Abstract
The epigenetic silencing of tumor suppressor genes is a crucial event during carcinogenesis and metastasis. Here, in a human genome-wide survey, we identified scavenger receptor class A, member 5 (SCARA5) as a candidate tumor suppressor gene located on chromosome 8p. We found that SCARA5 expression was frequently downregulated as a result of promoter hypermethylation and allelic imbalance and was associated with vascular invasion in human hepatocellular carcinoma (HCC). Furthermore, SCARA5 knockdown via RNAi markedly enhanced HCC cell growth in vitro, colony formation in soft agar, and invasiveness, tumorigenicity, and lung metastasis in vivo. By contrast, SCARA5 overexpression suppressed these malignant behaviors. Interestingly, SCARA5 was found to physically associate with focal adhesion kinase (FAK) and inhibit the tyrosine phosphorylation cascade of the FAK-Src-Cas signaling pathway. Conversely, silencing SCARA5 stimulated the signaling pathway via increased phosphorylation of certain tyrosine residues of FAK, Src, and p130Cas; it was also associated with activation of MMP9, a tumor metastasis–associated enzyme. Taken together, these data suggest that the plasma membrane protein SCARA5 can contribute to HCC tumorigenesis and metastasis via activation of the FAK signaling pathway.
Introduction
The genetic inactivation of tumor suppressor genes is considered a crucial event in the development and progression of tumors. In addition, increasing evidence has shown that epigenetic silencing of these genes as a result of aberrant hypermethylation of CpG islands in promoters and histone modification is essential to carcinogenesis and metastasis (1). Although epigenetic events involving tumor suppressor genes have been observed in some tumors, such as colon, breast, and lung cancers, the events involved in the formation and progression of hepatocellular carcinoma (HCC), the fifth most common cancer worldwide, are less well understood. HCC causes more than 600,000 deaths annually, and it is a highly malignant cancer, with an inherited predisposition to infiltrate and metastasize (2). Previous investigations have implicated a number of tumor suppressor genes in HCC oncogenesis. These genes are frequently inactivated in human HCC as a result of promoter CpG methylation and include cyclin-dependent kinase inhibitor 2A (CDKN2A also known as p16INK4a) (3), E-cadherin (CDH1) (4), apoptosis-associated speck-like protein (ASC) (5), deleted in liver cancer 1 (DLC1) (6), and secreted frizzled-related protein 1 (SFRP1) (7, 8). Inactivation of these genes can contribute to carcinogenesis by promoting cell proliferation and blocking apoptosis (9, 10). However, these genes have yet to be directly implicated in HCC progression and metastasis.
Chromosome 8p, one of the regions most frequently involved in the loss of heterozygosity (LOH), is thought to be closely associated with HCC oncogenesis, progression, venous permeation, and metastasis (11–15). These findings suggest that one or more novel tumor suppressor genes reside in the loci implicated in these cases with allelic deletion. Previous investigations have focused on identifying somatic genetic mutations, including deletions and point mutations, of candidate genes; however, more than a decade of research has failed to turn up any recurrent somatic genetic mutations in candidate tumor suppressor genes located in this chromosomal region. Thus, many researchers have proposed that epigenetic aberrations may contribute to the silencing of tumor suppressor genes observed in HCC. In fact, some recent studies have indicated that epigenetic silencing of some candidate tumor suppressor genes in this region, because of promoter hypermethylation, may be associated with oncogenesis of HCC. Among these candidate suppressor genes, DLC1 is a particularly attractive target due to its allelic deletion, promoter methylation, and downregulated mRNA expression in HCC (6, 16). DLC1 encodes the RhoGAP protein, which, when overexpressed, catalyzes the conversion of active GTP-bound RhoGTPase (Rho) to the inactive GDP-bound form. This suppresses Rho activity, which, in turn, inhibits the growth of tumor cells and xenografts (17–19). Conversely, DLC1 knockdown promotes the oncogenesis of HCC in mice (20). SFRP1 encodes a Wingless-type (Wnt) signaling antagonist and is frequently inactivated in many human cancers, including HCC, as a result of promoter hypermethylation. Overexpression of SFRP1 can significantly inhibit HCC cell growth and colony formation, while SFRP1 knockdown can markedly promote the growth of these cells (7, 8).
It has been pointed out that in addition to the handful of identified genes, such as DLC1 at chromosome 8p22 and SFRP1 at 8p12-p11.1, there likely exist other tumor suppressors in the chromosomal region. Previous studies of allelic imbalance using high-density polymorphic markers suggest that one or more tumor suppressor loci are involved in HCC, including chromosome 8p21, 8p22, and 8p23 (21, 22). Recent studies using microarray-based comparative genomic hybridization (CGH) have indicated that allelic loses occur over more extended region, such as chromosome 8p11-p23, implying that more novel tumor suppressors remain to be involved in tumor initiation and progression (23–26).
In addition to HCC, the LOH on chromosome 8p has been implicated in other human cancers, including bladder cancer (27), breast cancer (28, 29), B cell lymphoma (30), prostate cancer (31, 32), and head and neck squamous cell carcinoma (33). Thus, the identification of candidate tumor suppressor genes on chromosome 8p and elucidation of their biological functions would be a significant advance in our understanding of tumorigenesis and progression.
To identify the candidate tumor suppressor genes that are epigenetically silenced by genomic DNA hypermethylation, some genome-wide approaches have been used to study the so-called cancer epigenome or methylome and then to identify the key tumor suppressor genes that can be shown convincingly to contribute to tumorigenesis. Among these genome-wide approaches for analyzing DNA methylation patterns, gene-expression profiling using microarrays is now widely used and may prove to be particularly useful in cancer epigenomics (34). This strategy involves comparing mRNA levels from cancer cell lines before and after treatment with a demethylating agent, and it has proven successful at identifying hypermethylated genes (35, 36).
In the present study, we first used such a genome-wide method to study the epigenomic patterns of 15 HCC cell lines. We compared gene-expression profiles before and after treatment using the demethylating agent 5-aza-2′-deoxycytidine (decitabine, herein referred to as DAC) in conjunction with trichostatin A (TSA), and we identified the upregulated genes that were located on chromosome 8p. We then analyzed mRNA levels and promoter methylation of these genes in human HCC specimens. The results indicate that scavenger receptor class A, member 5 (SCARA5), which encodes a membrane scavenger receptor, is subject to allelic loss and epigenetic DNA hypermethylation in HCC. It appears to be a tumor suppressor that contributes to tumor initiation and metastasis, possibly by increasing tyrosine phosphorylation and thereby activating focal adhesion kinase (FAK) signaling pathway.
Results
Gene expression pattern in HCC cells treated with a demethylating agent.
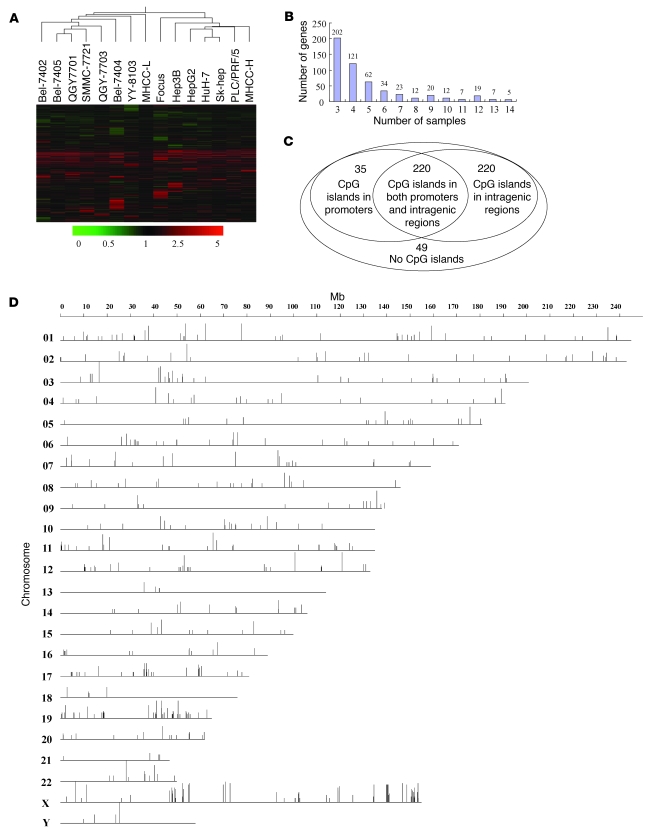
To screen tumor suppressor genes in HCC cells that are silenced by epigenetic modification, we tested all currently available 15 human HCC cell lines: Bel-7402, Bel-7404, Bel-7405, Focus, HepG2, Hep3B, Huh-7, MHCC-L, MHCC-H, PLC/PRF/5, QGY-7701, QGY-7703, SK-hep1, SMMC-7721, and YY-8103. The cell lines were treated with a demethylation agent, DAC, in conjunction with the histone deacetylase inhibitor TSA (35). Conditions were optimized to allow the minimum dose of agents that would be sufficient to reactivate well-known hypermethylation-silenced genes 72 hours after treatment, such as CDH1 (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI38012DS1), but not cause massive cell death (Supplemental Figure 1B). We analyzed gene-expression profiles of these 15 HCC cell lines after treatment with both DAC and TSA using an oligonucleotide microarray (Agilent G4112A) that allowed us to probe 16,891 genes and 5,795 potential transcripts. Clustering of the gene-expression changes indicated that some common genes were markedly upregulated in these HCC cell lines following demethylating treatment (Figure 1A), implying that the transcription of these genes was silenced by hypermethylation of CpG islands in their regulatory elements. Among the upregulated genes, 524 (2.3%) were found to be upregulated at least 3 fold in at least 3 (20%) of the 15 HCC cell lines (Figure 1B and Supplemental Table 1). These genes were assigned to a “markedly changed” group. Two genes well-known to be hypermethylated, CDH1 and GSTP1, were upregulated in 9 and 7 of the 15 HCC cell lines, respectively (Supplemental Table 1), which suggests that this experimental system is appropriate to screen for genes silenced by hypermethylation in HCC cells.
Figure 1. Gene expression profiling by demethylation treatment.
(A) Gene expression profiles of the 15 HCC cell lines that received treatment with or without DAC plus TSA were analyzed using an Agilent gene microarray. Clustering was performed with GeneSpring 7.5 software. Hierarchical clustering revealed 524 transcripts with marked upregulation, defined as an increase of more than 3 fold relative to those before treatment, in at least 3 cell lines, following drug treatment. (B) Distribution of the number of cell lines, showing marked upregulation (more than 3 fold) of the 524 genes after drug treatment. (C) Venn diagram of the distribution of the 524 upregulated genes, based on whether they do or do not possess CpG islands. (D) Chromosomal (chro) location of the 524 candidate genes. The vertical bars indicate the percentage of reexpression of the 524 upregulated genes following demethylation treatment in the 15 cell lines.
To identify genes potentially silenced by hypermethylation, we used bioinformatics tools to determine whether there were CpG islands in the regulatory elements of the genes in the markedly changed group. We defined the regulatory elements as sequences from 2,000 bp upstream to 200 bp downstream of the transcription start site; in addition, we examined the sequences of the intragenic regions. The results indicate that 475 (90.6%) of the 524 genes have CpG islands: 255 have CpG islands in their regulatory elements, while the remaining 220 have such CpG islands only in their intragenic regions (Figure 1C and Supplemental Table 1). No typical CpG islands were found in the remaining 49 (9.4%) genes, within either the regulatory elements or intragenic regions, implying that other mechanisms may contribute to their reexpression in HCC cells following treatment with both DAC and TSA.
