Abstract
Pancreatic β cell failure is thought to underlie the progression from glucose intolerance to overt diabetes, and ER stress is implicated in such β cell dysfunction. We have now shown that the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) accumulated in the islets of diabetic animal models as a result of ER stress before the onset of hyperglycemia. Transgenic overexpression of C/EBPβ specifically in β cells of mice reduced β cell mass and lowered plasma insulin levels, resulting in the development of diabetes. Conversely, genetic ablation of C/EBPβ in the β cells of mouse models of diabetes, including Akita mice, which harbor a heterozygous mutation in Ins2 (Ins2WT/C96Y), and leptin receptor–deficient (Lepr–/–) mice, resulted in an increase in β cell mass and ameliorated hyperglycemia. The accumulation of C/EBPβ in pancreatic β cells reduced the abundance of the molecular chaperone glucose-regulated protein of 78 kDa (GRP78) as a result of suppression of the transactivation activity of the transcription factor ATF6α, thereby increasing the vulnerability of these cells to excess ER stress. Our results thus indicate that the accumulation of C/EBPβ in pancreatic β cells contributes to β cell failure in mice by enhancing susceptibility to ER stress.
Introduction
The prevalence of type 2 diabetes is increasing rapidly in many industrialized nations, with the total number of people with this condition projected to reach 300 million worldwide by 2025. It is thus important to characterize the pathogenesis of type 2 diabetes with a view to the effective prevention of this disease. During the development of type 2 diabetes, pancreatic β cells initially compensate for insulin resistance (often associated with obesity or changes in lifestyle) by upregulating insulin secretion. At some point, however, this period of β cell compensation is followed by β cell failure — in which the pancreas fails to secrete sufficient insulin, as a result either of inadequate expansion of β cell mass or of a loss of the ability of existing β cells to respond to glucose — and diabetes ensues (1–4). Identification of the key molecules that contribute to β cell failure should thus provide important insight into the pathogenesis of type 2 diabetes.
The ER performs several important functions, including the posttranslational modification, folding, and assembly of newly synthesized proteins destined for secretion. Pancreatic β cells possess a highly developed and active ER because of their insulin secretory function. Disequilibrium between the protein load of the ER and its folding capacity is referred to as ER stress (5). In general, cells have the capacity to adapt to substantial ER stress, but they undergo apoptosis when this capacity is exceeded. Given that the period of β cell compensation during the development of type 2 diabetes is associated with increased demand on the ER for insulin production, it might be expected that ER stress is a contributing factor to β cell failure.
The CCAAT/enhancer-binding protein (C/EBP) family of basic leucine-zipper (bZIP) transcription factors includes C/EBPα, -β, -γ, -δ, and -ε as well as C/EBP homology protein (CHOP) (6). C/EBPβ performs diverse functions, participating in the regulation of genes that contribute to the acute phase response, glucose metabolism, and tissue differentiation, including adipogenesis and hematopoiesis (7). C/EBPβ expression was found to be increased in β cell lines exposed to high glucose concentrations as well as in pancreatic islets of diabetic animals (8, 9). Furthermore, C/EBPβ was shown to contribute to inhibition of insulin gene transcription in β cell lines exposed to supraphysiological glucose concentrations (8). These observations prompted us to investigate the role of C/EBPβ in the pathogenesis of β cell failure, especially in relation to ER stress.
In the present study, we demonstrated that C/EBPβ (encoded by Cebpb) accumulated in the pancreatic islets of mouse models of diabetes even during the prediabetic stage. We also showed that C/EBPβ expression was induced by ER stress in a β cell line. C/EBPβ accumulation in pancreatic β cells blocked the induction of the molecular chaperone glucose-regulated protein of 78 kDa (GRP78, also known as BiP) as a result of suppressed transactivation activity of activating transcription factor 6α (ATF6α), thereby increasing the vulnerability of these cells to excess ER stress. Such a mechanism may thus contribute to the pathogenesis of pancreatic β cell failure.
Results
Upregulation of insulin expression in β cell–specific C/EBPβ knockout mice.
