Abstract
The E3 ubiquitin ligase human murine double minute (HDM2) is overexpressed in 40%–80% of late-stage metastatic cancers in the absence of gene amplification. Hdm2 regulates p53 stability via ubiquitination and has also been implicated in altering the sensitivity of cells to TGF-β1. Whether TGF-β1 signaling induces Hdm2 expression leading to HDM2-mediated destabilization of p53 has not been investigated. In this study, we report that TGF-β1–activated SMA- and MAD3 (Smad3/4) transcription factors specifically bound to the second promoter region of HDM2, leading to increased HDM2 protein expression and destabilization of p53 in human cancer cell lines. Additionally, TGF-β1 expression led to Smad3 activation and murine double minute 2 (Mdm2) expression in murine mammary epithelial cells during epithelial-to-mesenchymal transition (EMT). Furthermore, histological analyses of human breast cancer samples demonstrated that approximately 65% of late-stage carcinomas were positive for activated Smad3 and HDM2, indicating a strong correlation between TGF-β1–mediated induction of HDM2 and late-stage tumor progression. Identification of Hdm2 as a downstream target of TGF-β1 represents a critical prosurvival mechanism in cancer progression and provides another point for therapeutic intervention in late-stage cancer.
Introduction
TGF-β1 is a pleiotropic factor that plays a physiological role in regulating proliferation, differentiation, development, wound healing, and angiogenesis. In early neoplasia, TGF-β1 can be a potent suppressor of proliferation (1). Conversely, TGF-β1 can promote the migration and proliferation of tumor cells in late-stage metastatic cancer (2, 3). In late-stage malignancies, key components of the TGF-β1 signaling cascade have been reported to be inactivated by gene mutations, which are predominantly found in a fraction of gastrointestinal and pancreatic cancers and a small proportion of other cancers. Additional epigenetic events maintain functional TGF-β1 receptor(s), and TGF-β1’s downstream signaling pathways provide a selective advantage to tumors by disrupting cytostatic and apoptotic programs. The nature of such a pathway that circumvents TGF-β1–mediated growth arrest while simultaneously stimulating phenotypic changes that alter the tumor microenvironment and tumor migration remains to be determined.
TGF-β1 signals by first binding to TGF-β1 type II receptor (TβRII), which then recruits the TGF-β1 type I receptor (TβRI or Alk5) to form a heterodimer serine/threonine kinase complex. This activated heterodimeric complex transphosphorylates TβRI and enables TβRI to directly phosphorylate 2 carboxyterminal serines on Smad2 and Smad3, leading to activation of these transcription factors (4). Smad2 or Smad3 dimerizes with Smad4 and translocates to the nucleus, where the activated complexes associate with Smad-binding elements (SBEs) in promoters of numerous genes (5–7). Some of the transcriptional targets are genes whose products regulate cellular processes that impede proliferation (e.g., p21) and, under certain conditions, cell death (8). Gene targets that are specifically regulated by Smad2 or Smad3 are still being defined. Additionally, Smad-targeted gene expression varies depending on the duration and/or dose of TGF-β1. During the development of lower organisms, alterations in smad gene family members lead to phenotypic changes in developmental programs (9). Smad2 and Smad3 have nonredundant roles. This is best illustrated by the embryonic lethality of smad2-knockout embryos versus altered proliferation rates of T cells and epithelial cells in smad3-knockout mice (10, 11). The alteration of T cells in smad3-knockout mice limits their adult life span, as they succumb to the development of abscesses related to defective mucosal immunity (10). Furthermore, these mice are runted, largely infertile, and exhibit delayed mammary gland development (11). These findings suggest that Smad3 is important for proliferation. Furthermore, smad3 is not mutated or deleted in human solid tumors (3).
While Smad3 is not related to apoptosis, Smad2 may play such a role. There appears to be a requirement for a stoichiometric balance of p53 and Smad2 expression in the regulation of mesoderm development (9). An increase in p53 levels appears to augment the induction of gene targets common to both p53 and Smad2. Additional observations increase the complexity of Smad2/p53 interplay, as knockout of murine double minute 2 (Mdm2) gene, an antagonist of p53, is embryonic lethal at E6.5 (12, 13). This time point of development coincides with the approximate window for mesoderm formation. These data suggest a relationship among Smad2, p53, and Mdm2 in the context of early development.
Mdm2 (human homolog Hdm2) is an E3 ubiquitin ligase that conjugates ubiquitin to p53. This ubiquitin conjugation alters the cellular localization of p53 and tags it for degradation by the proteasome (14). Hdm2 may also alter the function and stability of the retinoblastoma tumor suppressor (Rb) and the cell-cycle regulator p21 (15, 16). In the TGF-β1 pathway, overexpression of Hdm2 alters the activity of Smad2 and Smad3 through an indirect mechanism (17). Moreover, a forward genetic-screening approach was used to identify genes involved in TGF-β1 resistance. Several genes were identified, but the most potent gene to confer resistance was mdm2, which appeared in this study to influence the Rb tumor suppressor (18). In human cells, elevated Hdm2 expression correlates with loss of TGF-β1 sensitivity in several breast cancer cell lines (18). Further work demonstrated that the E3 ubiquitin ligase domain of Hdm2 was required to confer loss of the TGF-β1 cytostatic response (19). The HDM2 gene is a transcriptional target of p53. However, in late-stage metastatic disease, Hdm2 is elevated, typically independent of p53 status. Thus, it seems that other mechanisms are involved in elevating Hdm2 protein. One such observation comes from a subset of patients harboring a polymorphism in the promoter of HDM2 (SNP309). This SNP increases the recruitment of specific transcription factors to the HDM2 promoter, thus augmenting Hdm2 expression (20).
Considering that late-stage metastatic cancer is refractory to TGF-β1 and Hdm2 levels are elevated by some unknown mechanism, we examined the fundamental mechanism involved in regulating Hdm2 levels in response to TGF-β1. The HDM2 gene has 2 promoters, and we identified the second promoter of HDM2 as having an SBE, which is bound by the Smad3/Smad4 complex in response to TGF-β1. Since p53 is known to induce Hdm2 expression, we found that the binding of Smad3/4 complex led to the induction of Hdm2, which was independent of p53 activity. The induction of Hdm2 in response to TGF-β1 stimulation resulted in destabilization of p53. Additionally, the alteration in p53 levels was permissive for epithelial-to-mesenchymal transition (EMT) in NMuMG cells, a normal murine mammary epithelial cell line. Importantly, Nutlin3, an antagonist that binds to Hdm2 and leads to the stabilization of p53 by preventing the formation of the Hdm2/p53 complex, reengaged p53 activity in response to TGF-β1 and induced cell death. Finally, examination of active Smad3 in human breast cancers that had migrated to distal and regional sites showed a strong correlation with elevated Hdm2 levels. This represents what we believe is a novel mechanism in human breast cancer that shows that TGF-β1 can induce HDM2, leading to EMT and possible migration of tumor cells.
Results
TGF-β1 regulates p53 stability.
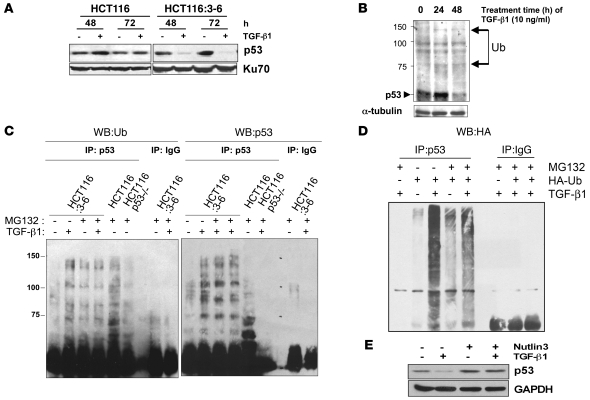
To examine the ability of TGF-β1 to regulate p53 stability, we utilized HCT116 colorectal cells. HCT116 cells are deficient in DNA mismatch repair and TβRII (21). HCT116:3-6 cells were produced by reintroduction of chromosome 3 into cells by microcell fusion, which restored TβRII protein expression and activity (22). HCT116 and HCT116:3-6 cells were used as a model system to analyze the effect of the TGF-β1 pathway on the regulation of p53. Treatment of HCT116 cells with TGF-β1 for 48 and 72 hours did not alter p53 levels (Figure 1A). Conversely, decreased levels of p53 were observed at 24 and 48 hours in HCT116:3-6 cells treated with TGF-β1 (Figure 1A). To determine whether this loss was due to Hdm2-mediated destabilization of p53, HCT116:3-6 cells were treated with TGF-β1 for 24 and 48 hours and a Western blot was prepared to examine higher molecular weight forms of p53 that would be consistent with ubiquitination. The data in Figure 1B show formation of higher molecular weight bands of p53 in HCT116:3-6 cells after TGF-β1 treatment. To demonstrate that ubiquitin was conjugated to p53, HCT116:3-6 or HCT116 p53–/– cells were pretreated with the proteasome inhibitor MG132, followed by TGF-β1 exposure. We observed that immunopurified p53 from HCT116:3-6 cells was conjugated with ubiquitin in response to TGF-β1 treatment by probing for ubiquitin (Figure 1C). In addition, we also transiently transfected HA-tagged ubiquitin (HA-Ub) into HCT116:3-6 cells and treated them with TGF-β1 and in the absence or presence of MG132 (Figure 1D). These data further confirm that p53 was conjugated with ubiquitin in response to TGF-β1 exposure.