To identify hypermethylated tumor suppressors located in the regions that are associated with frequent LOH, we mapped the 475 genes containing CpG islands onto human chromosomes (GeneMap’99). These genes were found to be clustered within certain gene-rich regions, such as 1p21, 1q36, 6p21, 8p21, 10q22-23, 11p15, 16p13, 16q13, 17p13, 19p13, 19q13, 20q13, 22q13, Xp11, and Xq28 (Figure 1D). In addition to CDH1 at 16q22, the screening detected several other hypermethylated tumor suppressors located in several different chromosomal regions and known to be associated with human cancers: SFRP1 (7, 8), DLC1 (6), and tumor suppressor candidate 3 (TUSC3) (37) on chromosome 8p; ASC on 16p12-p11.2 (5); RASSF2 on 20pter-p12.1 (38, 39); and tissue factor pathway inhibitor-2 (TFPI-2) on 7p22 (40). These results suggest that our screen is a reliable method for identifying candidate tumor suppressor genes that we believe to be novel associated with HCC.
The transcription of SCARA5 on chromosome 8p is associated with promoter methylation.
We focused our attention on chromosome 8p, which is one of the most frequent LOH regions and which is the site of potential tumor suppressor genes closely associated with HCC oncogenesis, progression, venous permeation, and metastasis (11–15). Interestingly, 9 upregulated genes were mapped to chromosome 8p (Figure 2A). Among the mapped genes, DLC1 at 8p22 and SFRP1 at 8p12 have previously been shown to be candidate tumor suppressors, since they are both downregulated in human HCC specimens as a result of hypermethylation of CpG islands within their promoters (6–8). ANGPT2 was shown to be highly overexpressed in hypervascular HCCs, resulting in rapid tumor growth and hemorrhage in an animal model of HCC (41–43). TUSC3 at 8p22 has also been reported to be significantly downregulated in ovarian, larynx, and pharynx carcinomas (37, 44). The remaining genes detected in our screen have not yet been associated with human cancers.
Figure 2. Genomic structure and methylation status of SCARA5 on chromosome 8p.
(A) Hierarchical clustering shows 9 genes located on chromosome 8p that were markedly upregulated more than 3 fold, in at least 3 cell lines following drug treatment. (B) The reexpression of the SCARA5 gene on 8p was evaluated by RT-PCR in the HCC cell lines treated with no drug, DAC, TSA, or DAC plus TSA. β-actin was used as a loading control. (C) Schematic representations of the location of CpG islands within the promoter and intragenic regions of SCARA5 and of the primers designed against the promoter region for PCR amplification. PCR fragments were amplified from the bisulfite-treated DNA from the HCC cell lines and specimens used as templates. The numbers in parentheses indicate the distance from these CpG islands within promoter (CG1) and intragenic regions (CG2 and CG3) to the SCARA5 transcription start sites in base pairs, and the corresponding numbers indicate the length of these CpG islands. (D) Representative results from the sequencing bisulfite-treated genomic DNA to detect the methylation level of the CG1 region in Bel-7404, Bel-7405, YY-8103, SMMC-7721, QGY-7701, and QGY-7703 cells after treatment with DAC, compared with the methylation level in control cells (P < 0.05). The numbers indicate the CG dinucleotide within the CpG island in the promoter.
To confirm whether expression of these genes was altered by combined treatment with DAC and TSA, we used an RT-PCR assay to assess their expression levels in the 15 HCC cell lines after treatment. The results show that transcription of SCARA5 was markedly upregulated in 6 of the 15 HCC cell lines (Figure 2B); whereas transcription of DLC1, SFRP1, TUSC3, NEFL, and NEF3 was induced in fewer than 5 of the cell lines (Supplemental Figure 2). These findings imply that, although all of the genes were hypermethylated, transcription of SCARA5 may be more sensitive than transcription of the other genes by treatment with a demethylation agent and/or histone deacetylase inhibitor.
To evaluate whether these genes are silenced in human primary HCC, we used RT-PCR to measure mRNA levels of these genes in HCC specimens relative to the levels in corresponding adjacent nontumor livers. The results indicate that only 3 of the genes (SCARA5, DLC1, and SFRP1) were markedly downregulated in HCC samples but the other genes were not (data not shown). Thus, similar to DLC1 and SFRP1, SCARA5 may be a novel candidate tumor suppressor gene downregulated in HCC as a result of promoter hypermethylation.
To attempt to correlate SCARA5 transcription with the methylation status of the CpG islands within its promoter, DNA sequencing of the bisulfite-treated SCARA5 promoter was performed in the 6 HCC cell lines, in response to DAC treatment. The results indicate that methylation level of the CpG island (Figure 2C) in the SCARA5 promoter (–150 bp to +100 bp) was significantly reduced in Bel-7404, Bel-7405, YY-8103, SMMC-7721, QGY-7701, and QGY-7703 cell lines after DAC treatment (Figure 2D), consistent with the gene’s reexpression under these conditions. This implies that transcription of SCARA5 could be regulated by promoter methylation.
SCARA5 is frequently downregulated in HCCs, and it is associated with HCC progression.
SCARA5 has recently been recognized as a class A scavenger receptor on the plasma membrane, as validated on PLC/PRF/5 cells by an immunofluorescence assay (Figure 3A). As predicted the protein product consists of an N-terminal cytoplasmic tail, transmembrane domain, spacer region, collagen-like domain, and C-terminal scavenger receptor cysteine-rich (SRCR) domain that is possibly responsible for the ligand binding (Figure 3B), and it is thought to play an important role in innate immunity by binding bacteria (45). Since the gene encoding this plasma membrane protein maps to chromosome 8p, a genomic region closely associated with HCC oncogenesis, progression, venous permeation, and metastasis, a hypothesis was proposed that this gene may be a novel tumor suppressor.
Figure 3. Expression pattern of SCARA5 in HCC.
(A) Endogenous SCARA5 (red) in PLC/PRF/5 cells was detected by immunofluorescence. Original magnification, ×1,000. (B) Schematic diagram of the SCARA5 membrane protein, consisting of multiple functional domains. (C) Real time RT-PCR analysis of SCARA5 was carried out on 40 paired HCC samples and adjacent noncancerous tissue. For each sample, the relative SCARA5 mRNA level was normalized to that of β-actin. The vertical line within each box represents the median –ΔCt value. The upper and lower edges of each box represent the 75th and 25th percentile, respectively. The upper and lower horizontal bars indicate the highest and lowest values determined, respectively. (D) Representative immunohistochemical staining of a pair of HCC specimens and corresponding noncancerous tissue, using the anti-SCARA5 antibody on a tissue array containing 78 pairs of HCC specimens. The nuclei were counterstained with hematoxylin. Original magnification, ×40 (left); ×400 (right). (E) Statistical analysis was performed using the GraphPad Prism 5 program to compare the relative levels of SCARA5 in HCC with PVTT with cells without PVTT (P = 0.02169, Mann-Whitney Test). Black triangles indicate the HCC sample, and the transverse lines indicate the mean.
To investigate whether SCARA5 contributes to HCC progression, real time RT-PCR was performed to analyze mRNA levels of SCARA5 in 40 HCC patients. Of the samples from these 40 cases, 32 (80%) HCC samples had a SCARA5 mRNA level at least 2-fold lower than the corresponding nontumorous livers (P < 0.01) (Figure 3C and Supplemental Table 2). Furthermore, the immunohistochemical staining using an anti-SCARA5 antibody was carried out on a tissue array containing an additional 78 pairs of HCC specimens and their corresponding adjacent noncancerous livers (Supplemental Table 3), which consisted of 38 HCCs with portal vein tumor thrombosis (PVTT) and 40 cases without PVTT. As expected, SCARA5 levels were significantly lower in 62 (79.5%) of the 78 HCC specimens compared with the adjacent noncancerous livers (P < 0.01; Figure 3D). Surprisingly, SCARA5 levels were significantly lower in the HCC specimens with PVTT than in those without PVTT (P < 0.05; Figure 3E), suggesting that SCARA5 downregulation may be associated with cellular invasion, venous permeation, and perhaps even metastasis in HCC.
In addition, we used a similar immunohistochemical staining procedure on a gastric tissue array containing 81 cancerous samples, as well as normal gastric epithelium. We found SCARA5 to be markedly downregulated in 62 (76.5%) of the cancer samples (Supplemental Figure 3). This suggests that downregulation of SCARA5 expression contributes to multiple human cancers and not only HCC.
Genetic and epigenetic aberrations of the SCARA5 locus in HCC.
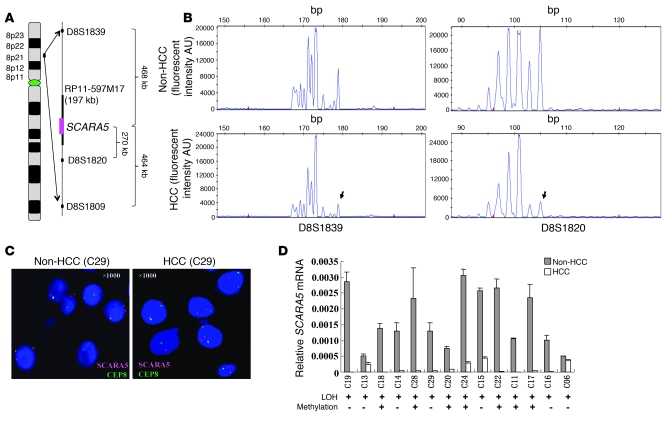
SCARA5 is located at chromosome 8p21.1, one of the most frequent LOH regions in HCC (13, 46) as well as in other human cancers (17, 47–50). To address whether genetic aberrations exist near the SCARA5 locus in HCC specimens, we detected allelic imbalances using 3 microsatellite markers, D8S1839, D8S1820, and D8S1809, which flank the SCARA5 locus (Figure 4A). Allelic imbalance was found in 14 (35%) of the 40 HCCs (Figure 4B and Supplemental Table 2), suggesting that LOH occurs at the SCARA5 locus.
Figure 4. LOH analysis of the SCARA5 locus in HCC samples.
(A) Schematic representation of the microsatellite markers D8S1839, D8S1820, and D8S1809 located on chromosome 8p21.1; these markers flank the SCARA5 locus. The BAC clone RP11-597M17 spanning the SCARA5 locus was used as a probe in the FISH assay. (B) The markers were used to analyze LOH in 40 pairs of HCC samples. The figure shows representative results of LOH from 1 pair of primary HCC tissues and the corresponding adjacent noncancerous tissue. The arrows indicate the heterozygous allelic loss in the tumor DNA. (C) HCC tissue samples were analyzed by FISH using the BAC clones RP11-597M17 (red) and CEP8 (green) for the centromere of chromosome 8 as probes. Typical FISH results are shown for the CEP8 (green) and SCARA5 (red) probes, while the genomic structure of chromosome 8p and the probe positions are illustrated in A. C29, case 29. Original magnification, ×1,000. (D) Correlation between mRNA levels and the corresponding methylation level of the SCARA5 promoter in 14 primary HCC samples with LOH (mean ± SD). + indicates an increase in SCARA5 promoter methylation.
To confirm this idea, we carried out FISH using the BAC clone RP11-597M17, which spans the SCARA5 locus at 8p21 (Figure 4A), as the region-specific probe (red). We attempted to detect allelic imbalance by hybridizing the probe to interphase chromosomes from 4 paired samples of HCC tissue and adjacent noncancerous tissue selected randomly from the 14 HCC samples with LOH. As a control, a probe specific to the centromere of chromosome 8 (CEP8, green) was used. We determined the ratio of SCARA5-positive chromosomes (red) to CEP8-positive chromosomes (green) in 250 cells. The results showed that all 4 HCC samples had allelic loss of the SCARA5 locus; Figure 4C shows representative FISH data. This finding agrees with the results obtained using microsatellite markers.
