Abstract
Oxysterols from steroid autooxidation have numerous harmful effects, but their biodegradation is poorly understood. Microarrays were used to study mineralization of the most common oxysterol, 7-ketocholesterol (7KC), by Rhodococcus jostii RHA1. Growth on 7KC versus growth on cholesterol resulted in 363 differentially expressed genes, including upregulation of two large gene clusters putatively encoding steroid catabolism. Despite this difference, 7KC degradation required key genes involved in cholesterol degradation, indicating a common catabolic route.
Cholesterol autooxidation preferentially generates C-7-oxidized sterols, such as 7-ketocholesterol (7KC), which may alter the biophysical properties of membranes in which they are incorporated (1, 10). Elevated oxysterol concentrations are also associated with disruption of cellular homeostasis, decreased cell viability, and increased cell death (19). This may lead to dysfunction and injury at the histological level, resulting in the development or exacerbation of several pathological conditions. Indeed, oxysterols have been implicated in atherosclerosis (3, 7, 20), Alzheimer's disease (14, 18), age-related macular degeneration (8, 9, 15), and Parkinson's disease (2).
Although considerable research has examined the biological effects and fates of oxysterols in mammalian systems, little has been reported on their microbial degradation. Knowledge of the bacterial transformation of oxysterols could identify enzymes capable of controlling oxysterol levels in vivo and in dietary sources. We recently identified bacteria, including Rhodococcus jostii RHA1, that mineralize 7KC (11). The current study compared cholesterol and 7KC catabolism by RHA1, with the aim of identifying enzymes particular to 7KC metabolism. We used transcriptomic analysis to compare gene expression in RHA1 cells grown separately on pyruvate, cholesterol, or 7KC as the sole organic substrate. To assess the roles of selected enzymes, gene deletion strains were tested for growth and metabolite accumulation. Methods are detailed in the supplemental material.
Differentially expressed genes.
RHA1 was grown on each of 2 mM 7KC, 2 mM cholesterol, or 20 mM pyruvate, as previously described (17), with specific growth rates of 0.018 ± 0.006, 0.023 ± 0.002, and 0.23 ± 0.02 h−1, respectively. RNA was isolated from mid-log-phase cells and used for microarray experiments, as previously described (6). When growing on 7KC versus pyruvate, RHA1 differentially expressed a large suite of genes, most of which (911 genes) were distinct from those differentially expressed during growth on cholesterol versus pyruvate (see Fig. S1 and Table S4 in the supplemental material). In a direct comparison, RHA1 differentially expressed 363 genes during growth on 7KC versus cholesterol.
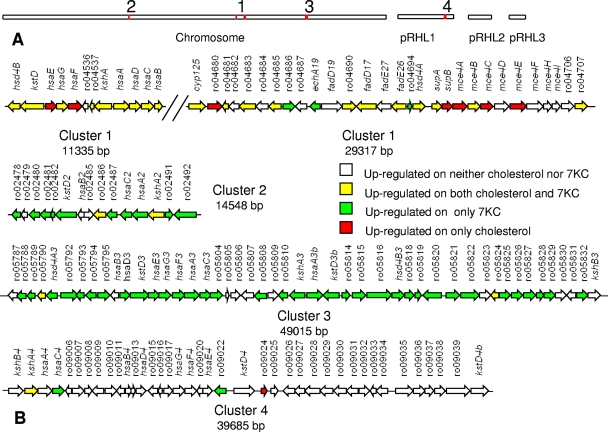
The most highly upregulated genes during growth on 7KC are located mainly in three of four large clusters of putative steroid catabolism genes (Fig. 1) (see Table S5 in the supplemental material). These clusters each include genes postulated to encode degradation of steroid rings A and B (12), including kstD, kshA, hsaABCDEFG, or their homologs, except that cluster 2 lacks hsaDEFG. During growth on 7KC, genes in clusters 1, 2, and 3 were upregulated, with average 7KC/pyruvate expression ratios in each cluster of 3.2, 8.6, and 28.7, respectively. In contrast, during growth on cholesterol, only cluster 1 genes were significantly upregulated, with an average cholesterol/pyruvate expression ratio of 4.2. The direct microarray comparison of 7KC versus cholesterol confirmed that most genes in clusters 2 and 3 were expressed at higher levels on 7KC than on cholesterol (expression ratios of >2.0), while most genes in cluster 1 had similar levels of expression on both steroids (expression ratios between 0.5 and 2.0). Reverse transcriptase quantitative PCR (RT-qPCR) analysis confirmed the general trends of the microarray results (Table 1). The PCR analysis is probably more accurate than microarray analysis, suggesting that the magnitude of steroid catabolism gene upregulation was in the range of 10- to 600-fold. Overall, 7KC induces a much broader range of steroid catabolism genes than does cholesterol, with those genes uniquely induced by 7KC also being the most highly induced.