Figure 1. p53 destabilization by TGF-β1 stimulation.
(A) HCT116 and HCT116:3-6 cells were treated with vehicle or 10 ng/ml of TGF-β1 for 48 and 72 hours. Cellular extracts were prepared for Western blot, and p53 and Ku70 (internal control) levels were detected. (B) Western blot of p53 laddering, an indication of ubiquitination (Ub) (arrows) in cellular extracts of HCT116:3-6 cells treated with 10 ng/ml of TGF-β1 for 0, 24, and 48 hours. (C) HCT116, Hct116 p53–/–, and HCT116:3-6 cells were treated with 10 ng/ml of TGF-β1 with or without the proteasome inhibitor MG132 (30 μM). Cellular extracts were immunoprecipitated with control IgG or p53 antibodies. Precipitates were separated on an SDS-PAGE gel, and a Western blot was prepared. The left side was probed for ubiquitin. The right side was probed for p53. (D) HCT116:3-6 cells were transfected with HA-Ub. Cells were then treated with MG132 (30 μM) with or without TGF-β1 (10 ng/ml); cellular extracts were immunoprecipitated with p53 or IgG antibodies and prepared for Western blot analysis of HA. (E) Western blot of p53 in extracts isolated from HCT116:3-6 cells treated with TGF-β1 (10 ng/ml) with or without Nutlin3 (10 μM).
Ubiquitination of p53 requires an E3 ubiquitin ligase. There are several E3 ubiquitin ligases that may conjugate ubiquitin to p53, including Cop1, Mule, Pirh2, and Mdm2 (human and murine) (23–27). Through overexpression and knockdown studies, Mdm2 (or human homolog Hdm2) is the best-known E3 ubiquitin ligase for p53. The regulation of p53 stability requires the induction of HDM2 gene expression by p53 in response to genotoxic stress (28). Hdm2 associates with p53 via van der Waal forces within the first 52 amino acids of p53; this site serves as a docking site for subsequent conjugation of ubiquitin to the carboxyterminal lysines of p53 (29). To determine whether Hdm2 played a role in the destabilization of p53 in response to TGF-β1, we used Nutlin3, a small molecule inhibitor that sterically inhibits the association of p53 to Hdm2 by binding the hydrophobic pocket in the amino terminus of Hdm2 (30). The data in Figure 1E demonstrate that 10 μM Nutlin3 protected p53 from degradation mediated by the TGF-β1 signaling pathway. Thus, the observed stabilization of p53 by Nutlin3 implicates Hdm2 in the destabilization of p53 in response to TGF-β1.
Induction of HDM2 gene expression by TGF-β1.
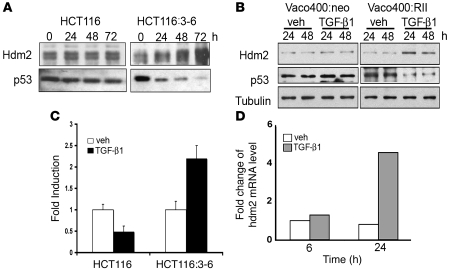
It has been reported that an increase in Hdm2 levels causes the formation of Hdm2 homodimers, which in turn function to destabilize p53 (31). Therefore, it was necessary to further characterize levels of Hdm2 protein in response to TGF-β1. HCT116:3-6 and HCT116 cells were treated with TGF-β1, and whole-cell extracts were isolated between 0 and 72 hours. Treatment of HCT116 cells with TGF-β1 did not alter the levels of Hdm2 over 72 hours, while HCT116:3-6 cells showed a dramatic induction in Hdm2 levels with a concomitant decrease in p53 over the same time course (Figure 2A).
Figure 2. TGF-β1 increases Hdm2 mRNA and protein levels.
(A) Western blot analysis of Hdm2 and p53 in HCT116 and HCT116:3-6 treated with TGF-β1 (10 ng/ml) for 0, 24, 48, and 72 hours. (B) Vaco400 cells with control plasmid (Vaco400:neo) or expressing the functional TGF-β1 receptor II (Vaco400:RII) cells were treated with vehicle (veh) or TGF-β1 (10 ng/ml) for 24 or 48 hours. Western blot was prepared from the extracts, and Hdm2, p53, and tubulin were detected. (C) HCT116 and HCT116:3-6 were transfected with the HDM2 promoter-luciferase reporter construct and renilla expression plasmid; then cells were treated with TGF-β1 (10 ng/ml) or vehicle. After 48 hours, extracts were prepared for analysis. Fold induction represents vehicle to TGF-β1 treatments and error bars represent SD generated from the mean. (D) Real-time PCR was performed on HCT116:3-6 cells treated with vehicle or TGF-β1 (10 ng/ml) at 6 or 24 hours.
We next examined another colorectal carcinoma cell line, Vaco400, which is also deficient in mismatch repair and lacks a functional TβRII. Control vector (Vaco400:neo) or Vaco400 cells that stably express TβRII (Vaco400:RII) were used to determine whether the observations in HCT116:3-6 were exclusively dependent on TβRII. Both Vaco400:neo and Vaco400:RII cells were treated with TGF-β1 for 24 or 48 hours. The data in Figure 2B are similar to the results from the isogenic HCT116 model system, whereby stimulation with TGF-β1 caused an increase in Hdm2 levels and a subsequent decrease in p53 only in cells that possess the TβRII. Both HCT116 and Vaco400 cell systems provided reproducible data of increased Hdm2 protein levels and the subsequent loss of p53.
These experiments led us to examine the mechanism by which Hdm2 was increased. In order to determine whether it was a transcriptional event, a HDM2 reporter assay was used to determine whether the P2 promoter of HDM2 was activated in response to TGF-β1 stimulation. The HDM2 reporter was constructed by linking the HDM2-P2 promoter upstream of a luciferase reporter gene. In addition to reporter assays, we examined changes in HDM2 mRNA levels by real-time PCR to determine whether the endogenous gene was induced in response to TGF-β1 treatment. The HDM2 promoter was reproducibly induced in response to TGF-β1 treatment in TβRII-reconstituted HCT116:3-6 cells, as determined by reporter assay (Figure 2C) and by real-time PCR (Figure 2D). These experiments demonstrate that TGF-β1 increased Hdm2 levels through a transcription-dependent event.
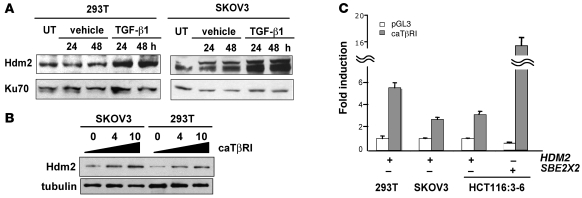
Induction of the Mdm2 gene (human and murine) through its P2 promoter is largely regulated by p53 in response to genotoxic stress. Therefore, it became important to characterize the involvement of p53 in the induction of the HDM2 gene in response to TGF-β1. Two cell lines, 293T cells transformed with large T antigen that predominantly neutralizes p53 and SKOV3 ovarian cancer cells that lack p53 expression, were used to assess whether induction of Hdm2 was independent of functional p53 when cells were subjected to TGF-β1 treatment. Treatment of either 293T or SKOV3 cells with TGF-β1 resulted in increased levels of Hdm2 (Figure 3A). Additionally, titration of a constitutively activated TβRI (caTβRI) plasmid in 293T and SKOV3 cells caused an increase in Hdm2 levels (Figure 3B). Furthermore, utilizing a luciferase reporter assay with the second promoter of HDM2 or the TGF-β1 responsive SBE2X2 (a synthetic promoter consisting of 4 GTCTs with AGAC linkers) as a positive control, we demonstrated that caTβRI mediates induction of HDM2 in 293T, SKOV3, and HCT116:3-6 cells (Figure 3C). Thus, activation of the TGF-β1 pathway by ligand or a constitutively active receptor resulted in induction of the HDM2 gene promoter and increased Hdm2 protein expression independent of p53 activity.