Interestingly, SCARA5 transcription was found to be downregulated in all 14 cases of HCC with LOH, while such downregulation occurred only in 18 of the remaining 26 cases without LOH (P < 0.05) (Supplemental Table 2). This suggests that genetic aberration at the SCARA5 locus contributes to the downregulation of SCARA5 in some cases of HCC.
We also performed DNA sequencing of bisulfite-treated DNA to evaluate the methylation status of the CpG island within SCARA5 promoter in the 14 cases of HCC with LOH. The results show that the methylation level of the CpG island (CG1) in the SCARA5 promoter, but not the CpG islands in the intragenic regions (CG2 and CG3), was significantly higher in 7 (50%) of the 14 HCC cases with LOH than in corresponding adjacent noncancerous livers (P < 0.01; Figure 4D and Supplemental Figure 4). This indicates that hypermethylation of the SCARA5 promoter can occur in the remaining single allele as a result of heterozygous allelic deletion in these cells. These findings suggest that both genetic and epigenetic events at the SCARA5 locus can contribute to downregulation of the gene in some HCC specimens.
Furthermore, to evaluate the relationship between methylation status and SCARA5 expression levels, we employed the methylation-specific PCR (MSP) assay to detect the methylated and unmethylated CpG islands within the SCARA5 promoter (CG1) in all 15 available HCC cell lines and 20 HCC specimens without LOH. The data revealed that the methylated CpG island within the promoter occurs in 13 (87%) out of the 15 HCC cell lines examined (Supplemental Figure 5) although not in the PLC/PRF/5 and WRL68 cells with high SCARA5 expression. This result implies that the frequent hypermethylation of the SCARA5 promoter may be associated with the decreased expression of this gene in HCC cell lines. However, the same methylated CpG island was detected in only 6 (30%) of the 20 HCC specimens without LOH, along with decreased SCARA5 expression (Supplemental Table 2 and Supplemental Figure 6), suggesting that epigenetic events other than promoter DNA methylation (e.g., histone modification) may also contribute to the downregulation of SCARA5 in HCC specimens. This observation is consistent with the finding that TSA treatment alone seems to trigger the transcription of SCARA5 in some HCC cell lines, including YY-8103, SMMC-7721, and QGY-7701 (Figure 2B).
SCARA5 inhibits cell proliferation, colony formation, and tumorigenicity.
To evaluate whether SCARA5 functions as a tumor suppressor in HCC cells, we observed the effect of SCARA5 expression on cell proliferation, colony formation, and tumorigenicity. Based on the SCARA5 expression pattern in HCC-derived cell lines (Supplemental Figure 5A), we transiently transfected mammalian expression vectors containing SCARA5 into Huh-7, Hep3B, and MHCC-H cells, all of which lack endogenous SCARA5 expression. SCARA5 overexpression suppressed the growth and colony formation of these cells relative to cells transfected with empty pcDNA3.1 vector (P < 0.01; Figure 5, A–E).
Figure 5. The effect of SCARA5 on cell growth, colony formation, and tumorigenicity.
(A–C) Exogenous SCARA5 was expressed in Huh7 (A), Hep3B (B), and MHCC-H (C) cells transfected with the pcDNA3.1 vector. Parental cells with empty vector were used as a control. The growth of these cells was analyzed using the CCK-8 kit. The experiments were repeated at least 3 times, and the symbols represent the mean values of triplicate tests (mean ± SD). Western blot analysis in the insets indicated the expressed SCARA5 in these cell lines. A t test was used to show significant differences between 2 groups (P < 0.05). (D and E) To observe the effect of SCARA5 on colony formation, pcDNA3.1-SCARA5 was transfected into Huh7 (D) and Hep3B (E) cells, and SCARA5 overexpression was confirmed by immunoblotting. After transfection for 24 hours, the cells were scraped and plated on dishes and cultured in G418 for 3 weeks. The representative dishes show the inhibitory effect of SCARA5 on colony formation. The lower histogram shows that colony formation was significantly suppressed by SCARA5, compared with the vector-only control (P < 0.01), where the numbers are the mean value of 3 independent experiments with SD. (F–H) Increased exogenous SCARA5 expression inhibits xenograft tumor growth of Huh7 (F), Hep3B (G), and MHCC-H (H) cells infected with a recombinant adenovirus carrying SCARA5. These cells were injected subcutaneously into nude mice, while the cells carrying empty vector or parental cells were used as controls. Tumor growth was monitored for 3 days by measuring the tumor diameters (mean ± SD).
Furthermore, we transfected an adenoviral vector containing SCARA5 into Huh-7, Hep3B, and MHCC-H cells, and then subcutaneously injected these cells into athymic mice in order to assess their tumorigenicity in this xenograft model. The results showed that SCARA5 overexpression can delay the occurrence of visible tumors and inhibit tumor growth in all 3 HCC cell lines. In fact, in our experiments the size and weight of tumors formed in cells overexpressing SCARA5 were significantly lower than those of tumors formed from control cells transfected with empty vector in Huh-7 and MHCC-H cells, and the tumorigenicity disappeared in Hep3B cells overexpressing SCARA5 (P < 0.05; Figure 5, F–H, and Supplemental Figure 7). This suggests that SCARA5 plays an important role in preventing cell overgrowth in vivo.
SCARA5 knockdown enhances tumorigenicity in vivo.
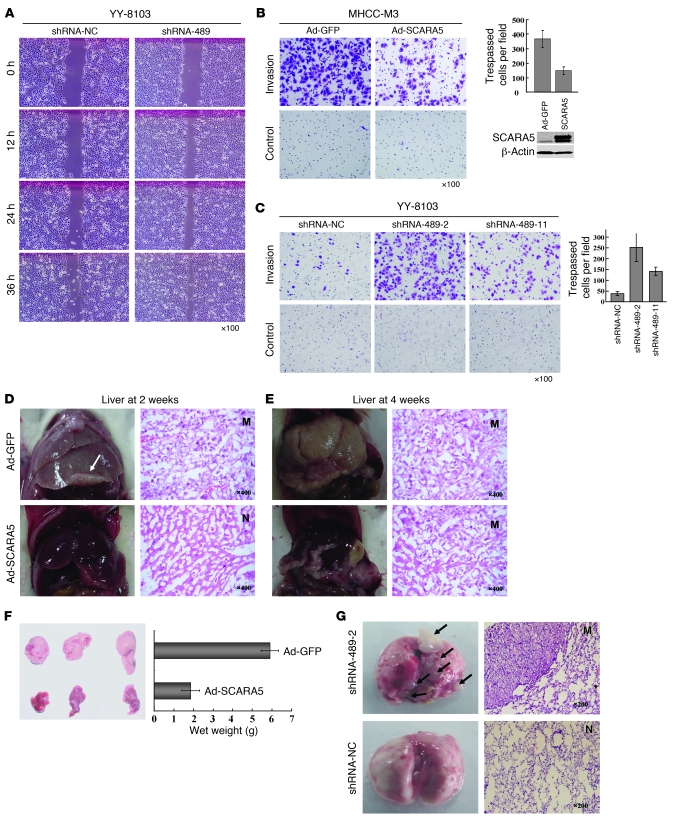
To determine whether SCARA5 downregulation contributes to HCC oncogenesis, we used 2 siRNAs that target SCARA5, siRNA-489 and siRNA-1515. As expected, these siRNAs efficiently knocked down the endogenous SCARA5 expression in WRL-68 and YY-8103 cells, as shown by real-time PCR (Figure 6, A and B). Interestingly, the silencing of SCARA5 significantly promoted the growth of WRL-68 and YY-8103 cells, as compared with the control groups transfected by a negative control siRNA (siRNA-NC) (P < 0.05; Figure 6, A and B). Moreover, both siRNAs enhanced the anchorage-independent growth of WRL-68, YY-8103, and PLC/PRF/5 cell lines in soft agar, as well as significantly increased the number of larger colonies relative to the number obtained from cells transfected with a siRNA-NC (Figure 6, C and D, and Supplemental Figure 8A). These results suggest that knockdown of SCARA5 disrupts contact inhibition among these cells. In this way, downregulation of SCARA5 may contribute to both tumor oncogenesis and progression.
Figure 6. Effect of SCARA5 silencing on cell growth, colony formation, and tumorigenicity of HCC cells.
(A and B) Both siRNA-489 and siRNA-1515 were used to knockdown SCARA5 in WRL-68 (A) and YY-8103 (B) cells, as demonstrated by real time RT-PCR, where siRNA-NC was used as control. Cell growth was measured, and each symbol represents a mean value of triplicate experiments (mean ± SD). (C and D) To observe the effect of SCARA5 silencing on colony formation in soft agar, pSUPER containing shRNA-489 and shRNA-1515 was transfected into WRL-68 (C) and YY-8103 (D) cells, respectively. Representative results show the increase in anchorage-independent colony formation. The numbers of colonies in the histogram represent the mean of 3 independent experiments (mean ± SD) (P < 0.01, compared with control). Original magnification, ×40. (E) Two stable SCARA5 knockdown subclones (shRNA-489-2 and shRNA-489-11) from YY-8103 cells were injected subcutaneously into mice; each group contained 8 mice. YY-8103 cells with either empty vectors or shRNA-NC were used as controls. A Kaplan-Meier survival plot 8 weeks after injection indicates that the mice injected with the shRNA-489 cells survived for a significantly shorter period of time than the controls (P < 0.001). All xenograft tumors were removed from the experimental mice (upper panel). (F) Silencing of SCARA5 enhances the tumorigenicity of WRL-68 cells with shRNA-489. The cells transfected with shRNA-NC were used as a negative control. Tumor growth was monitored for 4 days by measuring the diameter (mean ± SD).
To test this hypothesis, we established offspring subclones of the YY-8103 cell line containing a stable knockdown of endogenous SCARA5 by transfecting a pSUPER vector containing shRNA-489. As demonstrated with the transient lines above, the 2 stable knockdown subclones, shRNA-489-2 and shRNA-489-11, showed significantly greater cell growth than the parental YY-8103 cells and a subclone line with a stable shRNA-NC (P < 0.05; Supplemental Figure 8B). To extend these results to an in vivo context, we then injected YY-8103 cells (1 × 106 cells) from each of these stable subclones subcutaneously into 8 athymic mice and assessed tumorigenicity. By 8 weeks, no tumors were detected in any of the 8 mice injected with parental YY-8103 cells, and only 1 small tumor was formed in a mouse injected with the shRNA-NC cells. In contrast, multiple large tumors were clearly visible in the mice injected with either the shRNA-489-2 or shRNA-489-11 subclone. The former caused tumor formation in all 8 mice, while the latter caused tumor formation in 5 of the 8 mice (Figure 6E). In addition, we also observed that larger tumors were formed when SCARA5 expression was transiently silenced in a WRL-68 cells using shRNA-489, as compared with that of the control cells with shRNA-NC (P < 0.05; Figure 6F). These results suggest that downregulation of SCARA5 could contribute to tumorigenesis of HCC.
Downregulation of SCARA5 promotes cell migration and invasion.
The above tissue array data show that downregulation of SCARA5 may be associated with cellular invasion, venous permeation, and perhaps even metastasis in HCC (Figure 3E). Here, we employed wound-healing and Matrigel assays to assess the effects of SCARA5 expression on cell migration and invasion. The wound-healing assay revealed that cell migration was inhibited by SCARA5 overexpression in MHCC-LM6 cells (Supplemental Figure 9A), and it was enhanced by SCARA5 silencing in YY-8103 (Figure 7A) and WRL-68 cells (Supplemental Figure 9B). Next, we transiently transfected an adenoviral vector containing a SCARA5 expression construct into MHCC-LM3 cells (a well-documented cell line that is highly invasive) to evaluate the effect of the gene on cell invasion. Compared with cells transfected with empty vector, SCARA5 overexpression significantly inhibited cell invasion through a Matrigel barrier when fibronectin was used as an attractant (P < 0.01; Figure 7B). Interestingly, overexpression of SCARA5 had no effect on the cell proliferation and in vivo tumorigenicity of MHCC-LM3 cells (Supplemental Figure 10), suggesting that the inhibitory effect of SCARA5 on cell invasion is independent of cell proliferation.