FIG. 1.
Gene clusters encoding enzymes involved in steroid catabolism in RHA1. (A) Locations of clusters (red) in the genome. (B) Genes with expression ratios greater than 2.0 on 7KC or cholesterol versus pyruvate.
TABLE 1.
Comparison of RT-qPCR and microarray gene expression ratios for RHA1 grown on either cholesterol or 7KC relative to growth on pyruvate
| Gene name | RT-qPCR expressiona |
Microarray expressionc |
||
|---|---|---|---|---|
| Cholesterol | 7KC | Cholesterol | 7KC | |
| hsaA2 | 47.0 (15.5-142.0) | 302.0 (42.7-2140.0) | 0.8 | 10.6* |
| kshA | 54.2 (31.7-92.7) | 23.3 (8.6-63.0) | 4.6* | 4.0* |
| hsaA | 124.0 (90.7-172.0) | 64.8 (30.3-139.0) | 20.3* | 12.0* |
| hsaC | 79.8 (54.1-117.0) | 56.9 (23.5-138.0) | 2.1* | 2.2 |
| cyp125 | 53.9 (22.6-128.0) | 40.8 (13.9-119.0) | 11.0* | 5.7* |
| mce4A | 6.0 (4.2-8.6) | 10.3 (4.1-25.9) | 2.3 | 1.6 |
| ro05068b | 10.0 (4.8-20.9) | 4.6 (1.9-11.1) | 2.0 | 2.8* |
| hsaA3 | 12.6 (2.9-55.4) | 244.0 (75.8-788.0) | 1.0 | 5.8 |
| hsaA3b | 59.4 (9.5-371.0) | 603 (181.8-2000.0) | 1.8 | 5.4* |
| kshB | 8.3 (2.3-29.4) | 77.1 (18.5-321.0) | 1.5 | 1.0 |
Values are average ratios (ranges) relative to expression with pyruvate.
Homologous to hsaA.
*, P < 0.05.
Other genes differentially expressed on 7KC versus pyruvate tend to occur in putative operons and belong to only a few functional groups (see Table S6 in the supplemental material). Most of these genes were not differentially expressed on 7KC versus cholesterol, suggesting similar physiological conditions in cells growing on either steroid. Functional groups downregulated on 7KC versus pyruvate include electron transport, an H+-driven ATPase, tricarboxylic acid cycle, glycolysis, translation, and cell wall synthesis groups, all consistent with the slower growth on 7KC than on pyruvate.
Neither cluster 2 nor cluster 3 genes encode a complete 7KC degradation pathway.
Mutagenesis was used to determine the roles of several genes in 7KC catabolism. These genes are in cluster 1 and were previously implicated in cholesterol catabolism. Two mutants have in-frame deletions of supAB and mce4ABCDEF, respectively, all encoding the Mce4 steroid uptake system essential for cholesterol uptake (13). Both mutants grew on pyruvate but lost the ability to grow on 7KC, indicating that the Mce4 system is also essential for uptake of 7KC. Growth on 7KC was also completely abolished by replacement of hsaC or in-frame deletion of cyp125. The former encodes a dioxygenase involved in cleavage of ring A of cholesterol and the latter a cytochrome P450 enzyme essential for initiating side chain degradation (16, 17). Both enzymes appear to be essential for degradation of 7KC. In contrast, deletion of hsaA hindered but did not prevent growth on 7KC, suggesting that HsaA contributes to 7KC catabolism but is not essential. HsaA encodes the oxygenase of a two-component enzyme that hydroxylates ring A of cholesterol (unpublished).