Figure 3. Increased Hdm2 in response to TGF-β1 is independent of p53.
(A) 293T and SKOV3 cells were untreated (UT) or treated with vehicle or TGF-β1 (10 ng/ml) for 24 and 48 hours. Cellular extracts were prepared for Western blot analysis of Hdm2 and Ku70. (B) SKOV3 and 293T cells were transiently transfected with 0, 4, or 10 μg of constitutively active TβRI (caTβRI) expression plasmid. Cellular extracts were prepared for Western blot analysis of Hdm2 and α-tubulin. (C) Transient transfection of 293T, SKOV3, and HCT116:3-6 cells with the caTβRI and HDM2 reporter or the SBE2X2 reporter construct. All samples were transfected with a renilla construct. Reporter activity was determined relative to renilla to generate relative activity. Fold induction was determined relative to vehicle control and SD was calculated relative to the mean.
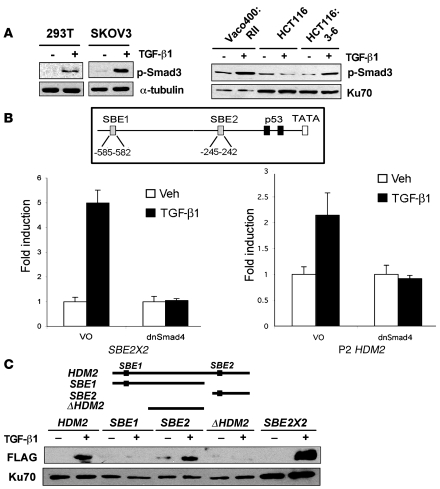
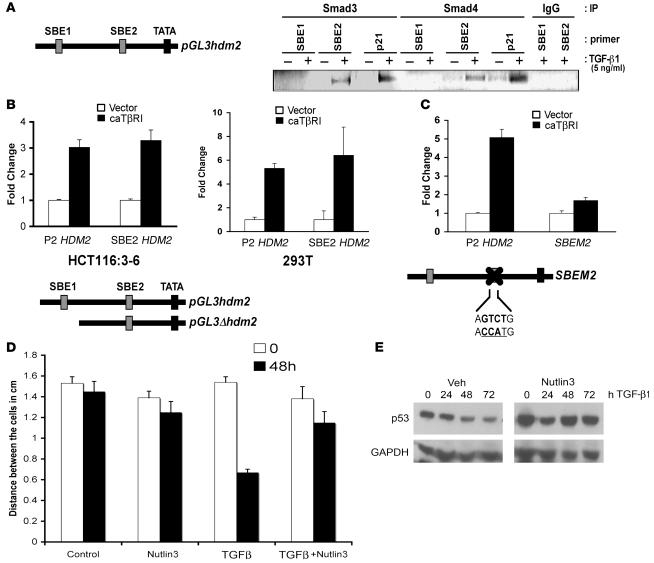
Induction of HDM2 gene expression has been reported in response to stimuli other than genotoxic stress. Transcription factors such as N-Myc, Ets family members, Sp1, and ERα can bind and induce HDM2 gene expression through the second promoter (P2) (32–34). After an in silico survey of the P2 promoter of HDM2 for possible transcription factor–binding elements that are activated in response to TGF-β1, 2 possible binding elements in the promoter conformed to the GTCT Smad3 DNA–binding element (SBE). One binding element (SBE2) was located at nucleotide –245 in the P2 promoter region of HDM2 and a second site (SBE1) was located at nucleotide –585 in the P2 promoter region of HDM2. To explore the possibility of Smad3 inducing HDM2 gene expression, we determined whether Smad2 and Smad3 were active after TGF-β1 exposure at times when Hdm2 levels were elevated. Dimerization of TβRI/RII in response to TGF-β1 formed an active serine/threonine kinase that phosphorylates and activates Smad2 and Smad3. We observed Smad3 activation using a phospho-antibody to Smad3 to probe a Western blot of cellular extracts from 293T, SKOV3, Vaco400:RII, and HCT116:3-6 cells treated with TGF-β1 for 48 hours. As expected, Smad3 activation was not observed in parental HCT116 cells treated with TGF-β1 (Figure 4A). Importantly, Smad3 activation occurred and corresponded with induction of the HDM2 gene. To further explore the involvement of Smads in the activation of HDM2 gene induction, we transiently transfected a dominant negative Smad4 (dnSmad4) in transient reporter assays. Smad4 is a binding partner of Smad3, and a dominant negative form would presumably impede the activation of the HDM2 promoter. Using the HDM2 promoter and SBE2X2 as a control, cells transiently transfected with the respective promoters were treated with or without TGF-β1. As predicted, dnSmad4 blocked induction of the synthetic Smad reporter SBE2X2 and also prevented the activation of HDM2 promoter (Figure 4B). Thus, these data support a role for the Smads in the induction of Hdm2 in response to TGF-β1 treatment.
Figure 4. Smad activation and induction of the HDM2 promoter.
(A) Western blot of p-Smad3 in 293T, SKOV3, Vaco400:RII, HCT116, and HCT116:3-6 cells. Tubulin and Ku70 were used as internal controls for loading. (B) A schematic of 2 putative SBEs designated SBE1 and SBE2 and the p53-binding elements in the P2 promoter of HDM2. Reporter assays were conducted using dominant negative Smad4 (dnSmad4) or control vector only (VO) in transient transfection assays. HCT116:3-6 cells were transfected with an SBE2X2 reporter as a positive control or HDM2 P2 reporter. All samples were transfected with renilla to use as an internal control. Cells were treated with vehicle or TGF-β1 (10 ng/ml). Reporter activity was determined relative to renilla to generate relative activity. Fold induction was determined relative to vehicle control and SD was calculated relative to the mean. (C) Western blot of FLAG-Smad3 and Ku70 that bound to the HDM2 promoter. 293T cells were transfected with FLAG-Smad3 and treated with vehicle or TGF-β1 (10 ng/ml). Nuclear extracts were prepared from the latter mentioned cell treatments. Biotinylated PCR fragments of the HDM2 promoter, SBE1, SBE2, ΔHDM2, or a positive control, SBE2X2, were mixed with the nuclear extracts. Biotinylated DNA fragments bound to streptavidin beads were used to purify FLAG-Smad3 and Ku70 (internal control).
We next sought to identify the transcription factors responsible for, and the DNA binding elements involved, in HDM2 gene induction by Smads in response to TGF-β1 stimulation. DNA pulldown analyses using full-length and selected truncations of the HDM2 promoter were amplified by PCR with 5′ biotinylated oligos. Biotin-labeled DNA fragments were gel purified, then bound to streptavidin beads, and nuclear extracts from 293T cells transfected with Flag-Smad3 treated with TGF-β1 were mixed. The full-length promoter region of HDM2 purified Flag-Smad3 (Figure 4C). Truncated promoter regions lacking the SBE2 or the Δ element, lacking both SBE sites, did not purify Flag-Smad3. The promoter fragment that contained the SBE2 element purified Flag-Smad3. The SBE2X2 element was used as a positive control. As an internal control, we probed our blots for Ku70. Ku70 is a DNA repair protein that binds to free ends of double-stranded DNA and can be used to normalize nuclear complexes (35). Thus, Smad3 bound to the SBE2 element in the HDM2 promoter region.