Figure 7. SCARA5 modulates HCC cell migration, invasion, and metastasis.
(A) The migration of YY-8103 cells transfected with shRNA-489 in wound-healing experiment. (B) Cell invasion of MHCC-M3 cells infected with an adenovirus vector containing SCARA5 was evaluated with Matrigel assay. (C) The same approach was used for 2 stable YY-8103 subclones (shRNA-489-2 and shRNA-489-11) with SCARA5 knockdown. Counts of trespassed cells represent mean values per field (from at least 5 fields) from 3 independent experiments (right) (mean ± SD). (D–F) Effect of SCARA5 overexpression on liver metastasis of MHCC-LM3 cells with adenovirus-SCARA5, from spleen through the portal vein. No metastatic nodules were found in livers of any of SCID mice (n = 8) at 2 weeks after injection (D), but a small metastatic nodule formed at 4 weeks (n = 3). (E). However, cells with empty vector formed metastatic nodules (n = 6) at 2 weeks and larger ones at 4 weeks (n = 3). Representative images show livers and H&E-stained sections. (F) Tumors were weighed at 4 weeks; the weight is indicated (mean ± SD). (G) Representative images of metastases that formed in lungs of each nude mouse (n = 7) at 16 weeks after tail vein injection of YY-8103 cells with shRNA-489-2 or shRNA-NC. The sections show the lung tissues of mice injected with these cells. N, normal lung; M, metastatic nodule. White and black arrows indicate metastatic tumors in liver and lung, respectively. Original magnification, ×100 (A–C); ×400 (D and E, right panels); ×200 (G, right panels).
To confirm this observation, we repeated the Matrigel experiments using the 2 YY-8103 offspring subclones, shRNA-489-2 and shRNA-489-11 (described above). Both of these subclones showed significantly greater cell invasion than the cells transfected with the shRNA-NC (P < 0.01; Figure 7C). Moreover, YY-8103 cells transiently transfected with SCARA5 siRNAs also showed greater cell invasion than these control cells (Supplemental Figure 11). Thus, these findings indicate that downregulation of SCARA5 contributes to HCC progression, such as PVTT, by promoting cell migration and invasion.
SCARA5 contributes to tumor metastasis in vivo.
To further explore the effect of SCARA5 on tumor metastasis in vivo, we injected MHCC-LM3 cells transfected with an adenoviral vector containing SCARA5 (1 × 106 cells) into the spleen of NOD/SCID mice and observed their metastasis to the liver through the portal vein. After 2 weeks, no liver metastases were visible in any of the 8 mice injected with the cells expressing SCARA5; in contrast, 5 of the 6 mice injected with the control cells carrying empty vector developed macroscopic metastatic liver tumors (Figure 7D). The presence of liver tumors in these mice was confirmed by histological analysis. These results indicate that SCARA5 expression can significantly inhibit liver metastasis of MHCC-LM3 cells (P < 0.05). Furthermore, although metastatic liver tumors were observed after 4 weeks in 3 of the mice injected with the cells expressing SCARA5, these tumors were significantly smaller than the tumors in mice injected with control cells (P < 0.01; Figure 7, E and F).
To obtain solid evidence to support the notion that downregulation of SCARA5 contributes to HCC metastasis, we evaluated the effects of SCARA5 knockdown on long-distance tumor metastasis in vivo. We injected YY-8103 cells in which SCARA5 expression was stably knocked down (shRNA-489-2) or MHCC-LM3 cells overexpressing SCARA5 into the tail veins of athymic nude mice, and we checked for lung metastasis after 16 weeks. No visible metastatic tumors were found in the lungs of the mice injected with the MHCC-LM3 cells, regardless of whether or not they were overexpressing SCARA5. It is likely that this cell type is not suitable for use in distant lung metastasis analysis under these conditions. However, we found that all 7 of the mice injected with the YY-8103 cells with SCARA5 knockdown had significant numbers of metastatic lung tumors (Figure 7G). In contrast, the smaller, visible lung tumor was found only in 1 of the 7 mice injected with the control cells with shRNA-NC (P < 0.01). These data suggest that SCARA5 downregulation plays a crucial role in the malignant aggression and metastasis of certain HCC cells.
SCARA5 regulates activation of the FAK signaling pathway by affecting tyrosine phosphorylation.
We believe that the above data suggest that SCARA5 is a novel tumor suppressor, the inhibition of which potentiates oncogenesis, cell invasion, and metastasis. To explore the molecular mechanisms by which SCARA5 contributes to these malignant features, we performed colocalization experiments to determine whether SCARA5, as a membrane protein, interacts with other membrane proteins or their partners that are known to be involved in HCC progression and metastasis. In particular, we focused on 2 proteins, E-cadherin and FAK. The former is a well-known membrane protein associated with HCC progression and metastasis, and the latter is a key regulator of survival, proliferation, migration, and invasion in a wide variety of human cancers (51, 52). We found that SCARA5 and E-cadherin did not colocalize (data not shown), although we did observe colocalization of SCARA5 and FAK in PLC/PRF/5 cells, as indicated by immunofluorescence confocal microscopy (Figure 8A).
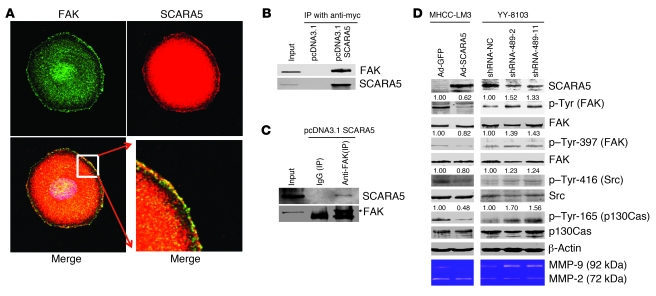
Figure 8. SCARA5 physically associates with FAK and modulates the tyrosine phosphorylation of FAK, Src, and p130Cas.
(A) Colocalization of both SCARA5 (red) and FAK (green) in PLC/PRF/5 cells by immunofluorescence. The bottom right panel is enlarged from the boxed region of the bottom left image. Original magnification, ×1,000 (top row and bottom left panel); ×5,000 (bottom right panel). (B and C) Co-IP assays were performed in MHCC-LM3 cells transiently transfected with a pcDNA3.1 vector containing myc-tagged SCARA5. The cells transfected with empty vector were used as a control. Endogenous FAK was immunoprecipitated with the anti-myc antibody (B), while the myc-tagged SCARA5 was reciprocally immunoprecipitated using the anti-FAK antibody (C). Native mouse IgG was used as the negative control, and 5% of the total MHCC-LM3 cell lysate was used for input. (D) Overexpression of SCARA5 inhibits the tyrosine phosphorylation of FAK (Tyr-397), Src (Tyr-416), and p130Cas (Tyr-165) in MHCC-LM3 cells. SCARA5 knockdown by shRNA promotes phosphorylation in YY-8103 cells. The total levels of these proteins were assessed by immunoblotting with the corresponding antibodies. β-actin was used as a loading control. Quantification of FAK, Src, and p130Cas phosphorylation levels, as indicated by the numbers above the corresponding panels, was performed by normalizing the total FAK, Src, and p130Cas concentrations to the β-actin loading control. The activity of 2 MMPs, MMP-2 and MMP-9, was determined by the gelatin-based zymography assay. SCARA5 overexpression inhibits the activity of MMP-9 but not MMP-2 in MHCC-LM3 cells. SCARA5 knockdown by shRNA promotes the activity of MMP-9 in YY-8103 cells.
To determine whether SCARA5 and FAK physically interact, we performed coimmunoprecipitation (Co-IP) assays in cells expressing SCARA5. Because it is difficult to examine the physical interactions of endogenous proteins in PLC/PRF/5 cells, we transiently transfected a pcDNA3.1 vector containing myc-tagged SCARA5 into MHCC-LM3 cells and then performed the Co-IP assay using an anti-myc antibody. Subsequent probing of the Co-IP reactions showed that endogenous FAK was immunoprecipitated by the anti-myc antibody (Figure 8B), while the myc-tagged SCARA5 was also immunoprecipitated by the anti-FAK antibody (Figure 8C). These reciprocal Co-IP data suggest that SCARA5 physically associates with FAK.
To assess the effect of SCARA5 expression on FAK activity, we examined the tyrosine phosphorylation of FAK, which has been proposed to contribute to cell growth and invasive tumor cell movement (53). Surprisingly, the phosphorylation of FAK was markedly suppressed in MHCC-LM3 cells transfected with SCARA5 (Figure 8D, left); conversely, SCARA5 knockdown markedly enhanced FAK phosphorylation in YY-8103 cells (Figure 8D, right). Furthermore, we assessed the effect of SCARA5 on tyrosine phosphorylation levels at given sites of certain molecules involved in the FAK signaling pathway in additional HCC cell lines (including MHCC-LM3, YY-8103, Huh-7, and WRL-68 cells) via SCARA5 overexpression or knockdown. Interestingly, SCARA5 overexpression in MHCC-LM3 and Huh-7 cells significantly inhibited the phosphorylation of the FAK residue (Tyr-397) that facilitates the autophosphorylation and intramolecular phosphorylation of FAK (53), Tyr-416 on Src and Tyr-165 on p130Cas, which are key components of the FAK protein complex. By contrast, SCARA5 knockdown markedly activated the FAK signaling cascade in YY-8103 and WRL-68 cells (Figure 8D and Supplemental Figure 12).
In addition, we also assessed the activity of 2 MMPs, MMP-2 and MMP-9, which are crucial downstream molecules in the FAK signaling pathway and are well documented to be associated with tumor invasion and metastasis (54). We used the gelatin-based zymography assay to measure the activity of MMP-2 and MMP-9 as a function of SCARA5 overexpression or knockdown. SCARA5 overexpression in MHCC-LM3 cells markedly inhibited the secretion of MMP-9, whereas SCARA5 knockdown in YY-8103 cells substantially enhanced the activity of MMP-9 but not that of MMP-2 (Figure 8D). This result is interesting in light of the fact that MMP-9 is considered to be an indicator of tumor progression and metastasis (55).
It would be valuable to know which domains of SCARA5 are required for its ability to bind FAK and to inhibit FAK phosphorylation. To determine which SCARA5 region is required for FAK binding, a series of myc-tagged SCARA5 variants were constructed (Figure 9A) and transfected into HCC cells. Co-IP assays were then performed, and we found that endogenous FAK was immunoprecipitated by the myc-tagged SCARA5 variants containing the intracellular fragment of amino acids 1–59 but not the extracellular fragment (60–495 amino acids) (Figure 9B). This implies that the short intracellular region, residues 1–59, may be responsible for the binding of SCARA5 to FAK. Furthermore, the phosphorylation level of FAK (Tyr-397) was evaluated in MHCC-LM3 cells transfected with the SCARA5 variants. The data showed that, with the exception of full-length SCARA5, the SCARA5 variants lacking the SRCR domain could not significantly inhibit FAK phosphorylation (Figure 9C). This suggests that the SRCR domain responsible for ligand binding could be crucial to regulating FAK activity.
Figure 9. Key domains of SCARA5 responsible for tyrosine phosphorylation of FAK.
