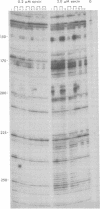
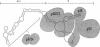
Abstract
We have used the nuclease alpha-sarcin to map the binding sites of the 19-kDa and the 68/72-kDa proteins of signal recognition particle (SRP) on SRP RNA. We found that the regions of protection to nuclease afforded by the two proteins were distinct. p19 protected primarily the two tips in the RNA secondary structure. p68/72 protected a large region extending across the center of the particle and altered the nuclease pattern in the regions that p19 would bind, suggesting that these two proteins may be in close proximity in the particle. The protection afforded by the two proteins in combination was equal to the sum of the individual protections. We have not observed cooperativity in the binding of these two proteins as assessed by the protection assay; nor do we have any evidence that the structure becomes more compact as it assembles. The map derived from this "footprint" analysis places the signal recognition domain (p54 bound to the RNA via the 19-kDa protein) and the elongation arrest domain (associated with the Alu end of the particle) on opposite ends of the particle. Thus, it is possible that SRP recognizes signals by the direct interaction of p54 with the signal sequence at the nascent chain exit site and simultaneously blocks elongation by the entrance of p9/14 into the aminoacyl tRNA site 16 nm away.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews D. W., Walter P., Ottensmeyer F. P. Evidence for an extended 7SL RNA structure in the signal recognition particle. EMBO J. 1987 Nov;6(11):3471–3477. doi: 10.1002/j.1460-2075.1987.tb02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D. W., Walter P., Ottensmeyer F. P. Structure of the signal recognition particle by electron microscopy. Proc Natl Acad Sci U S A. 1985 Feb;82(3):785–789. doi: 10.1073/pnas.82.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu C., Tobin E. M., Fowler A., Zabin I., Lake J. A. Nascent polypeptide chains exit the ribosome in the same relative position in both eucaryotes and procaryotes. J Cell Biol. 1983 May;96(5):1471–1474. doi: 10.1083/jcb.96.5.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Huber P. W., Wool I. G. The ribonuclease activity of the cytotoxin alpha-sarcin. The characteristics of the enzymatic activity of alpha-sarcin with ribosomes and ribonucleic acids as substrates. J Biol Chem. 1983 Feb 25;258(4):2662–2667. [PubMed] [Google Scholar]
- Endo Y., Mitsui K., Motizuki M., Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987 Apr 25;262(12):5908–5912. [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Krause E., Melli M., Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 1983 Nov 11;11(21):7363–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg U. C., Walter P., Johnson A. E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T. V., Wiedmann M., Girshovich A. S., Bochkareva E. S., Bielka H., Rapoport T. A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986 Apr 17;320(6063):634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988 Jan 15;52(1):39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol. 1985 Jun;100(6):1913–1921. doi: 10.1083/jcb.100.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986 Mar 6;320(6057):81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983 Sep;34(2):525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Gilmore R., Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984 Aug;38(1):5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- Zwieb C. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 1985 Sep 11;13(17):6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]