Identification of the ability of Smad3 to bind the HDM2 promoter (SBE2) by DNA pulldown assays led to the determination of whether the Smad3/4 complex could bind the genomic SBE elements in the HDM2 promoter. Smad3 and Smad4 ChIP assays were used on HCT116:3-6 cells treated with TGF-β1 to test for the binding to the SBE1 and SBE2 elements in the HDM2 genomic promoter. As a positive control, we performed ChIP on the p21 promoter, a known target of Smads (8) (Figure 5A). The data in Figure 5A show Smad3 and Smad4 bound to the p21 promoter in response to TGF-β1 exposure. Analysis of the HDM2 P2 promoter shows that Smad3 and Smad4 occupied the promoter region that contained the SBE2 element but not the SBE1 element. These data are consistent with the DNA pulldown experiments (Figure 4C). To definitively demonstrate the requirement of the SBE2 element as a downstream target of the TGF-β1 signaling pathway, we used a deletion construct or mutated the SBE2-binding element (SBEM2) in the P2 promoter of HDM2 in transient transfection assays with caTβRI. caTβRI was able to induce the P2 promoter independent of the SBE1 element (Figure 5B). In contrast, using the mutated SBEM2 promoter showed a diminished induction of promoter activity in the presence of caTβRI, similar to the vector control. Thus, the HDM2 gene is regulated by TGF-β1–mediated activation of Smad3/4 activation, leading to elevated Hdm2 protein levels.
Figure 5. Binding of Smad3 and Smad4 to the HDM2 SBE2 in vivo.
(A) HCT116:3-6 cells were treated with vehicle or TGF-β1 (10 ng/ml) for 48 hours. Smad3, Smad4, or IgG isotype control antibodies were used to immunoprecipitate Smads binding to the HDM2 promoter. Oligo sets were used to PCR amplify the SBE1, SBE2, or the p21 promoter SBE (a positive control for Smad3/4 binding) and resolved on an agarose gel. (B) HCT116:3-6 and 293T cells were transfected with caTβRI and either the full-length P2 HDM2-promoter driving luciferase or a deletion construct missing SBE1. Reporter activity was determined relative to renilla to generate relative activity. Fold induction was determined relative to vehicle control, and SD was calculated relative to the mean. (C) Cells were transfected with caTβRI and the P2 HDM2 promoter driving luciferase or a mutant whereby the SBE2 site was mutated. A renilla expression construct was used as an internal control for normalization. SEM was calculated as in B. (D) Wound healing assay using HCT116:3-6 cells, scored in a double-blind assay. Bar graph represents the distance between confluent cells after stimulation with TGF-β1 (10 ng/ml), Nutlin3 (10 μM), or TGF-β1 (10 ng/ml) and Nutlin3 (10 μM). (E) Western blot analysis of p53 in HCT116:3-6 cells treated with TGF-β1 (10 ng/ml) with or without Nutlin3 (10 μM) over a 72-hour time course. SD was calculated relative to the mean.
Biological effects of TGF-β1/HDM2/p53.
Clinically, the last event for permissive migration/metastatic nature of a tumor is loss of p53 function. Since induction of Hdm2 by TGF-β1 can decrease p53 levels, we determined whether preventing binding of Hdm2/p53 in cells using Nutlin3 would have a physiological effect. HCT116:3-6 cells were grown to confluency, and then some cells were removed by scraping to produce a “wound.” Wound-healing assays were scored in a double-blind method analyzing several areas of the wound. Treatment of cells with TGF-β1 stimulated the cells to migrate and close the wound, while coincubation with Nutlin3 reversed TGF-β1–mediated migration. Mock or Nutlin3 treatment alone did not dramatically affect cell migration (Figure 5D and Supplemental Figure 1C; supplemental material available online with this article; doi: 10.1172/JCI39194DS1). Extracts from HCT116:3-6 cells treated with TGF-β1 alone or in conjunction with Nutlin3 were examined by Western blot to assess levels of p53. As expected, Nutlin3 prevented Hdm2-mediated p53 destabilization (Figure 5E) and prevented cell migration mediated by TGF-β1. These data indicate that Nutlin3 can reverse the effects of TGF-β1–mediated downregulation of p53 by Hdm2.
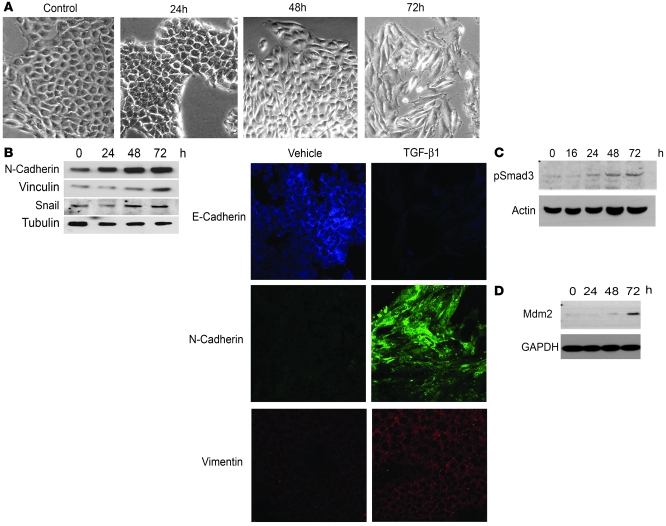
In response to TGF-β1, cancer cells can change biochemically and transition toward a metastatic phenotype (36). Some epithelial cells may adopt a mesenchymal phenotype, which is termed EMT. Murine mammary epithelial cell line, NMuMg, can undergo EMT in response to TGF-β1 in a cell-culture system (37, 38). We used NMuMg cells to determine whether the TGF-β1/p53/Mdm2 axis was engaged to regulate EMT. As NMuMg cells undergo EMT, there is a morphological change to the cell that causes it to have a “fibroblast-like appearance.” Figure 6A indicates that after 48 hours of treatment with TGF-β1, the cells begin to transition and acquire a mesenchymal phenotype; by 72 hours, the vast majority of these cells resemble a mesenchymal morphology. To confirm molecularly that these cells had undergone EMT, levels of N-cadherin, Snail, and Vinculin, all markers of mesenchyme, were elevated over the same 72-hour simulation with TGF-β1 (Figure 6B). In addition, confocal microscopy showed that E-cadherin was decreased upon exposure to TGF-β1, and Vimentin and N-cadherin were elevated following 24 and 48 hours of TGF-β1 treatment. As predicted, Smad3 was activated in response to TGF-β1 at 24 hours and increased over the 72-hour period (Figure 6C). Mdm2 levels began to increase at 48 hours and dramatically increased after 72 hours of TGF-β1 treatment (Figure 6D). Increased levels of Mdm2 in response to TGF-β1 corresponded with Smad3 activation and with the mesenchymal phenotype in NMuMg cells (Figure 6A).
Figure 6. Characterization of TGF-β1 treatment on cell migration and EMT.
(A) NMuMg mammary cells treated with 4 ng/ml of TGF-β1. Photographs were taken at 0, 24, 48, and 72 hours. (B) N-cadherin, Vinculin, Snail, and α-tubulin (loading control) were detected by Western blot of whole-cell lysates from NMuMg cells treated with TGF-β1 (4 ng/ml) over a 72-hour time course. Confocal microscopy of E-cadherin, N-cadherin, and Vimentin in NMuMG cells treated with 4 ng/ml. Original magnification, ×20. (C) Western blot of a time-course activation of p-Smad3 in NMuMg in response to TGF-β1 (4 ng/ml). Loading was normalized to actin. (D) Time-course analysis of Mdm2 induction as measured by Western blot after treatment of NMuMg cells with TGF-β1 (4 ng/ml).