(A) Determination of the domains of SCARA5 required for its ability to bind FAK and inhibit FAK phosphorylation. A series of myc-tagged SCARA5 variants with truncated fragments were constructed. (B) The plasmid constructs were transfected into MHCC-LM3 cells, and then immunoprecipitation was performed using an anti-myc antibody. Immunoblotting assays were subsequently carried out with the anti-FAK antibody; 5% of the total MHCC-LM3 cell lysate was used as input. (C) The phosphorylation level of FAK (Tyr-397) was also evaluated. Quantification of phosphorylated FAK levels, as indicated by the numbers above the corresponding panels, was performed by normalizing total FAK concentrations to β-actin as a loading control.
The above findings suggest that the physiological function of SCARA5 is to inhibit the activity of the FAK signaling pathway by physically associating with FAK. If that is indeed the case, downregulation of SCARA5 expression due to epigenetic and genetic events in HCC may contribute to tumorigenesis and progression by activating the FAK signaling pathway via initiation of the cascade by tyrosine phosphorylation of the FAK-Src-Cas complex, along with increased MMP-9 activity (Figure 10). However, some known secondary pathways, in the context of activating FAK signaling, may also be involved in cell proliferation, migration, invasion, and tumor metastasis and should be investigated further.
Figure 10. A hypothetical schematic of the contribution of SCARA5 to HCC via activation of the FAK signaling pathway.
The physiological function of SCARA5 is to inhibit the activity of the FAK signaling pathway by physically associating with FAK. In this way, downregulation of SCARA5 due to epigenetic and genetic events in HCC may contribute to tumorigenesis and progression, possibly by activating the FAK signaling pathway via initiation of the FAK-Src-Cas complex tyrosine phosphorylation cascade, along with increased MMP-9 activity. Based on the published data (52, 53, 67–71), the activating FAK-Src-Cas complex leads to tumor growth and metastasis, by promoting cell motility, invasion, cell cycle progression, survival, angiogenesis, and epithelial-to-mesenchymal transition, possibly through the secondary signaling pathways (indicated in bold). Cdc42, cell division cycle 42; CRK, v-crk sarcoma virus CT10 oncogene homolog (avian); DOCK180, dedicator of cytokinesis 1 (also known as DOCK1); Grb2, growth factor receptor-bound protein 2; PKL, paxillin kinase linker.
Discussion
The epigenetic silencing of tumor suppressor genes is considered to be a major event contributing to the development and progression of human cancers. Hypermethylation of the CpG islands in the promoter regions of these genes has been shown to be an important mechanism by which tumor suppressors are inactivated. DNA hypermethylation of promoters has been shown to affect the expression of genes involved in the cell cycle, DNA repair, metabolism of carcinogens, cell-cell interactions, apoptosis, and angiogenesis (1). Moreover, DNA hypermethylation can also occur at different stages in tumor development and in different cellular networks. Recently, some epigenomic approaches have indicated that in a given tumor, the promoter regions of genes can contain 100–400 hypermethylated CpG dinucleotides (34). The loss of function of numerous tumor suppressor genes as a result of the hypermethylation of CpG islands in promoters may contribute to both the genesis and progression of tumors.
In the present study, we applied a pharmacologic unmasking strategy, involving treatment with both a demethylation agent and a histone deacetylase inhibitor, to 15 HCC cell lines to identify tumor suppressor genes silenced by DNA hypermethylation in HCC, using a genome-wide screen (35, 36). In this system, we looked for genes that were markedly upregulated by the demethylation treatment. We found that although 524 were substantially upregulated, which we defined as an at least 3-fold upregulation relative to those before treatment, in at least 3 of 15 HCC cell lines, only 255 genes have typical CpG islands in their promoters. This result implies that the 255 upregulated genes with CpG islands in their promoters are likely to include tumor suppressor genes that are epigenetically silenced in HCC oncogenesis or progression.
Given that aberrant DNA methylation can result from prior genetic lesions (56), we focused our attention on chromosome 8p, one of the most frequent LOH regions in HCC and other cancers, which is associated with tumor progression and metastasis. Nine of the upregulated genes that we detected mapped to chromosome 8p. One of these genes, SCARA5, was reexpressed following demethylation in 6 of the 15 HCC cell lines, whereas the known candidate tumor suppressors DLC1, SFRP1, and TUSC3, were upregulated in only 5, 4, and 3 of the lines, respectively. This implies that SCARA5 could be easily governed by the methylation status at the CpG island in its promoter, although the epigenetic events other than DNA hypermethylation, such as histone modification, also could be involved in the reexpression of SCARA5. As it turns out, only DLC1, SFRP1, and SCARA5, but not TUSC3, have been shown to be downregulated in HCC in clinical specimens. Increasing evidence points to DLC1 and SFRP1 as particularly attractive candidate tumor suppressors (6–8, 20, 57). However, the fact that SCARA5 is subject to various aberrations — LOH, hypermethylation of a CpG island in the promoter, and downregulated mRNA levels in HCC specimens — suggests that it is a tumor suppressor. In fact, the functions of SCARA5, which include inhibiting tumorigenicity, colony formation, cell invasion, tumor metastasis, and tyrosine phosphorylation of FAK, Src, and p130Cas, are typical of tumor suppressors. We therefore propose that SCARA5, like the known cases of DLC1 and SFRP1, is a tumor suppressor gene.
However, unlike DLC1 and SFRP1, SCARA5, which is found at chromosome 8p21.1, encodes a putative class A membrane scavenger receptor that localizes to the plasma membrane (Figure 3, A and B) (45). SCARA5 is one of several scavenger receptors, yet genes for other such receptors, such as macrophage scavenger receptor 1 (MSR1, also known as SCARA1) at 8p22 and scavenger receptor class A, member 3 (SCARA3) at 8p21, were not upregulated following demethylation in any of the 15 HCC cell lines tested (Supplemental Table 1).
To determine whether SCARA5 is mutated in HCC, we performed a mutational screen of SCARA5 in 40 paired samples of HCC tissue and adjacent noncancerous tissue. No mutations were found in these samples. However, 35% (14 of 40) of the HCC specimens showed an allelic imbalance or LOH (not homozygous deletion), suggesting that hemizygous deletion of the SCARA5 locus, along with hypermethylation of the CpG island in the promoter of the remaining allele, contributes to oncogenesis and progression of some HCC cases by downregulating the gene.
Most interestingly, SCARA5 expression is associated with HCC progression, such as PVTT, suggesting that the lack of the plasma membrane protein SCARA5 contributes to tumor invasion and metastasis. The in vitro and in vivo experimental data show that SCARA5 overexpression can inhibit tumorigenicity, cell invasion, and metastasis; conversely, SCARA5 knockdown can enhance tumorigenicity, cell invasion, and tumor metastasis in vivo. These results are consistent with previous reports that chromosome 8p contains one or more tumor suppressor genes that are closely associated with HCC progression and metastasis (11–15). Among the known candidate tumor suppressors located on chromosome 8p, SFRP1 is frequently downregulated in HCC, but studies so far do not link it with HCC progression or metastasis (58). In contrast, the known tumor suppressor DLC1 is considered to be associated with HCC carcinogenesis and metastasis (20, 59). DLC1 significantly inhibits cell proliferation, anchorage-independent growth, in vivo tumorigenicity, cell motility, and invasiveness as well as long-distance dissemination of HCC cells (18, 60). DLC1 plays important roles in signal transduction pathways regulating cell proliferation, cell morphology, and cell migration, by affecting the Rho family GTPases and the focal adhesion proteins (17, 18, 20, 61). SCARA5, which is located in the metastasis-related LOH region of 8p21.3, may prove to be a more potent metastasis suppressor than DLC1, because it encodes a transmembrane protein and can therefore link the extracellular matrix directly with cytoplasmic molecules. Indeed, in addition to inhibiting colony formation and tumorigenicity, SCARA5 overexpression can suppress MHCC-LM3 cell invasion and metastasis from the spleen to the liver; conversely, reducing SCARA5 expression can promote colony formation, cell invasion, tumorigenicity, and long-distance lung metastasis via the tail vein injection, suggesting that SCARA5 plays a crucial role in preventing HCC carcinogenesis and metastasis.
The results of the present study show that SCARA5 appears to participate in focal adhesion by physically associating directly with FAK, a non-receptor tyrosine kinase, and modulating tyrosine phosphorylation. FAK is known to be important in the regulation of focal adhesion dynamics and disassembly during cell migration (62). It is activated in a range of tumor cells, and its increased activity correlates with the malignancy and invasiveness of human HCC and other tumors (63–65). Phosphorylation of FAK at Tyr-397 and the subsequent phosphorylation of other focal adhesion complex–associated proteins (e.g., paxillin and p130Cas) are required for focal adhesion formation and cell migration (66). The activated FAK forms a complex with Src and p130Cas in many tumor cells, which leads to tumor growth and metastasis by promoting cell motility, invasion, cell cycle progression, survival, angiogenesis, and epithelial-to-mesenchymal transition, via secondary signaling pathways, as summarized in Figure 10 (52, 53, 67–71). Surprisingly, our work indicates that SCARA5 can inhibit the FAK tyrosine phosphorylation cascade and the downstream signaling pathways, which involve Tyr-397 of FAK, Tyr-416 of Src, and Tyr-165 of p130Cas. Conversely, SCARA5 knockdown stimulates the focal adhesion complex–associated signaling transduction cascade. This suggests that downregulation of SCARA5 in HCC specimens (as a result of allelic deletion, DNA hypermethylation of the CpG island in the promoter, and/or possible histone modifications) contributes to oncogenesis and the progression of this carcinoma by promoting cell proliferation, migration, invasion, and metastasis. Furthermore, this occurs via activation of the FAK-Src-Cas signaling pathways, which can lead to MMP9-mediated degradation of the extracellular matrix (Figure 10).
However, it should be pointed out that although SCARA5 overexpression can inhibit cell invasion and the FAK signaling pathway in MHCC-LM3 cells (Figure 8D), it did not have the same effect on cell proliferation, including xenograft tumor growth, in this line compared with the other HCC lines (Supplemental Figure 10). These interesting data suggest that SCARA5 serves as one of many contributors to tumor growth and/or progression in HCC specimens, with distinct genetic and epigenetic aberrations, in which the activation of the FAK-Src-p130Cas pathway due to the SCARA5 silencing, cooperation with signaling pathways responsible for cell proliferation, and other malignant features could have a synergic role in HCC initiation and progression.
Methods
Tissue specimens.
All HCC specimens were obtained from patients who underwent surgical resection of their tumors and signed informed consent before their liver operations. The primary tumor specimens were immediately frozen at –80°C until DNA/RNA extraction. Specimens (~1 cm3) of both tumor and adjacent nontumor tissue were taken from each patient, and the HCC diagnosis was confirmed by pathological examination. The HCC specimens presented in this work were grouped according to the differentiation grades II–III according to the Edmondson grading system. The clinical characteristics of the patients and tumors are summarized in Supplemental Table 2. This project and the protocols for the investigation involving human and animal tissues were approved by the ethics committee of the Chinese National Human Genome Center.
Liver cancer cell lines.
The following liver tumor-derived cell lines were used in this study: PLC/PRF/5, QGY-7703, QGY-7701, Bel-7402, Sk-hep1, MHCC-H, Bel-7404, YY-8103, Bel-7405, SMMC7721, MHCC-L, Focus, HepG2, Hep3B, Huh-7, SNU398, MHCC-LM6, and MHCC-LM3. All of these cell lines were grown under standard cell culture conditions in the following media: minimum essential medium Eagle (Sigma-Aldrich) supplemented with 10% FBS (Life Technologies), 1% l-glutamine, and 1% nonessential amino acids in a 5% CO2-humidified chamber.
Extraction of DNA and RNA.
Genomic DNA was extracted from all specimens using the DNeasy Tissue kit (Qiagen) according to the protocols recommended by the manufacturer. RNA was extracted using TRIzol solution (Invitrogen) according to the protocols recommended by the manufacturer. RNAse-free DNase I was used to remove DNA contamination. The total RNA concentration and quantity were assessed by absorbance at 260 nm, using a DNA/Protein Analyzer (DU 530; Beckman).