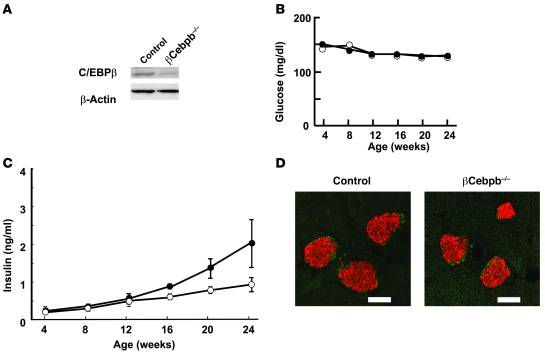
To examine the role of C/EBPβ in pancreatic β cell function, we generated pancreatic β cell–specific C/EBPβ knockout mice (βCebpb–/– mice) by breeding mice homozygous for a floxed allele of Cebpb (Cebpbfl/fl mice; Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI39721DS1) with mice that express Cre recombinase under the control of the rat Ins2 gene promoter (Ins-Cre mice; ref. 10). We first examined whether the introduction of the Cre gene or loxP sequences into the mouse genome affected glucose metabolism or β cell function. There were no significant differences in blood glucose or plasma insulin concentrations among WT, Ins-Cre, and Cebpbfl/fl mice in the fed state at 8 weeks of age (Supplemental Table 1). We therefore used Cebpbfl/fl mice as control animals. Immunoblot analysis showed that the amount of C/EBPβ in islets was reduced by approximately 90% in βCebpb–/– mice compared with that in control mice (Figure 1A). The abundance of C/EBPβ in the brain, hypothalamus, lung, liver, adipose tissue, and muscle was similar in βCebpb–/– and control mice (data not shown), which indicates that βCebpb–/– mice are indeed deficient in C/EBPβ specifically in pancreatic β cells.
Figure 1. Effects of β cell–specific ablation of C/EBPβ on glucose metabolism and islet morphology.
(A) Pancreatic islets isolated from 24-week-old Cebpbfl/fl control or βCebpb–/– mice were lysed and subjected to immunoblot analysis with antibodies to C/EBPβ and to β-actin (loading control). (B and C) Fed blood glucose (B) and plasma insulin (C) concentrations of control (open circles) or βCebpb–/– (filled circles) mice at the indicated ages. Data are mean ± SEM from at least 6 mice per genotype. (D) Immunostaining of pancreatic sections from 24-week-old control or βCebpb–/– mice with antibodies to insulin (red) and to glucagon (green). Scale bars: 100 μm.
The rate of increase in body weight did not differ between βCebpb–/– and control mice (data not shown). There was also no difference in blood glucose concentration between βCebpb–/– and control mice in the fed state up to 24 weeks of age (Figure 1B). Plasma insulin concentration tended to be higher in βCebpb–/– mice than in control animals (Figure 1C), and the insulin content of the pancreas was significantly increased in βCebpb–/– mice compared with that in controls (see below). Insulin tolerance tests did not reveal a significant difference in the insulin sensitivity of peripheral tissues between control and βCebpb–/– mice, excluding the possibility that βCebpb–/– mice are insulin resistant (Supplemental Figure 2).
The insulin secretory response of islets isolated from βCebpb–/– mice was significantly greater than that of control islets at 11.8 or 16.8 mM glucose; however, after normalization for insulin content, there was no significant difference in insulin secretion between βCebpb–/– and control mice (data not shown). Immunostaining of pancreatic sections from 24-week-old animals with antibodies to insulin and to glucagon revealed normal islet architecture in βCebpb–/– mice (Figure 1D). Quantitative analysis also revealed no difference in total β cell mass between βCebpb–/– and control mice (data not shown).
Induction of C/EBPβ in pancreatic β cells by ER stress.
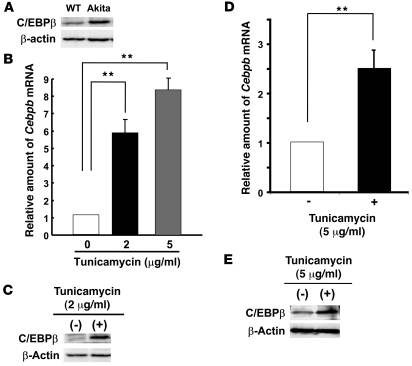
The Akita mouse is an animal model of diabetes in which β cell failure results from a reduction in β cell mass caused by apoptosis (11). The abundance of C/EBPβ in islets of Akita mice at 8 weeks of age increased compared with that in WT animals (Figure 2A). Akita mice are heterozygous for a mutation in Ins2 that results in substitution of tyrosine for cysteine 96 (Ins2WT/C96Y), which participates in the formation of a disulfide bond between the A and B chains of insulin. Expression of this mutant form of insulin induces ER stress, which in turn triggers β cell apoptosis (11, 12). We therefore examined the effect of ER stress on C/EBPβ expression in cultured MIN6 mouse insulinoma cells and islets isolated from C57BL/6J mice. Tunicamycin induces ER stress by inhibiting N-linked glycosylation in the ER. Incubation of MIN6 cells and isolated islets with tunicamycin for 6 hours resulted in an increase in both Cebpb mRNA and C/EBPβ protein expression (Figure 2, B–E). Together, these results suggested that C/EBPβ might be upregulated in pancreatic β cells as a result of ER stress.
Figure 2. Accumulation of C/EBPβ in pancreatic β cells in response to ER stress.