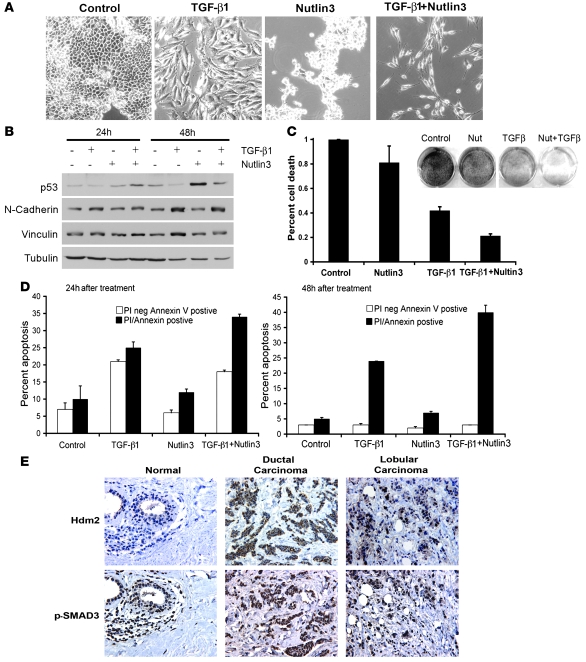
Since Mdm2 was induced upon TGF-β1 treatment and its elevated levels correlated with EMT, we examined one possible mechanism whereby Mdm2 might mediate destabilization of p53, which would be permissive of EMT. If so, then the utilization of Nutlin3 might protect p53 from Mdm2 destabilization. This would reengage p53 activity and possibly affect EMT progression. To test this, NMuMg cells were incubated with vehicle, TGF-β1, Nutlin3, or Nutlin3 and TGF-β1 cells for 48 hours. Treatment with vehicle or Nutlin3 alone did not grossly affect cells after 2 days (Figure 7A). This is consistent with previous reports, as Nutlin3 exposure to these normal mammary epithelial cells does not promote apoptosis and adherent cells are still present (39). While incubation with TGF-β1 alone induced the formation of mesenchymal cells, the combination of Nutlin3 and TGF-β1 resulted in significantly fewer mesenchymal cells. The remaining cells resembled mesenchymal cells, yet did not appear healthy. We confirmed the presence of mesenchymal cells by detecting Vinculin and N-cadherin by Western blot of cells treated with vehicle, TGF-β1, Nutlin3, or TGF-β1 and Nutlin3 together after 24 and 48 hours (Figure 7B). Western blot analysis of p53 levels showed that, as predicted, Nutlin3 rescued TGF-β1–mediated destabilization of p53 (Figure 7B).
Figure 7. Induction of apoptosis in response to TGF-β1 and Nutlin3 exposure.
(A) Morphological changes to NMuMg cells after incubation with 4 ng/ml TGF-β1, Nutlin3 (10 μM), or TGF-β1 and Nutlin3. Original magnification, ×20. (B) Western blot of p53, N-cadherin, Vinculin, and Tubulin under the same treatment conditions as A. (C) Survival assay of NMuMg cells. Cell were treated as in A and stained with methylene blue at day 5. Methylene blue was liberated from the cells and quantified by OD595. Percentage of cell death was calculated from control; data represent 3 independent experiments completed in triplicate. SD was calculated relative to the mean. (D) Flow cytometry of NMuMg cells treated as in A and stained for annexin V and PI at 24 and 48 hours. Data represent triplicates of experiments. White bars represent PI negative and annexin V positive, and black represents PI and annexin V positive. SD was calculated relative to the mean. (E) Immunohistochemistry of Hdm2 and p-Smad3 (serine 425) in normal breast tissue, ductal carcinoma, and lobular carcinoma. Original magnification, ×16.
The observation that fewer cells were present with treatment of TGF-β1 and Nutlin3 suggested that cells were either cytostatic or undergoing cell death. It did not appear that cells were cytostatic, as the cells treated with Nutlin3 or TGF-β1 alone did not demonstrate the same outcome as the combination of both TGF-β1 and Nutlin3. To determine the long-term survival effects of Nutlin3 and TGF-β1 cotreatment on NMuMg cells, a colony-forming assay was used. As shown in Figure 7C, staining of cells cotreated with TGF-β1 and Nutlin3 resulted in a 79% decrease in colonies relative to control. In contrast, Nutlin3 or TGF-β1 treatment alone caused a 21% and 58% decrease in colonies, respectively. Control and Nutlin3-treated cells were not statistically different, while treatment with TGF-β1 alone, or in conjunction with Nutlin3, was statistically different as analyzed from 3 independent experiments performed in triplicate (Figure 7C). The data generated in these assays suggest that TGF-β1 alone, and in conjunction with Nutlin3, has an affect on the viability and/or proliferation of cells.
The limitation of colony forming assays is that one cannot determine whether the cells are cytostatic or undergoing apoptosis in the presence of TGF-β1 for 7 days. To address this question, we performed a short-term study by utilizing flow cytometry analysis of annexin V and propidium iodide (PI) staining to examine the mechanism of cell death. This assay allowed for the segregation of apoptosis versus other cell death mechanisms at the early stages of programmed cell death. NMuMg cells were treated with vehicle, TGF-β1, Nutlin3, or TGF-β1 and Nutlin3 for 24 and 48 hours. In Figure 7D, early (PI negative/annexin V positive) and late apoptotic/necrotic (PI positive/annexin V positive) cells were detected at 24 hours. Similar levels of apoptotic cells were observed in control and Nutlin3-exposed cells. At 48 hours, while TGF-β1 alone induced low levels of apoptotic cells, the combination of TGF-β1 and Nutlin3 resulted in a much more dramatic induction of apoptosis. These data show that Nutlin3 alone had a minimal effect on cell survival, which is consistent with other reports using various cell lines (30, 39, 40). Moreover, although TGF-β1 treatment caused cytostatic responses as well as cell death, the combination of TGF-β1 and Nutlin3 was more effective at inducing cell death than either agent alone.
Further experiments were conducted to determine whether knockdown of mdm2 in NMuMG cells via siRNA would confer results similar to those of Nutlin3. Earlier studies have shown that knockdown of mdm2 in proliferating cells is lethal (41–43). Furthermore, Mdm2–/– mice are embryonic lethal (12, 13). As predicted, knockdown of mdm2 significantly decreased proliferation and quickly caused cell death in our NMuMG cell system (Supplemental Figure 1, A and B). This prevented us from approaching the question of Mdm2 dependence in regard to TGF-β1–induced EMT through a genetic approach. The use of an Mdm2 inhibitor induced much less stress, as shown by the low percentage of cell death (Figure 7C) and minimal apoptosis in response to Nutlin3 (Figure 7D).
Clinical correlation of Smad3 activation and HDM2 levels.
Since we mechanistically showed that TGF-β1 activation of Smad3 induced HDM2 gene expression, we next determined whether there was a clinical correlation between Smad3 activation and detection of Hdm2 in human breast cancer patient samples. We examined a total of 45 patient samples: (a) normal tissue from breast reductions; (b) benign tissues; and (c) ductal and lobular carcinomas. We examined the activated Smad3 and Hdm2 levels in tumor tissue and normal tissue (Figure 7E). We found that 65% of the samples were positive for both Smad3 activation and elevated levels of Hdm2 (P = 0.0014) (Table 1). Furthermore, 73% of ductal or lobular carcinomas that presented as extensions to either regional lymph nodes or metastasis to other organ systems costained for Hdm2 and activated Smad3. We did not detect staining of Hdm2 or activated Smad3 in the surrounding stroma or normal epithelia cells, showing the specificity of the detection of these proteins in breast carcinomas. In normal breast tissue, we observed modest staining for Hdm2 in the ducts. This staining may reflect some ductal hyperplasia, which is often observed. In addition, since Hdm2 may be regulated by the estrogen receptor, we stratified the ductal carcinoma samples into estrogen and progesterone receptor positive (ER+/PR+) or negative (ER–/PR–) expression (when the annotation was available). There was no significant difference in ER/PR status and Hdm2 levels (P = 0.26) (Table 1). Thus, selective costaining of activated Smad3 and elevated Hdm2 levels in infiltrative and metastatic breast cancer patient samples strongly supports a role for an activated TGF-β1/Smad3/Hdm2 pathway in metastatic breast cancer.
Table 1 .
Clinical samples stained for Hdm2 and p-Smad3
Discussion
The present work reveals what we believe is a novel pathway in which TGF-β1 signaling induces the HDM2 gene. Elevated levels of Hdm2 lead to the loss of p53, which is a key component in the process of EMT. Selectively targeting p53 protein for disruption by SV40 T antigen in the mammary epithelia of a mouse produces a ductal carcinoma, similar to those examined in our study (44). This highlights the importance of p53 in governing the progression of breast cancer. Our work provides evidence for another mechanism that tumor cells may use to promote metastasis, especially in tumors where genes mutated in the TGF-β1 pathway are rare. Moreover, using small molecules that are effective at neutralizing Hdm2 oncogenic activities may bring about effective cellular responses to engage cytostasis or even cell death.
Induction of HDM2 by TGF-β1.
The HDM2 gene is well characterized as a transcriptional target of p53 in response to genotoxic stress. There are a few instances that show induction of the HDM2 gene independent of p53. Elevated Hdm2 protein is associated with destabilization of p53, which is attributed to the Ring finger/E3 ubiquitin ligase domain. Thus, loss of p53 levels by elevated Hdm2 forms an autoregulatory feedback loop responsible for entry into the cell cycle and survival. Elimination of p53 through induction of Hdm2 independent of p53 transcriptional activity may provide a mechanism to rapidly promote cellular proliferation in the context of physiological processes such as wound healing, a process that draws parallels with tumor progression and metastasis.