RT-PCR.
RT was performed in a 20-μl reaction, with a total of 2 μg of RNA. Each PCR reaction was run for 35 cycles, and the PCR products were observed by electrophoresis on a 2% agarose gel and visualized after staining with ethidium bromide, where band intensities were normalized to that of β-actin. The following primers were used to amplify a 245-bp PCR product for SCARA5: forward, 5′-CAGCTGGTTTCTTACCACGTAT-3′; reverse, 5′-GCACAAGTTCTCCCACACTTAG-3′. The following primers were used to amplify a 295-bp PCR product for ACTB: forward, 5′-TCACCCACACTGTGCCCATCTACGA-3′; reverse, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′. To perform further quantitative analysis of the expression of the SCARA5 gene in HCC specimens, the relative mRNA level of SCARA5 was measured by quantitative real-time RT-PCR, using the TaKaRa PCR Thermal Cycler Dice Detection System and SYBR green dye (TaKaRa) in 40 paired HCC specimens, according to the protocols recommended by the manufacturer. A housekeeping gene, ACTB, was used as an endogenous control. Measurements were repeated at least 3 times to ensure the reproducibility of the results. The mRNA level for each gene in each HCC sample was compared with the level in the paired nontumorous tissue (72). The mean Ct value for the ACTB gene was subtracted from the mean Ct value for SCARA5 for each sample, using the following formula: SCARA5ΔCt = (mean SCARA5 Ct – mean ACTB Ct), SCARA5ΔΔCt = (SCARA5ΔCt_HCC – SCARA5ΔCt_non-HCC). The fold change (2–SCARA5ΔΔCt) of the SCARA5 expression level relative to the β-actin expression level was calculated for each HCC sample. The significance level was defined as a P value of less than 0.01 (73).
Microarrays.
Total RNA was isolated using the RNeasy RNA isolation kit (Qiagen) according to the supplier’s protocol. Possible genomic DNA contamination was removed with a column DNase treatment step, using the RNase-free DNase set (Qiagen). Purified RNA was quantified using a Nanodrop spectrophotometer (ND-1000; Wilmington), and RNA quality was evaluated using the Agilent 2100 Bioanalyzer. cDNA synthesis was carried out according to the manufacturer’s instructions, using the Agilent Fluorescent Direct Label kit to allow incorporation of nucleotides containing either cyanine 5 (Cy5-dCTP, for untreated samples) or cyanine 3 (Cy3-dCTP, for samples treated with drugs) (Perkin-Elmer LifeSciences). Labeled cDNA was purified using QIAquick PCR purification columns (Qiagen), then concentrated by vacuum centrifugation. Labeled cDNA was resuspended in hybridization buffer and hybridized for 18 hours at 68°C to an Agilent G4112A microarray containing 41,000 probes (Agilent Technologies), according to Agilent protocols. The microarray image file was processed using the Agilent’s Feature Extraction software (version A.9.5.3; Agilent Technologies) and subjected to Lowess normalization. The log ratio of the red to green intensities for each signal was used for statistical analyses, with all subsequent analyses carried out using SPSS statistical software (version 11.5). We selected a fold change of 3 as the threshold for significant upregulation.
Immunohistochemistry and immunofluorescence assays.
Immunohistochemical staining for the target genes was carried out on sections of the formalin-fixed samples on the tissue microarray, including 100 paired liver cancer samples and adjacent noncancerous tissue. Briefly, the sections (4-μm thickness) were deparaffinized in xylene and rehydrated by transfer through graded concentrations of ethanol to distilled water, and endogenous peroxidase activity was blocked by incubation with 30 ml/l H2O2 in methanol for 10 minutes at room temperature. Then, sections were submitted to antigen retrieval in a pressure cooker containing 0.01 mmol/l sodium citrate buffer for 10 minutes. Slides were subsequently incubated in 100 ml/l normal goat serum for 20 minutes at room temperature. Sections were permeabilized in PBS-Triton and incubated overnight with primary antibody at 4°C. The antibodies were used in PBS-Triton at different dilutions. Rabbit anti-human polyclonal antibodies (made in our laboratory) to SCARA5 (1:100) were incubated at 37°C for 2 hours and placed at 4°C overnight. Then, the slides were incubated with a horseradish peroxidase–conjugated mouse anti-rabbit secondary antibody (Dako Japan Ltd.) at 37°C for 1 hour. Finally, the sections were reacted with 0.02% 3,3′-diaminoberzidine and 0.005% H2O2 in 0.05 mmol/l Tris-HCl buffer, and counterstaining was performed with hematoxylin. Negative control slides were treated by incubation with PBS instead of the primary mouse antibody, whereas normal livers, in which SCARA5 is known to be expressed, served as positive controls in each experiment. In addition, the slides with HCC samples and adjacent non-HCC tissue samples were simultaneously subjected to immunohistochemical staining. Stained slides were observed under light microscopy. Positivity was ascertained based on the presence and intensity of brown granules. All slides were reviewed independently by 2 pathologists who were blinded to each other’s readings. Tissues were graded on the following scale: 0, negative; 1, low expression; 2, moderate expression; 3, high expression; 4, strong expression.
An immunofluorescence assay was performed to detect endogenous SCARA5 expression in PLC/PRF/5 cells. The cells were plated on polylysine-treated slides and incubated at 37°C for 60 hours. The cells were fixed, blocked with PBS buffer containing 5% BSA, and then stained with rabbit anti-human polyclonal antibodies to SCARA5 (1:200) at 4°C overnight, followed by incubation with a Cy3-conjugated (red) anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories Inc.) at 4°C for 2 hours. After rinsing, the slides were analyzed using immunofluorescence microscopy. For colocalization of both SCARA5 and FAK, the cells were incubated with a polyclonal antibody against SCARA5 and a monoclonal antibody against FAK (1:1,000; Santa Cruz Biotechnology Inc.). Fluorescein isothiocyanate Cy3-coupled (red) anti-rabbit IgG antibody and Alexa Fluor 488–coupled (green) anti-mouse IgG antibody (Molecular Probes Inc.) were used as secondary antibodies, respectively. The stained cells were analyzed by Zeiss confocal microscopy and ZEISS LSM Image Browser version 2.80 software (Carl Zeiss).
SCARA5 expression vector.
The full-length SCARA5 ORF (nt 441–1928; GenBank accession number NM_173833) was amplified by PCR from the cDNA of an adjacent nontumor tissue. The primers contained EcoR I and BamH I linkers. The primers were as follows: forward, 5′-CGGAATTCCGATGGAGAACAAAGCTATGTACCTACACAC-3′; reverse, 5′-CGGGATCCGCGTGTCTGTTGCATGTCACGCTG-3′. After digestion with EcoR I and BamH I, the PCR product was inserted into the EcoR I and BamH I sites of the multiple cloning site of the pcDNA3.1(B) plasmid expression vector (Clontech). DNA sequencing and restriction enzyme digestion were used to confirm that the sequence and orientation of the construct were correct. Similarly, to determine which domains are responsible for the binding of SCARA5 to FAK or for inhibition of FAK activity, recombinant pcDNA3.1 vectors containing different fragments of SCARA5 were further constructed (Figure 9A). To construct SCARA5 recombinant adenovirus vector, full-length SCARA5 cDNA was amplified by PCR from human liver cDNA. The primers contained Bgl II and Hind III linkers. The primers were as follows: forward, 5′-GAAGATCTATGGAGAACAAAGCTATGTA-3′; reverse, 5′-CCCAAGCTTTCAGTGTCTGTTGCATGTCA-3′. After digestion with Bgl II and Hind III, the PCR product was inserted into the multiple cloning site of pShuttle-IRES-hrGFP-1 (Stratagene), a shuttle vector that contains a CMV promoter with a GFP. Next, pShuttle-IRES-hrGFP-1-SCARA5 and pAdEasy-1 (Stratagene) were homologously recombined in E. coli BJ5183. The newly recombined plasmid, Ad-SCARA5, was verified by restriction endonuclease digestions and sequencing. Ad-SCARA5 was then propagated in 293 cells and a viral stock was harvested from these cells. After 6 cycles of freezing and thawing, cell debris was removed by subjecting the lysed cells to 12,000 g centrifugation. The virus stock was stored at –80°C.
Cell transfection.
To observe cell proliferation, recombinant pcDNA3.0 vectors containing the SCARA5 gene were transfected into target cells using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. The transiently transfected cells were seeded in a 96-well plate at a cell density of 2 × 103 and then cultured at 24-hour intervals for 7 days. Cell viability was measured using the Cell Counting Kit-8 (CCK-8; Dojindo Laboratories) according to the instructions of the manufacturer. Briefly, 10 μl of CCK-8 solution was added to each well of the plate, and the plate was incubated at 37°C for 1 hour. The absorbance was measured at 450 nm as an indicator of cell viability. All experiments were independently repeated at least 3 times.
Colony formation.
To assay the effect of SCARA5 on colony formation, recombinant pcDNA3.0 vectors containing the SCARA5 were transfected into targeted HCC cells (empty vector as a control) in 35-mm dishes by Lipofectamine 2000 (Invitrogen) for 24 hours, and then stripped and plated onto 100-mm tissue culture dishes. Then, G418 (Life Technologies Inc.) was added to the medium at a final concentration of 700 μg/ml. After 3 weeks of selection, the remaining colonies were washed twice with PBS, stained with crystal violet, and counted on crystal violet–stained dishes. All experiments were independently repeated at least 3 times. For the soft agar colony formation assay in 24-well plates, 2 × 104 cells were plated and grown on a plate containing 1% base agar and 0.5% top agar. Plated cells were incubated at 37°C for 21 days. Plates were stained with 0.005% crystal violet for 1 hour. Colonies were counted under a dissecting microscope. All experiments were independently repeated at least 3 times.
Co-IP.
MHCC-LM3 cells transfected with pcDNA3.1 vectors encoding myc-tagged SCARA5 variants were resuspended in 1 ml of lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1.0% Triton X-100, 1 mM EDTA and protease inhibitor cocktail). Immunoprecipitation of lysates from MHCC-LM3 cells transfected with myc-tagged SCARA5 was conducted using the anti-myc antibody (1:1,000; Santa Cruz Biotechnology Inc.), followed by immunoblotting with antibodies against FAK (1:1,000; Santa Cruz Biotechnology Inc.) or SCARA5 (1:500; made in our laboratory). The MHCC-LM3 cell lysates transfected with empty vector (pcDNA3.1) served as a control.
Immunoblotting.
The cells were collected using a 2× loading lysis buffer (2× concentrations, 50 mmol/l Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue). Total protein extracts from cultured cells were subjected to protein gel electrophoresis using 12% SDS-PAGE and were then transferred to a Hybrid-PVDA membrane (Amersham Life Sciences) by a semidry electrophoretic transfer method and treated using 20% methanol in a Tris-glycine buffer (25 mM Tris-HCl, pH 8.0, 0.2 M glycine, 0.1% SDS). After blocking with PBS containing 5% BSA, the membrane was incubated for immunoblotting analysis with a rabbit anti-SCARA5 polyclonal antibody (1:100; made in our laboratory) or anti-myc (1:1,000) at room temperature for 2 hours, followed by incubation with an IRDye 800DX–conjugated, affinity-purified goat anti-rabbit secondary antibody (1:1,000; Rockland). The signals were detected using the Odyssey Infrared Imaging System (LI-COR Biosciences). After stripping, the same membrane was reprobed with other antibodies, including anti-FAK (1:1,000), anti-p130CAS (1:1,000; Santa Cruz Biotechnology Inc.), anti-Src (1:1,000; Santa Cruz Biotechnology Inc.), anti–phospho-FAK (Y397) (1:1,000; ABCAM Inc.), anti–phospho-Src (Y416) (1:500; Cell Signaling Technology Inc.), anti–phospho-p130CAS (Y165) (1:500; Cell Signaling Technology Inc.), and anti-actin (1:500; Sigma-Aldrich) at room temperature for 2 hours, followed by incubation with an IRDye 800DX-conjugated, affinity-purified goat anti-mouse secondary antibody (1:1,000; Rockland). The signals were detected using the Odyssey Infrared Imaging System (LI-COR Biosciences). β-actin was used as a loading control.
