PubMed] [Google Scholar]
- 10.Gyorgy, P., R. F. Norris, and C. S. Rose. 1954. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 48:193-201. [DOI] [PubMed] [Google Scholar]
- 11.Gyorgy, P., R. Kuhn, C. S. Rose, and F. Zilliken. 1954. Bifidus factor. II. Its occurrence in milk from different species and in other natural products. Arch. Biochem. Biophys. 48:202-208. [DOI] [PubMed] [Google Scholar]
- 12.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, T., K. Fujita, and K. Yamamoto. 2005. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J. Biosci. Bioeng. 99:457-465. [DOI] [PubMed] [Google Scholar]
- 14.Katayama, T., J. Wada, K. Fujita, M. Kiyohara, H. Ashida, and K. Yamamoto. 2008. Functions of novel glycosidases isolated from bifidobacteria. J. Appl. Glycosci. 55:101-109. [Google Scholar]
- 15.Kitaoka, M., J. Tian, and M. Nishimoto. 2005. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 71:3158-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knol, J., P. Scholtens, C. Kafka, J. Steenbakkers, S. Gro, K. Helm, M. Klarczyk, H. Schopfer, H. M. Bockler, and J. Wells. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 40:36-42. [DOI] [PubMed] [Google Scholar]
- 17.Kunz, C., S. Rudloff, W. Baier, N. Klein, and S. Strobel. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20:699-722. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd, K. O., J. Burchell, V. Kudryashov, B. W. Yin, and J. Taylor-Papadimitriou. 1996. Comparison of O-linked carbohydrate chains in MUC-1 mucin from normal breast epithelial cell lines and breast carcinoma cell lines. Demonstration of simpler and fewer glycan chains in tumor cells. J. Biol. Chem. 271:33325-33334. [DOI] [PubMed] [Google Scholar]
- 19.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsuoka, T. 1969. Vergleichende Untersuchungen über die Bifidobakterien aus dem Verdauungstakt von Menschen und Tieren (Zugleich die Beschreibung von B, thermophilum nov. spec. und B. pseudolongum nov. spec.). Zentralbl. Bakteriol. Orig. 210:52-64. [PubMed] [Google Scholar]
- 21.Mitsuoka, T., and C. Kaneuchi. 1977. Ecology in the bifidobacteria. Am. J. Clin. Nutr. 30:1799-1810. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima, M., and M. Kitaoka. 2008. Identification of lacto-N-biose I phosphorylase from Vibrio vulnificus CMCP6. Appl. Environ. Microbiol. 74:6333-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima, M., T. Nihira, M. Nishimoto, and M. Kitaoka. 2008. Identification of galacto-N-biose phosphorylase from Clostridium perfringens ATCC1312. Appl. Microbiol. Biotechnol. 78:465-471. [DOI] [PubMed] [Google Scholar]
- 24.Nishimoto, M., and M. Kitaoka. 2007. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211). Biosci. Biotechnol. Biochem. 71:1587-1591. [DOI] [PubMed] [Google Scholar]
- 25.Nishimoto, M., and M. Kitaoka. 2007. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 73:6444-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimoto, M., and M. Kitaoka. 2007. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci. Biotechnol. Biochem. 71:2101-2104. [DOI] [PubMed] [Google Scholar]
- 27.Podolsky, D. K. 1985. Oligosaccharide structures of human colonic mucin. J. Biol. Chem. 260:8262-8271. [PubMed] [Google Scholar]
- 28.Sakata, S., T. Tonooka, S. Ishizeki, M. Takada, M. Sakamoto, M. Fukuyama, and Y. Benno. 2005. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 243:417-423. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, R., J. Wada, T. Katayama, S. Fushinobu, T. Wakagi, H. Shoun, H. Sugimoto, A. Tanaka, H. Kumagai, H. Ashida, M. Kitaoka, and K. Yamamoto. 2008. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 283:13165-13173. [DOI] [PubMed] [Google Scholar]
- 30.Tissier, H. 1905. Repartition des microbes dans l'intenstin du nourrinson. Ann. Inst. Pasteur 19:109-123. [Google Scholar]
- 31.Urashima, T., T. Saito, T. Nakamura, and M. Messer. 2001. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj. J. 18:357-371. [DOI] [PubMed] [Google Scholar]
- 32.Wada, J., R. Suzuki, S. Fushinobu, M. Kitaoka, T. Wakagi, H. Shoun, H. Ashida, H. Kumagai, T. Katayama, and K. Yamamoto. 2007. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:751-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada, J., T. Ando, M. Kiyohara, H. Ashida, M. Kitaoka, M. Yamaguchi, H. Kumagai, T. Katayama, and K. Yamamoto. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 74:3996-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, R. E., M. Ninonuevo, D. A. Mills, C. B. Lebrilla, and J. B. German. 2007. In vitro fermentation of human milk oligosaccharides by several strains of bifidobacteria. Mol. Nutr. Food Res. 51:1398-1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