The HDM2 gene is composed of 2 promoters designated P1 and P2. P1 is considered the housekeeping promoter, while P2 is induced by transcription factors in response to various stimuli including p53. Our data show a striking decrease in p53 after 24 hours of TGF-β1 exposure in TGF-β1–responsive cells that corresponds closely with induction of Hdm2 protein. We show a causal relationship between lower levels of p53 protein and increased Hdm2 levels. Using real-time PCR, Hdm2 reporter assays, and ChIP analysis, we demonstrate that the elevation of Hdm2 levels is regulated at the transcriptional level in response to TGF-β1 exposure.
To determine whether a TGF-β1 signaling pathway enabled p53 to transactivate the HDM2 gene, we used cells devoid of p53 activity and demonstrated that induction of HDM2 was independent of p53 (Figure 3). Further analysis revealed that induction of HDM2 gene expression was dependent on the Smad3/4 complex binding to the Smad3 consensus site (SBE2) located proximal to the TATA box in the P2 promoter of HDM2. ChIP analyses demonstrated that the Smad3/4 heterodimer was able to bind to the SBE2 element in response to TGF-β1. Additionally, mutation of SBE2 in the P2 diminished TGF-β1–mediated induction, demonstrated by luciferase reporter assays. These data conclusively demonstrate that TGF-β1–mediated induction of the HDM2 promoter is mediated by Smad3/4 and results in elevated Hdm2 protein.
TGF-β1–mediated destabilization of p53 by HDM2.
In response to DNA damage, p53 can induce the HDM2 gene, and the subsequent elevation of Hdm2 protein facilitates destabilization of p53. This is achieved by the E3 ubiquitin ligase activity of Hdm2, which is able to conjugate multiple mono-ubiquitin moieties to p53, which then signals p53 to exit the nucleus. Additional studies suggest that p53 is then polyubiquitinated by p300 or possibly other enzymes (45). Both Hdm2 and p53 must localize to the same cellular compartment to initiate the inactivation of p53 by Hdm2. Hdm2 undergoes nuclear translocation in response to IGF-1/EGF/insulin through the activation of Akt, which phosphorylates Hdm2 in its nuclear localization sequence (46, 47). Upon nuclear localization, a second signal causes the nuclear export of p53 and Hdm2, which is mediated by the Mapk/RSK pathway (48). These pathways are also activated in response to TGF-β1 (37). Our work shows that p53 is destabilized when Hdm2 levels are increased in response to TGF-β1. In addition to signaling transduction pathways, a recent study argues that alterations in the stoichiometry of Hdm2 to p53 protein levels is a key factor in the regulation of p53 stability (31).
TGF-β1 is involved in cellular processes that evoke cytostasis or even apoptosis in primary cells, while during cancer progression, tumor cells become refractory to cell death and more aggressive. TGF-β1 can also influence cell migration. In Figure 6, HCT116:3-6 cell migration stimulated by TGF-β1 was halted when combined with the Hdm2 antagonist, Nutlin3. While these are transformed cells, they maintain wild-type p53 status, and their response to Nutlin3 correlated with altered p53 stability. We show a marked increase in both Smad3 activation and Mdm2 in NMuMg cells, which are not transformed and can be induced to undergo EMT. Activation of Smad3 caused elevation of Mdm2 levels, which resulted in the subsequent loss of p53. This is consistent with the data we generated in transformed and immortalized human cell lines. If p53 plays a pivotal role in suppressing cellular growth and EMT, it would be necessary to neutralize its activity in order for cells to undergo EMT and proliferate. Clinically, it has been suggested that the genetic or functional loss of p53 is the last event necessary for tumor cells to metastasize. Thus, prolonged exposure to TGF-β1 as seen in the tumor microenvironment may elicit p53 downregulation and permit tumor progression.
Hdm2 binds the amino terminus and conjugates ubiquitin to the carboxyl terminus of p53. Recently, several small molecules have emerged that alter the ability of Hdm2 to bind to p53 (49). To determine whether reengaging p53 by neutralizing Mdm2 would reverse EMT in response to TGF-β1, we treated NMuMg cells with Nutlin3. To our surprise, instead of just reversing EMT, cells underwent apoptosis with the cotreatment of TGF-β1 and Nutlin3. NMuMg cells treated with Nutlin3 alone resulted in what appeared to be static cells with no gross levels of cell death, which is consistent with a prior report (32). Thus, treatment of cells with Nutlin3 transitions their response to TGF-β1 from a factor able to induce EMT to a tumor suppressor through the activation of p53. Our work strongly suggests a connection between p53 and the TGF-β1 signaling pathway that is sufficient to induce cell death.
TGF-β1 plays a convoluted role in cancer progression. During early stages of cancer progression, TGF-β1 functions as a tumor suppressor by promoting cytostasis and in some cases apoptosis. In contrast, TGF-β1 functions to promote or select a more aggressive phenotype in later stages of carcinogenesis. Many tumors produce and secrete TGF-β1, which may affect the tumor microenvironment by altering the extracellular matrix and immune system activity, and/or stimulating angiogenesis. A correlation may be drawn with Hdm2 in that it is detected in late-stage carcinogenesis and may alter the sensitivity of cells to TGF-β1 in favor of proliferation. Mechanistically, Hdm2 oncogenic activity can be complex, as it can destabilize the Rb tumor suppressor and E-cadherin, a protein that maintains cell-cell contact to prevent migration. Furthermore, it alters the activity of the cell-cycle arrest protein p21 (16, 50, 51). Hdm2 may also indirectly alter Smad transcriptional activity, offering yet another level of regulation in the TGF-β1 pathway. Additionally, Hdm2 forms a complex with HIF-1α, and this complex is responsible for the induction of VEGF (52–54). Drawing this parallel with the described activity of TGF-β1, the gain in function of TGF-β1 may result from the downstream increase in the levels of Hdm2, thereby leading to TGF-β1 resistance.
In support of this supposition, staining of breast cancer patient samples for Hdm2 and phosphorylated Smad3 showed a correlation with late-stage infiltrative and metastatic disease. It is worth noting that increased levels of Hdm2 protein in patient samples may also be a result of HDM2 gene induction by other transcription factors. We are currently investigating additional pathways that may contribute to HDM2 induction. Here, we provide yet another molecular pathway, in which induction of HDM2 by TGF-β1 alters the stability of p53. Furthermore, we present evidence for therapeutic intervention of this pathway using the small molecule inhibitor Nutlin3. This therapy could possibly terminate downstream mediators of EMT and alter the tumor microenvironment. This does not preclude the use of small molecules that target the TGF-β1 receptor or its signaling pathway, but identifies multiple points for therapeutic intervention.
Methods
Cell culture, transfections, and induction of EMT
Mammalian cells were cultured at 37°C in a humidified incubator with 5% CO2. HCT116, HCT116:3-6, SKOV3, and 293T cells were cultured in DMEM in high glucose with 10% FBS and the addition of 10 μg/ml of insulin for NMuMg cells. Vaco400 with or without TβRII, gifts from J.K.V. Willson (University of Texas Southwestern) and S. Markowitz (Case Western Reserve University), were cultured in MEM supplemented with 2% FBS, 2 mM l-glutamine, 50 μg/ml gentamicin, 1 μg/ml hydrocortisone, 2 μg/ml human transferrin, 5 nM sodium selenite, 10 μg/ml bovine insulin, and 300 μg/ml G418 (55). For induction of EMT, cells were plated the previous day. Medium was changed and 4 ng/ml TGF-β1 (R&D systems) was added; cells were examined at various times after treatment.