RNAi.
Two siRNAs against SCARA5 were designed at the Whitehead Institute Web Server (http://jura.wi.mit.edu/bioc/siRNAext/) and chemically synthesized (Shanghai GenePharma Co.) for targeting different coding regions of the gene as follows: siRNA-489 (5′-GCUCCAUCUGUGAGGAUUCdTdT-3′ and 5′-GAAUCCUCACAGAUGGAGCdTdT-3′) for nt 489–507 of SCARA5 and siRNA-1515 (5′-UGGGCAUGCGUGGGUUCAAdTdT-3′ and 5′-UUGAACCCACGCAUGCCCAdTdT-3′) for nt 1515–1533 of SCARA5. In addition, siRNA-NC (5′-UUCUCCGAACGUGUCACGUdTdT-3′ and 5′-ACGUGACACGUUCGGAGAAdTdT-3′) was also synthesized. All the above siRNAs were transfected into HCC cell lines, and cell growth was observed. For siRNA transfection, 3 × 103 HCC cell lines were seeded in 96-well plates. Cells grown to 30%–50% confluence were transfected with synthetic siRNAs at final concentrations of 50 nM, using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. The cells were then cultured at 24-hours intervals for 7 days. Cell viability was measured using the CCK-8 according to the instructions of the manufacturer. Briefly, 10 μl of CCK-8 solution were added to each well of the plate, and the plate was incubated at 37°C for 1 hour. The absorbance was measured at 450 nm to assess cell viability. All experiments were independently repeated at least 3 times.
In addition, for the construction of the RNAi plasmid, the oligonucleotides for the double-stranded shRNA were inserted into the expression plasmid pSUPER containing the polymerase-III H1-RNA gene promoter (74); where pSUPER was provided by R. Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). The oligonucleotides for shRNA were synthesized as follows: shRNA-489, forward, GATCCCCGCTCCATCTGTGAGGATTCTTCAAGAGAGAATCCTCACAGATGGAGCTTTTTGGAAA; shRNA-489, reverse, AGCTTTTCCAAAAAGCTCCATCTGTGAGGATTCTCTCTTGAAGAATCCTCACAGATGGAGCGGG for pSUPER SCARA5; shRNA-NC, forward, GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTGGAAA; shRNA-NC, reverse, AGCTTTTCCAAAAATTCTCCGAACGTGTCACGTTCTCTGAACGTGACACGTTCGGAGAAGGG. The negative control oligos served as the nonspecific control.
LOH analysis.
LOH analysis was performed by detecting the DNA of 40 pairs of HCC specimens and adjacent noncancerous tissue using polymorphic microsatellite markers D8S1839 (forward, GGAGAATGGGTCAGAAC; reverse, TCCAGGGTTGCTACAGT), D8S1820 (forward, AGCTGTGCTCACTGGAAA; reverse, TGGATACCGAGGGACAAC), and D8S1809 (forward, ATGGGACTTTTGACTGG; reverse, TTTGGCAATCTCTGGAGAC), all of which locate closely to the SCARA5 locus. PCR products were analyzed using a program on the ABI 3730 sequencer. LOH was analyzed by determining the fluorescence intensity of each allele and calculating the ratio using the peak height, where the longer or shorter alleles were considered to be substantially lost in HCC specimens with an LOH value of less than or equal to 0.5 or more than or equal to 1.5, respectively.
FISH.
FISH to detect SCARA5 deletion was carried out using the BAC clone RP11-597M17 (ntLocus 8: 6050539–6248157), which was verified by PCR to contain the SCARA5 gene. Sections (5 μm in thickness) were cut from freshly frozen liver cancer tissue and adjacent noncancerous livers using a standard microtome. The BAC clone was labeled by nick translation using SpectrumOrange-dUTP (no. 30-803000; Vysis Inc.). The Spectrum Green–labeled CEP8 centromere (Vysis Inc.) was used as control. Hybridization and evaluation of the results were performed as described previously with minor modifications (75). In brief, the slides were pretreated with 0.01% pepsin solution (1 mg Pepsin in 10 ml of 0.01 N HCl). The slides and probes were then denatured using the HYBrite Denaturation/Hybridization System (no. 30-144010; Vysis Inc.) at 85°C for 10 minutes. After incubation for more than 48 hours at 37°C in a humidified chamber, the slides were washed with 0.4× SSC/0.3% NP-40 for 2 minutes at 72°C ± 1°C. The slides were placed in 2× SSC/0.1% NP-40 for 1 minute at room temperature. DAPI I (1,000 ng/ml; Vysis Inc.) was then applied to each spot, which was then coverslipped. The slides were observed under a fluorescence microscope (BX-51; Olympus) equipped with epifluorescence filters and a photometric CCD camera and an image analyzer system (CytoVision; Applied Imaging Ltd.). Two observers were independently involved in the FISH analysis, and investigators responsible for the FISH analysis were blinded to all parameters of the specimens, except for the date of collection. The score was calculated as the mean between the 2 observers’ scores. The number of FISH signals per cell was counted for a total of more than 200 intact and nonoverlapping cell nuclei. Furthermore, to prevent counting of “artificial loss” as a result of applying the same method to noncancerous tissue, the cutoff value was intentionally set at a higher percentage of 15% (75). A loss of more than 15% in cancerous tissue, corresponding to 2 green and 1 red signals was considered a true loss.
MSP and bisulfite sequencing.
Genomic DNA was treated with bisulfite according as previously described (76). Genomic DNA (1 μg) was denatured by incubation with 0.2 M NaOH. Aliquots of 10 mM hydroquinone and 3 M sodium bisulfite (pH 5.0) were added and the solution was incubated at 50°C for 16 hours. To analyze the DNA methylation status of the CpG islands of SCARA5 promoter in HCCs and cell lines, MSP was performed with genomic DNA treated by bisulfite and specific primers for unmethylated and methylated DNA were designed within the CpG island of the SCARA5 gene as following: CpG island I, methylation (forward, GTTTAGCGGGCGTTTTTATACG; reversal, AACAAAACCACAAAAACCACTTC) and without methylation (forward, AGTGGGTGTTTTTATATGGGGTT; reversal, ACTCCCTATCCTTAATACCTAATCCT). In order to analyze the DNA methylation status of the CpG islands of the SCARA5 gene in HCCs and cell lines, regions enriched in CpG islands were amplified from bisulfite-treated genomic DNA using the following primers: CpG island I, forward, 5′-GGGGTGTTGGAGTTTTAAGG-3′; reverse, 5′-CACAACCAACCAAATCCAAA-3′; CpG island II, forward, 5′-GGTAGGTTTGGGGATTATTGG-3′; reverse, 5′-CAACTCCTACTTCAACTACAACTCCA-3′; CpG island III, forward, 5′-GAGTAGGTGTGGAAGGTGTAGGA-3′; reverse, 5′-CCACCAAACTCCCATTAACC-3′. Then, the PCR product was subcloned into a pMD 18-T vector (TaKaRa Inc.) for DNA sequencing on an ABI 3730 sequencer.
Tumorigenicity in vivo.
A total of 2 × 106 targeted cells, showing either knockdown or overexpression of SCARA5, were injected subcutaneously into the right flank or both flanks of nude mice. Growth curves were plotted based on mean tumor volume within each experimental group at the indicated time points. The tumor dimensions were measured every 3 days using a digital caliper, and the tumor volume calculated using the following formula: V = π/6 × (larger diameter) × (smaller diameter)2. Tumor growth was observed for at least at 8 weeks. The tumorigenic experiments in vivo were performed with 8 mice in each treatment group.
Wound-healing assay.
Cells were grown to 80%–90% confluence in 60-mm plates, which were transfected by shRNA-489 for silencing SCARA5 or plasmid containing SCARA5. A wound was made by dragging a plastic pipette tip across the cell surface after 48 hours. The phase contrast images of the wounds were recorded at 37°C for incubations of 0, 12, 24, 36 hours, and 3 separate experiments were performed. The cells were transfected by shRNA-NC or empty vector as the controls.
Cell invasion assay.
Cell invasion assays were performed using 24-well transwells (8-μm pore size; BD Biosciences), coated with Matrigel (Falcon 354480; BD Biosciences). HCC cells were starved overnight in serum-free medium, trypsinized, and washed 3 times in DMEM containing 1% FBS. A total of 1 × 105 cells was then suspended in 500 μl DMEM containing 1% FBS and added to the upper chamber, while 750 μl DMEM containing 10% FBS and 10 μg/ml fibronectin (catalog no. 356008, BD Biosciences) was placed in the lower chamber. For the control, medium containing 1% FBS was added to the lower chamber. After 48 hours of incubation, Matrigel and cells remaining in the upper chamber were removed by cotton swabs. Cells on the lower surface of the membrane were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet. Cells in at least 6 random microscopic fields (magnification, ×100) were counted and photographed. All experiments were performed in duplicate and repeated 3 times.
Metastasis in vivo.
We employed the subcutaneous split-spleen reservoir model and tail vein injection assay to assess the effect of SCARA5 on tumor metastasis. The operations to establish split-spleen reservoirs were conducted as follows: 8- to 10-week-old NOD/SCID mice were anesthetized with 2.5% sodium pentobarbital (40 mg/kg; Sigma-Aldrich), the hair on the left side of the abdomen was removed, and the area was sterilely prepped. The abdomen was entered through a small opening in the muscle and peritoneum. The upper part of each spleen was then injected with 200 μl MHCC-LM3 cells (5 × 106) transfected with adenovirus vector containing the SCARA5 ORF, while those with empty adenovirus vector were used as control. Then, the skin was closed. Liver tumor weights were compared using the t test in SPSS (version 11.5; SPSS).
In addition, a total of 2 × 106 MHCC-LM3 cells previously transfected with either recombinant adenovirus vector containing full-length SCARA5 or empty adenovirus vector was injected into the tail veins of athymic nude mice. A total of 2 × 106 YY-8103 cells transfected with either shRNA-489-2 or the shRNA-NC control were injected into the tail veins of athymic nude mice. The mice were assessed for long-distance lung metastasis at 16 weeks.
Statistics.
Statistical differences and variances were evaluated using the χ2 test in SPSS. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the China National Key Projects for Infectious Disease (2008ZX10002-021 and 2008ZX10002-020), the Chinese National Key Program on Basic Research (2010CB529200 and 2006CB910402), International Scientific Collaborative Project (20072901), the Shanghai Commission for Science and Technology (08JC1416400, 06ZR14069, and 06DZ22903), the China Postdoctoral Science Foundation (20070420097), and the Shanghai Postdoctoral Science Foundation (07R214138).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:223–241 (2010). doi:10.1172/JCI38012.