The above results do not exclude the possibility that one or more of the 62 genes in clusters 2 and 3 are essential for growth on 7KC. However, neither cluster encodes a complete 7KC catabolic pathway. 7KC appears to gratuitously induce many steroid catabolism genes not essential for its metabolism. This is in marked contrast to cholesterol (17), which induces only cluster 1. Thus, the 7-keto substituent has a major impact on expression of steroid catabolism genes, either directly by affecting the function of the molecule as a regulatory effector or indirectly by affecting the occurrence of metabolites influencing gene regulation. Because of its low abundance, 7KC is unlikely to be a major substrate for soil bacteria, but it may structurally resemble other steroids that are important substrates.
Removal of the 7-keto substituent.
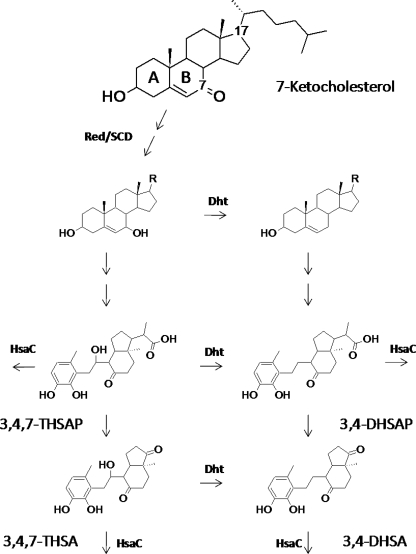
The hsaC mutant cells accumulated several potential metabolites from 7KC, seven of which were not produced by wild-type RHA1. These metabolites accumulated during the first 20 h of incubation, after which there were no major changes in metabolites. Purified HsaC from Mycobacterium tuberculosis H37Rv, a close homolog of the RHA1 enzyme, was used to transform the mixture of metabolites to identify catecholic by-products likely to be intermediates of 7KC catabolism. HsaC completely removed four metabolites, which were identified by mass spectrometry (Fig. 2; see also the supplemental material).
FIG. 2.
Proposed scheme for 7KC metabolism based on metabolites accumulated by the RHA1 hsaC mutant. Degradation of the C-17 side chain, that of the 7-keto substituent, and that of rings A and B all appear to occur concurrently. R, side chain intact or in various possible stages of degradation; Red/SCD, 7-keto reductase plus partial or complete side chain degradation; Dht, dehydratase; 3,4,7-THSAP, 3,4,7-trihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione propionic acid; 3,4-DHSAP, 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione propionic acid; 3,4,7-THSA, 3,4,7-trihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione; 3,4-DHSA, 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione.
The metabolites indicate a common pathway for 7KC and cholesterol metabolism up to the HsaC step (Fig. 2). Both 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4-DHSA) and 3,4-DHSA propionic acid (3,4-DHSAP) were previously found as cholesterol metabolites of the hsaC mutant (17). The production of these metabolites from 7KC by the hsaC mutant plus their 7-hydroxy analogs, 3,4,7-trihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4,7-THSA) and 3,4,7-THSA propionic acid (3,4,7-THSAP), indicates that the 7-keto substituent is reduced to a hydroxyl group prior to the HsaC ring cleavage step. Removal of the 7-hydroxy substituent and complete side chain degradation are not required for cleavage of ring A to occur. Either or both the 7-keto and 7-hydroxyl substituents, as well as the C-17 propionic acid side chain, can be transformed by enzymes preceding the HsaC step.
No enzymes unique to 7KC metabolism were identified. However, removal of the 7-keto substituent is putatively catalyzed via a reductase plus a dehydratase (Fig. 2). The genes ro06637 and ro06638, respectively, may encode these enzymes. Both were significantly upregulated on 7KC, and the latter encodes a protein similar to bile acid 7α-dehydratase (4, 5). Enzymes capable of 7-keto substituent removal could facilitate testing of the effects of 7KC in cellular disease models and control of 7KC levels in various media. These genes and their protein products warrant further investigation for possible selective transformation of 7KC.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Methuselah Foundation and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada.