Reporter assays
Cells were plated in 12-well plates and transfected with various HDM2 promoter-luciferase reporter constructs (895 bp of pGL3-HDM2 promoter and 710 bp of deletion mutant of HDM2 promoter [ΔHDM2]; a gift from J.P. Blaydes, University of Southampton, Southampton, United Kingdom) or SBE2X2-tkluc along with pRL-TK renilla using the Lipofectamine Plus reagent (Invitrogen) according to manufacturer’s instructions. Mutations of SBE (SBE2) were introduced into HDM2-luc by the QuikChange II site-directed Mutagenesis System (Stratagene), and all constructs were verified by DNA sequencing (Gene Gateway LLC). The forward site-directed mutagenesis primer for SBEM2 was 5′-GAACGCTGCGCGTACCATGGGCGGGATTGG-3′ and its compliment. For TGF-β1 treatments, 24 hours after transfection, cells were treated with 10 ng/ml TGF-β1. At indicated times, cells were harvested in 1× reporter lysis buffer. In other cases, cells were cotransfected with either caTβRI or constitutively activated Smad3 (caSmad3) and the HDM2-luciferase along with pRL-TK renilla were assessed. Reporter activity was determined using the Luciferase Assay System (Promega Inc.) as described by the manufacture’s instructions and normalized by renilla activity. Fold induction was determined as TGF-β1–treated samples compared with vehicle-treated control samples. Each reporter assay was performed 3 times in triplicate and values represented as mean ± SEM determined by Student’s t tests.
Western blot and reagents
Nutlin3 compound was purchased from Cayman Chemical. Whole-cell lysates were prepared after TGF-β1 treatment or caTβRI expression in RIPA buffer (0.1% SDS, 0.5% deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris, pH 8.0). Protein samples were size fractionated by SDS-PAGE and transferred to PVDF membrane. Antibodies used were p53 (DO-1), Ku70 (C19), and α-tubulin (TU-02) (Santa Cruz Biotechnology Inc.). The Flag M2-clone antibody (Sigma-Aldrich) and the phospho-Smad3 (p-Smad3) (ser433/435) were purchased from Abcam. Antibodies to Hdm2/Mdm2 were purchased from Calbiochem. Proteins were detected by chemiluminescence (PerkinElmer).
HA-Ub was expressed in HCT116:3-6 cells using Lipofectamine Plus. Twenty-four hours after transfection, cells were treated with TGF-β1 for 48 hours, followed by 20 μM MG132 treatment for 4 hours. Immunoprecipitation of p53 was performed as previously described (56). In brief, cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.4, 1 mM MgCl2, 0.2 % v/v of Tween 20, 10 mM sodium molybdate, protease inhibitor cocktail, and 1 mM PMSF) and collected by centrifugation. IP was performed with 2 mg of total protein with 6 μg of anti-p53 antibody (pAb421; Oncogene) or mouse IgG as a control. Protein G–sepharose (Pierce Biotechnology Inc.) was added to the mixture. After 1 hour incubation at 4°C, precipitates were collected, washed 4 times, and extracted into SDS loading buffer by boiling for 5 minutes. Samples were separated by SDS-PAGE, and HA-Ub was detected by Western blot using α-HA antibody.
DNA pulldown assay and chromatin immunoprecipitation
DNA pulldown assays were performed as described (57). In brief, cells were seeded onto 150-mm dishes and cultured overnight. FLAG-Smad3 plasmid was transiently transfected into HCT116:3-6 cells. The next day, cells were treated with TGF-β1 (5 ng/ml) for 48 hours, and nuclear extracts were prepared (58). Nuclear fractions from treated or control cells were incubated overnight at 4°C with a mixture of biotinylated oligonucleotide promoters, as indicated, and streptavidin agarose beads. Biotinylated oligonucleotide SBE2X2-tkluc and SBE2X2 were used as a positive control for Smad DNA–binding activities. Mixtures were then extensively washed and separated by 10% SDS-PAGE as described (59). TGF-β1–activated Smad-specific binding to HDM2 promoter regions or other control DNAs was detected by Western blots using antibodies to Smad4 or FLAG (FLAG-Smad3). Biotinylated oligonucleotides of various regions of the HDM2 promoter (HDM2, SBE1, SBE2, and ΔHDM2) or SBE2X2 were amplified by PCR, using specific primers purchased from Integrated DNA Technologies Inc. as follows: HDM2, forward: 5′-/5Bio/GCAGCGAGCGGTCACTTTTG-3′, reverse: 5′-TTCGGAACGTGTCTGAACTTGACC-3′; SBE1, forward: 5′-/5Bio/-GCAGCGAGCGGTCACTTTTG-3′, reverse: 5′-CAGCGTTCACACTAGTGAC-3′; SBE2, forward: 5′/5Bio/-TCGGGTCACTAGTGTGAACGCT-3′, reverse: 5′-TTCGGAACGTGTCTGAACTTGACC-3′; ΔHDM2, forward: 5′-/5Bio/-CTCTGACGGTGTCCCCTCTATC-3′, reverse: 5′-CAGCGTTCACACTAGTGAC-3′; SBE2X2, forward: 5′-/5Bio/-GATATCGAGATCTGCTAGTC-3′, reverse: 5′-CACGCTGTTGACGCTGTTAAG-3′. Biotinylated versions of the various HDM2 promoter regions were amplified from the pGL3-HDM2 reporter template. Biotinylated SBE(2X2), which contains 4 SBEs linked by AGAC, was amplified from the SBE2X2-tkluc plasmid.
ChIP assays to detect Smad3 and Smad4 protein-DNA interactions in vivo were performed as previously described (60). In brief, after 48 hours of TGF-β1 exposure (see above), cells were treated with formaldehyde at a final concentration of 1%. After incubation, cells were washed and resuspended in ice-cold lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1 mM PMSF, protease inhibitor cocktails). Lysates were sonicated, and soluble chromatin was collected by centrifugation (5000 g, 10 minutes, 4°C). Extracts were diluted in ice-cold dilution buffer (1% Triton X-100, 2 mM EDTA, 160 mM NaCl, 20 mM Tris, pH 8.1, protease inhibitor cocktails). Immunoprecipitations were performed overnight with specific antibodies (p-Smad3) or α-Smad4 (clone B8; Santa Cruz Biotechnology Inc.) or with secondary α-rabbit antibody (Santa Cruz Biotechnology Inc.) as a negative control. Complexes were then incubated with protein G sepharose beads (Pierce). Immunoprecipitates were washed with TE and eluted by incubation at 65°C overnight in 1% SDS (in 0.1 M NaHCO3 at 65°C, 4 hours). DNA was purified using a PCR purification kit (Roche) according to the manufacturer’s instructions and eluted with TE; templates were used in PCR reactions to detect HDM2 promoter regions bound by specific proteins. PCR primer sequences used were as follows: SBE1, forward: 5′-GCAGCGAGCGGTCACTTTTG-3′, reverse: 5′-CCCGTGACCTTTACCCTGAACT-3′; SBE2, forward: 5′- TCTCCGCGGGAGTTCAGGGTAA-3′, reverse: 5′-TTCGGAACGTGTCTGAACTTGACC-3′; p21, forward: 5′-CAAGGCTTCTGCAAATATGGACC-3′, reverse: 5′-CTCAGCATCAGTGTTACCAAC-3′. PCR products were resolved on a 2% agarose gel and visualized by ethidium bromide staining.
Immunohistochemistry
Tissue preparation.
The tissues were fixed overnight in 3% paraformaldehyde and then transferred to 70% ethanol prior to processing through to a paraffin block. Five-micron sections were microtomed, and the sections were placed on positive charged slides. The slides were then baked overnight at 60°C.
Immunostaining.
The slides were then deparaffinized in xylene and rehydrated through graded alcohols to water. Antigen retrieval was performed for p-Smad3 staining by immersing the slides in Target Retrieval Solution (DAKO Corporation) for 20 minutes in a pressure cooker, then cooling to room temperature for 10 minutes, washing in water, and then proceeding with immunostaining. Antigen retrieval was performed by immersing the slides in EDTA for 20 minutes in a pressure cooker for Hdm2, cooling at room temperature for 10 minutes, washing in water, and then proceeding with immunostaining. All subsequent staining steps were performed on the Dako Immunostainer; incubations were done at room temperature, and Tris-buffered saline plus 0.05% Tween 20, pH 7.4 (TBST; Dako), was used for all washes and diluents. Thorough washing was performed following subsequent incubations. Slides were blocked with protein-blocking solution (Dako) for 25 minutes; after washing, Hdm2 at a dilution of 1:50 for 2 hours and p-Smad3 at a 1:50 dilution for 1 hour were added to the slides. A biotinylated link antibody plus streptavidin–horseradish peroxidase kit (LSAB2; Dako) was then utilized along with a DAB chromagen and peroxide substrate to detect the bound antibody complexes. The slides were briefly counterstained with hematoxylin, removed from the autostainer, and dehydrated through graded alcohols to xylene. The slides were coverslipped with a permanent mounting media.