References
- 1.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Diaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5 Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. 1999;116(2):394–400. doi: 10.1016/S0016-5085(99)70137-X. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Downregulation of E-cadherin by hepatitis B virus X antigen in hepatocellullar carcinoma. Oncogene. 2006;25(7):1008–1017. doi: 10.1038/sj.onc.1209138. [DOI] [PubMed] [Google Scholar]
- 5.Kubo T, et al. Apoptotic speck protein-like, a highly homologous protein to apoptotic speck protein in the pyrin domain, is silenced by DNA methylation and induces apoptosis in human hepatocellular carcinoma. Cancer Res. 2004;64(15):5172–5177. doi: 10.1158/0008-5472.CAN-03-3314. [DOI] [PubMed] [Google Scholar]
- 6.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63(22):7646–7651. [PubMed] [Google Scholar]
- 7.Huang J, Zhang YL, Lin Y, Zheng DL, Han ZG. Down-regulation of SFRP1 as a tumor suppressor gene can contribute to human hepatocellular carcinoma. BMC Cancer. 2007;7:126–132. doi: 10.1186/1471-2407-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih YL, et al. Promoter methylation of the secreted frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma. Cancer. 2006;107(3):579–590. doi: 10.1002/cncr.22023. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31(4):339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163(4):1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emi M, et al. Allelic loss at chromosome band 8p21.3-p22 is associated with progression of hepatocellular carcinoma. Genes Chromosomes Cancer. 1993;7(3):152–157. doi: 10.1002/gcc.2870070307. [DOI] [PubMed] [Google Scholar]
- 12.Becker SA, Zhou YZ, Slagle BL. Frequent loss of chromosome 8p in hepatitis B virus-positive hepatocellular carcinomas from China. Cancer Res. 1996;56(21):5092–5097. [PubMed] [Google Scholar]
- 13.Pineau P, et al. Identification of three distinct regions of allelic deletions on the short arm of chromosome 8 in hepatocellular carcinoma. Oncogene. 1999;18(20):3127–3134. doi: 10.1038/sj.onc.1202648. [DOI] [PubMed] [Google Scholar]
- 14.Qin LX, et al. The association of chromosome 8p deletion and tumor metastasis in human hepatocellular carcinoma. Cancer Res. 1999;59(22):5662–5665. [PubMed] [Google Scholar]
- 15.Chan KL, Lee JM, Guan XY, Fan ST, Ng IO. High-density allelotyping of chromosome 8p in hepatocellular carcinoma and clinicopathologic correlation. Cancer. 2002;94(12):3179–3185. doi: 10.1002/cncr.10612. [DOI] [PubMed] [Google Scholar]
- 16.Yuan BZ, et al. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58(10):2196–2199. [PubMed] [Google Scholar]
- 17.Kim SW, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: Potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007;46(1):1–9. doi: 10.1002/gcc.20384. [DOI] [PubMed] [Google Scholar]
- 18.Wong CM, et al. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65(19):8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23(6):1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 20.Xue W, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22(11):1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pineau P, et al. Identification of three distinct regions of allelic deletions on the short arm of chromosome 8 in hepatocellular carcinoma. Oncogene. 1999;18(20):3127–3134. doi: 10.1038/sj.onc.1202648. [DOI] [PubMed] [Google Scholar]
- 22.Lu T, et al. Frequent loss of heterozygosity in two distinct regions, 8p23.1 and 8p22, in hepatocellular carcinoma. World J Gastroenterol. 2007;13(7):1090–1097. doi: 10.3748/wjg.v13.i7.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon TC, et al. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131(4):1262–1270. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Patil MA, et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26(12):2050–2057. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 25.Huang J, et al. Correlation between genomic DNA copy number alterations and transcriptional expression in hepatitis B virus-associated hepatocellular carcinoma. FEBS Lett. 2006;580(15):3571–3581. doi: 10.1016/j.febslet.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 26.Midorikawa Y, et al. Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene. 2006;25(40):5581–5590. doi: 10.1038/sj.onc.1209537. [DOI] [PubMed] [Google Scholar]
- 27.Adams J, Williams SV, Aveyard JS, Knowles MA. Loss of heterozygosity analysis and DNA copy number measurement on 8p in bladder cancer reveals two mechanisms of allelic loss. Cancer Res. 2005;65(1):66–75. [PubMed] [Google Scholar]
- 28.Morikawa A, et al. Allelic imbalances of chromosomes 8p and 18q and their roles in distant relapse of early stage, node-negative breast cancer. Breast Cancer Res. 2005;7(6):R1051–R1057. doi: 10.1186/bcr1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venter DJ, et al. Complex CGH alterations on chromosome arm 8p at candidate tumor suppressor gene loci in breast cancer cell lines. Cancer Genet Cytogenet. 2005;160(2):134–140. doi: 10.1016/j.cancergencyto.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Rubio-Moscardo F, et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. 2005;106(9):3214–3222. doi: 10.1182/blood-2005-05-2013. [DOI] [PubMed] [Google Scholar]
- 31.Chang BL. Integration of somatic deletion analysis of prostate cancers and germline linkage analysis of prostate cancer families reveals two small consensus regions for prostate cancer genes at 8p. Cancer Res. 2007;67(9):4098–4103. doi: 10.1158/0008-5472.CAN-06-4570. [DOI] [PubMed] [Google Scholar]
- 32.Lu W, et al. Allelotyping analysis at chromosome arm 8p of high-grade prostatic intraepithelial neoplasia and incidental, latent, and clinical prostate cancers. Genes Chromosomes Cancer. 2006;45(5):509–515. doi: 10.1002/gcc.20314. [DOI] [PubMed] [Google Scholar]
- 33.Ye H, et al. Genomic assessments of the frequent loss of heterozygosity region on 8p21.3-p22 in head and neck squamous cell carcinoma. Cancer Genet Cytogenet. 2007;176(2):100–106. doi: 10.1016/j.cancergencyto.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki H, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita K, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–495. doi: 10.1016/S1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 37.Pils D, et al. Five genes from chromosomal band 8p22 are significantly down-regulated in ovarian carcinoma: N33 and EFA6R have a potential impact on overall survival. Cancer. 2005;104(11):2417–2429. doi: 10.1002/cncr.21538. [DOI] [PubMed] [Google Scholar]
- 38.Cooper WN, et al. Epigenetic regulation of the ras effector/tumour suppressor RASSF2 in breast and lung cancer. Oncogene. 2008;27(12):1805–1811. doi: 10.1038/sj.onc.1210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endoh M, et al. RASSF2, a potential tumour suppressor, is silenced by CpG island hypermethylation in gastric cancer. Br J Cancer. 2005;93(12):1395–1399. doi: 10.1038/sj.bjc.6602854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong CM, et al. Tissue factor pathway inhibitor-2 as a frequently silenced tumor suppressor gene in hepatocellular carcinoma. Hepatology. 2007;45(5):1129–1138. doi: 10.1002/hep.21578. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka S, et al. Biologic significance of angiopoietin-2 expression in human hepatocellular carcinoma. J Clin Invest. 1999;103(3):341–345. doi: 10.1172/JCI4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimachi K, et al. Angiopoietin switching regulates angiogenesis and progression of human hepatocellular carcinoma. J Clin Pathol. 2003;56(11):854–860. doi: 10.1136/jcp.56.11.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsuhashi N, et al. Angiopoietins and Tie-2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology. 2003;37(5):1105–1113. doi: 10.1053/jhep.2003.50204. [DOI] [PubMed] [Google Scholar]
- 44.Guervós MA, et al. Deletions of N33, STK11 and TP53 are involved in the development of lymph node metastasis in larynx and pharynx carcinomas. Cell Oncol. 2007;29(4):327–334. doi: 10.1155/2007/635962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Oliver P, Davies KE, Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J Biol Chem. 2006;281(17):11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang LH, et al. Allelic imbalance regions on chromosomes 8p, 17p and 19p related to metastasis of hepatocellular carcinoma: comparison between matched primary and metastatic lesions in 22 patients by genome-wide microsatellite analysis. J Cancer Res Clin Oncol. 2003;129(5):279–286. doi: 10.1007/s00432-002-0407-5. [DOI] [PubMed] [Google Scholar]
- 47.Wistuba II, et al. Allelic losses at chromosome 8p21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59(8):1973–1979. [PubMed] [Google Scholar]
- 48.Swalwell JI, et al. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes Chromosomes Cancer. 2002;33(2):201–205. doi: 10.1002/gcc.10015. [DOI] [PubMed] [Google Scholar]
- 49.Coon SW, et al. Prognostic implications of loss of heterozygosity at 8p21 and 9p21 in head and neck squamous cell carcinoma. Int J Cancer. 2004;111(2):206–212. doi: 10.1002/ijc.20254. [DOI] [PubMed] [Google Scholar]
- 50.Lassus H, et al. Comparison of serous and mucinous ovarian carcinomas: distinct pattern of allelic loss at distal 8p and expression of transcription factor GATA-4. Lab Invest. 2002;81(4):517–526. doi: 10.1038/labinvest.3780260. [DOI] [PubMed] [Google Scholar]
- 51.Van Nimwegen MJ, van de Water B. Focal adhesion kinase: a potential target in cancer therapy. Biochem Pharmacol. 2006;73(5):597–609. doi: 10.1016/j.bcp.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Tilghman RW, Parsons JT. Focal adhesion kinase as a regulator of cell tension in the progression of cancer. Semin Cancer Biol. 2008;18(1):45–52. doi: 10.1016/j.semcancer.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692(2-3):77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Fingleton B. Matrix metalloproteinases: Roles in cancer and metastasis. Front Biosci. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]
- 55.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25(1):9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 56.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: Causes and implications. Cancer Lett. 2008;268(2):177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 57.Dahl E, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26(38):5680–5691. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 58.Shih YL, et al. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121(5):1028–1035. doi: 10.1002/ijc.22750. [DOI] [PubMed] [Google Scholar]
- 59.Song LJ, et al. Relationship between DLC-1 expressions and metastasis in hepatocellular carcinoma [In Chinese]. Zhonghua Gan Zang Bing Za Zhi. 2005;13(6):428–431. [PubMed] [Google Scholar]
- 60.Zhou X, et al. DLC1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and down-regulation of genes involved in metastasis. Int J Oncol. 2008;32(6):1285–1291. doi: 10.3892/ijo_32_6_1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66(17):8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- 62.Ilic D, et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377(6549):539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 63.Dai HY, et al. Carbonic anhydrase III promotes transformation and invasion capability in hepatoma cells through FAK signaling pathway. Mol Carcinog. 2008;47(12):956–963. doi: 10.1002/mc.20448. [DOI] [PubMed] [Google Scholar]
- 64.Fujii T, et al. Focal adhesion kinase is overexpressed in hepatocellular carcinoma and can be served as an independent prognostic factor. J Hepatol. 2004;41(1):104–111. doi: 10.1016/j.jhep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 65.Itoh S, et al. Role of expression of focal adhesion kinase in progression of hepatocellular carcinoma. Clin Cancer Res. 2004;10(8):2812–2817. doi: 10.1158/1078-0432.CCR-1046-03. [DOI] [PubMed] [Google Scholar]
- 66.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6(1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 67.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18(5):516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 68.McLean GW, et al. The role of focal-adhesion kinase in cancer — a new therapeutic opportunity. Nat Rev Cancer. 2005;5(7):505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28(1-2):35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 70.Cance WG, Golubovskaya VM. Focal adhesion kinase versus p53: apoptosis or survival? Sci Signal. 2008;1(20):pe22. doi: 10.1126/stke.120pe22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim ST, Mikolon D, Stupack DG, Schlaepfer DD. FERM control of FAK function: implications for cancer therapy. Cell Cycle. 2008;7(15):2306–2314. doi: 10.4161/cc.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoehr R, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84(4):465–478. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 73.Huang J, et al. Up-regulation of DLK1 gene could contribute to human hepatocellular carcinoma. Carcinogenesis. 2007;28(5):1094–1103. doi: 10.1093/carcin/bgl215. [DOI] [PubMed] [Google Scholar]
- 74.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 75.Zuo T, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129(7):1275–1286. doi: 10.1016/j.cell.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB.Methylation specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 199693189821–9826. . 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.