We thank Katherine Yam for providing purified HsaC from M. tuberculosis H37Rv.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bach, D., R. F. Epand, R. M. Epand, and E. Wachtel. 2008. Interaction of 7-ketocholesterol with two major components of the inner leaflet of the plasma membrane: phosphatidylethanolamine and phosphatidylserine. Biochemistry 47:3004-3012. [DOI] [PubMed] [Google Scholar]
- 2.Bosco, D. A., D. M. Fowler, Q. Zhang, J. Nieva, E. T. Powers, P. Wentworth, Jr., R. A. Lerner, and J. W. Kelly. 2006. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2:249-253. [DOI] [PubMed] [Google Scholar]
- 3.Brown, A. J., and W. Jessup. 1999. Oxysterols and atherosclerosis. Atherosclerosis 142:1-28. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, J. P., W. B. White, B. Egestad, J. Sjovall, and P. B. Hylemon. 1987. Biosynthesis of a novel bile acid nucleotide and mechanism of 7 alpha-dehydroxylation by an intestinal Eubacterium species. J. Biol. Chem. 262:4701-4707. [PubMed] [Google Scholar]
- 5.Dawson, J. A., D. H. Mallonee, I. Bjorkhem, and P. B. Hylemon. 1996. Expression and characterization of a C24 bile acid 7 alpha-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37:1258-1267. [PubMed] [Google Scholar]
- 6.Goncalves, E. R., H. Hara, D. Miyazawa, J. E. Davies, L. D. Eltis, and W. W. Mohn. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes, H., B. Mathews, M. L. Lenz, and J. R. Guyton. 1994. Cytotoxicity of oxidized LDL to porcine aortic smooth muscle cells is associated with the oxysterols 7-ketocholesterol and 7-hydroxycholesterol. Arterioscler. Thromb. 14:1177-1185. [DOI] [PubMed] [Google Scholar]
- 8.Javitt, N. B., and J. C. Javitt. 2009. The retinal oxysterol pathway: a unifying hypothesis for the cause of age-related macular degeneration. Curr. Opin. Ophthalmol. 20:151-157. [DOI] [PubMed] [Google Scholar]
- 9.Malvitte, L., T. Montange, C. Joffre, A. Vejux, C. Maiza, A. Bron, C. Creuzot-Garcher, and G. Lizard. 2006. Analogies between atherosclerosis and age-related maculopathy: expected roles of oxysterols. J. Fr. Ophthalmol. 29:570-578. (In French.) [DOI] [PubMed] [Google Scholar]
- 10.Massey, J. B., and H. J. Pownall. 2006. Structures of biologically active oxysterols determine their differential effects on phospholipid membranes. Biochemistry 45:10747-10758. [DOI] [PubMed] [Google Scholar]
- 11.Mathieu, J., J. Schloendorn, B. E. Rittmann, and P. J. Alvarez. 2008. Microbial degradation of 7-ketocholesterol. Biodegradation 19:807-813. [DOI] [PubMed] [Google Scholar]
- 12.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohn, W. W., R. van der Geize, G. R. Stewart, S. Okamoto, J. Liu, L. Dijkhuizen, and L. D. Eltis. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368-35374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson, T. J., and D. L. Alkon. 2005. Oxidation of cholesterol by amyloid precursor protein and beta-amyloid peptide. J. Biol. Chem. 280:7377-7387. [DOI] [PubMed] [Google Scholar]
- 15.Ong, J. M., A. M. Aoki, G. M. Seigel, I. Sacerio, R. Castellon, A. B. Nesburn, and M. C. Kenney. 2003. Oxysterol-induced toxicity in R28 and ARPE-19 cells. Neurochem. Res. 28:883-891. [DOI] [PubMed] [Google Scholar]
- 16.Rosłoniec, K. Z., M. H. Wilbrink, J. K. Capyk, W. W. Mohn, M. Ostendorf, R. van der Geize, L. Dijkhuizen, and L. D. Eltis. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Geize, R., K. Yam, T. Heuser, M. H. Wilbrink, H. Hara, M. C. Anderton, E. Sim, L. Dijkhuizen, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaya, J., and H. M. Schipper. 2007. Oxysterols, cholesterol homeostasis, and Alzheimer disease. J. Neurochem. 102:1727-1737. [DOI] [PubMed] [Google Scholar]
- 19.Wielkoszynski, T., K. Gawron, J. Strzelczyk, P. Bodzek, M. Zalewska-Ziob, G. Trapp, M. Srebniak, and A. Wiczkowski. 2006. Cellular toxicity of oxycholesterols. Bioessays 28:387-398. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, Q., E. Wasowicz, B. Handler, L. Fleischer, and F. A. Kummerow. 2000. An excess concentration of oxysterols in the plasma is cytotoxic to cultured endothelial cells. Atherosclerosis 149:191-197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.