Slide evaluation.
Three investigators using light microscopy evaluated the intensity and localization of the staining. Immunocytochemistry was scored as follows: percentage of cells staining, intensity of IHC stain (negative =0; borderline, minimal = 1; moderate = 2; or strong = 3); localization of stain in the cell (none, membrane, cytoplasm, nuclear, combination of membrane/cytoplasm, combination of cytoplasm/nuclear), and tumor distribution of the stain (homogenous, heterogeneous, focal, multifocal, and/or variable). P values were calculated using Fisher’s exact test. Discarded patient samples for this study were approved through the institutional review board at Indiana University School of Medicine.
Confocal microscopy
Confocal microscopy was performed as previously described (46). Antibodies that detect Vimentin (Santa Cruz Biotechnology Inc.), E-cadherin (Santa Cruz Biotechnology Inc.), and N-cadherin were incubated at a 1:50 dilution. Nuclear staining was detected by DAPI (Roche).
siRNA to Mdm2 and survival analysis
See Supplemental Methods.
Statistics
Statistical analyses were performed using 2-sample, 2-tailed, equal variance Student’s t test. Two-tailed Fisher’s exact test was used for binary data. P values of less than 0.05 were considered significant. SAS 9.1.3 Service Pack 3 was used for all the analyses.
Supplementary Material
Acknowledgments
The authors wish to thank members of the Boothman and Mayo laboratories for their technical assistance, George Sandusky for the pathology work, and Cheng-Ming Chiang and Julie Driscol for their critical reading of the manuscript. This work was supported by grants NIH R01 CA109262 (to L.D. Mayo); and grants NIH 5U19AI067773-04 (subcontract from D. Brenner, principle investigator, Columbia University) and Department of Energy DE-FG02-09ER64789 (to D.A. Boothman); and Immunohistochemistry and Biological Microscopy Cores at the Simon Cancer Center, Indiana University (NIH P30CA08709); as well as the Biostatistics Core, Simmons Comprehensive Cancer Center, University of Texas Southwestern (NIH P30CA142543). This is paper CSCN048.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:290–302 (2010). doi:10.1172/JCI39194.
References
- 1.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100(15):8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu SL, et al. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res. 2004;64(13):4405–4410. doi: 10.1158/0008-5472.CAN-04-1032. [DOI] [PubMed] [Google Scholar]
- 3.Gold LI. The role for transforming growth factor-beta (TGF-beta) in human cancer. Crit Rev Oncog. 1999;10(4):303–360. [PubMed] [Google Scholar]
- 4.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 5.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390(6659):465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 6.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383(6603):832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes Dev. 1997;11(23):3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci U S A. 1998;95(12):6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113(3):301–314. doi: 10.1016/S0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18(5):1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang YA, et al. Smad3 in the mammary epithelium has a nonredundant role in the induction of apoptosis, but not in the regulation of proliferation or differentiation by transforming growth factor-beta. Cell Growth Differ. 2002;13(3):123–130. [PubMed] [Google Scholar]
- 12.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 13.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 14.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2(9):569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 15.Xiao ZX, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375(6533):694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 16.Uchida C, et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24(1):160–169. doi: 10.1038/sj.emboj.7600486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yam CH, et al. MDM2 and MDMX inhibit the transcriptional activity of ectopically expressed SMAD proteins. Cancer Res. 1999;59(20):5075–5078. [PubMed] [Google Scholar]
- 18.Sun P, Dong P, Dai K, Hannon GJ, Beach D. p53-independent role of MDM2 in TGF-beta1 resistance. Science. 1998;282(5397):2270–2272. doi: 10.1126/science.282.5397.2270. [DOI] [PubMed] [Google Scholar]
- 19.Kannemeier C, Liao R, Sun P. The RING finger domain of MDM2 is essential for MDM2-mediated TGF-beta resistance. Mol Biol Cell. 2007;18(6):2367–2377. doi: 10.1091/mbc.E06-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Veigl ML, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95(15):8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan DP, et al. Antisense inhibition of hMLH1 is not sufficient for loss of DNA mismatch repair function in the HCT116+chromosome 3 cell line. Clin Cancer Res. 2000;6(10):3827–3831. [PubMed] [Google Scholar]
- 23.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 24.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 25.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112(6):779–791. doi: 10.1016/S0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121(7):1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 27.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 29.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 30.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 32.Ries S, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103(2):321–330. doi: 10.1016/S0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- 33.Phelps M, Darley M, Primrose JN, Blaydes JP. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. Cancer Res. 2003;63(10):2616–2623. [PubMed] [Google Scholar]
- 34.Slack A, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A. 2005;102(3):731–736. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Criswell T, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280(14):14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- 36.Muraoka RS, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109(12):1551–1559. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275(47):36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 38.Xie L, et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res. 2003;5(6):R187–R198. doi: 10.1186/bcr640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3(4):419–421. [PubMed] [Google Scholar]
- 40.Vassilev LT. p53 activation by small molecules: application in oncology. J Med Chem. 2005;48(14):4491–4499. doi: 10.1021/jm058174k. [DOI] [PubMed] [Google Scholar]
- 41.Izumi T, et al. MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology. 2009;6:1. doi: 10.1186/1742-4690-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaydes JP, Wynford-Thomas D. The proliferation of normal human fibroblasts is dependent upon negative regulation of p53 function by mdm2. Oncogene. 1998;16(25):3317–3322. doi: 10.1038/sj.onc.1201880. [DOI] [PubMed] [Google Scholar]
- 43.de Rozieres S, Maya R, Oren M, Lozano G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene. 2000;19(13):1691–1697. doi: 10.1038/sj.onc.1203468. [DOI] [PubMed] [Google Scholar]
- 44.Schulze-Garg C, Lohler J, Gocht A, Deppert W. A transgenic mouse model for the ductal carcinoma in situ (DCIS) of the mammary gland. Oncogene. 2000;19(13):1028–1037. doi: 10.1038/sj.onc.1203281. [DOI] [PubMed] [Google Scholar]
- 45.Grossman SR, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300(5617):342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 46.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98(20):11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3(11):973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 48.Jackson MW, et al. Hdm2 nuclear export, regulated by insulin-like growth factor-I/MAPK/p90Rsk signaling, mediates the transformation of human cells. J Biol Chem. 2006;281(24):16814–16820. doi: 10.1074/jbc.M511617200. [DOI] [PubMed] [Google Scholar]
- 49.Lehman JA, Eitel JA, Batuello CN, Mayo LD. Therapeutic considerations for Mdm2: not just a one trick pony. Expert Opin Drug Discov. 2008;3(11):1309–1321. doi: 10.1517/17460441.3.11.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol Cell Biol. 2008;28(4):1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bey EA, et al. Mornings with Art, lessons learned: feedback regulation, restriction threshold biology, and redundancy govern molecular stress responses. J Cell Physiol. 2006;209(3):604–610. doi: 10.1002/jcp.20783. [DOI] [PubMed] [Google Scholar]
- 52.Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278(16):13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 53.Bardos JI, Chau NM, Ashcroft M. Growth factor-mediated induction of HDM2 positively regulates hypoxia-inducible factor 1alpha expression. Mol Cell Biol. 2004;24(7):2905–2914. doi: 10.1128/MCB.24.7.2905-2914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaRusch GA, et al. Nutlin3 blocks vascular endothelial growth factor induction by preventing the interaction between hypoxia inducible factor 1alpha and Hdm2. Cancer Res. 2007;67(2):450–454. doi: 10.1158/0008-5472.CAN-06-2710. [DOI] [PubMed] [Google Scholar]
- 55.McBain JA, Weese JL, Meisner LF, Wolberg WH, Willson JK. Establishment and characterization of human colorectal cancer cell lines. Cancer Res. 1984;44(12 pt 1):5813–5821. [PubMed] [Google Scholar]
- 56.Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1998;18(3):1517–1524. doi: 10.1128/mcb.18.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Criswell TL, et al. Delayed activation of IGF-1R/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280(14):14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- 58.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Criswell T, Klokov D, Beman M, Lavik JP, Boothman DA. Repression of IR-inducible clusterin expression by the p53 tumor suppressor protein. Cancer Biol Ther. 2003;2(4):372–380. doi: 10.4161/cbt.2.4.430. [DOI] [PubMed] [Google Scholar]
- 60.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98(5):675–686. doi: 10.1016/S0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.