(A) Pancreatic islets isolated from Akita and WT mice at 8 weeks of age were lysed and subjected to immunoblot analysis with antibodies to C/EBPβ. (B–E) MIN6 cells (B and C) and isolated islets (D and E) were incubated with tunicamycin at the indicated concentrations for 6 hours and then subjected either to real-time RT-PCR analysis of Cebpb mRNA (B and D) or to immunoblot analysis with antibodies to C/EBPβ (C and E). Data in B and D are expressed relative to the value for cells incubated without tunicamycin and are means of triplicates from a representative experiment. **P < 0.01.
Amelioration of hyperglycemia in Akita mice by C/EBPβ ablation in β cells.
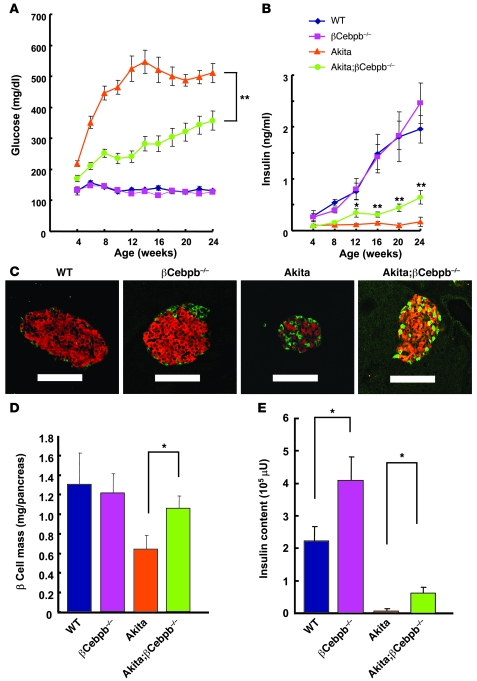
To examine the effects of C/EBPβ accumulation in Akita β cells, we generated Akita mice with β cell–specific ablation of C/EBPβ (Akita;βCebpb–/– mice). As described previously (11, 12), Akita mice developed hyperglycemia that progressed to a blood glucose concentration of approximately 500 mg/dl by 12 weeks of age (Figure 3A). Akita;βCebpb–/– mice exhibited a significant improvement in blood glucose levels compared with Akita mice, maintaining a blood glucose concentration of about 350 mg/dl in the fed state even at 20–24 weeks of age (Figure 3A). Akita mice manifested a low plasma insulin level at 4 weeks of age. The plasma insulin concentration in the fed state was moderately, but significantly, increased in Akita;βCebpb–/– mice compared with that in Akita mice at 12 weeks of age and thereafter (Figure 3B). Whereas β cell mass decreased by approximately 50% in Akita mice at 8 weeks of age compared with that in WT animals, C/EBPβ ablation in β cells of Akita mice resulted in a significant increase in β cell mass (Figure 3, C and D). On the other hand, we could not detect any apparent differences in α cell volume between Akita and Akita;βCebpb–/– mice (data not shown).The insulin content of the pancreas was also markedly decreased in Akita mice compared with that in WT mice, possibly correlating to reduced intensity of insulin staining, whereas loss of C/EBPβ resulted in an approximately 8-fold increase in the pancreatic insulin content of Akita mice (Figure 3E). Given that the transgene is integrated randomly in Ins-Cre mice, it might affect the expression of the endogenous mutant Ins2 gene of Akita;βCebpb–/– mice. We thus compared Akita mice with Akita;Ins-Cre mice in terms of glucose homeostasis. Blood glucose and plasma insulin concentrations in the fed state did not differ between these 2 genotypes (Supplemental Figure 3). These results thus suggested that C/EBPβ accumulation in pancreatic β cells might be responsible for the reduced β cell mass and insulin content of islets in Akita mice.
Figure 3. Effects of β cell–specific ablation of C/EBPβ in Akita mice.
(A and B) Blood glucose (A) and plasma insulin (B) concentrations in WT (n = 7), βCebpb–/– (n = 9), Akita (n = 7), and Akita;βCebpb–/– (n = 14) mice in the fed state and at the indicated ages. Data are mean ± SEM. *P < 0.05, **P < 0.01, Akita;βCebpb–/– versus Akita. (C) Pancreatic sections from 8-week-old WT, βCebpb–/–, Akita, and Akita;βCebpb–/– mice were stained with antibodies to insulin (red) and to glucagon (green). Scale bars: 100 μm. (D) Quantitation of β cell mass in 8-week-old WT, βCebpb–/–, Akita, and Akita;βCebpb–/– mice. Data are mean ± SEM from 5 mice per genotype. *P < 0.05. (E) Insulin content of the pancreas of WT, βCebpb–/–, Akita, and Akita;βCebpb–/– mice at 12 weeks of age. Data are mean ± SEM from 4 mice per genotype. *P < 0.05.
Loss of β cell mass induced by accumulation of C/EBPβ.
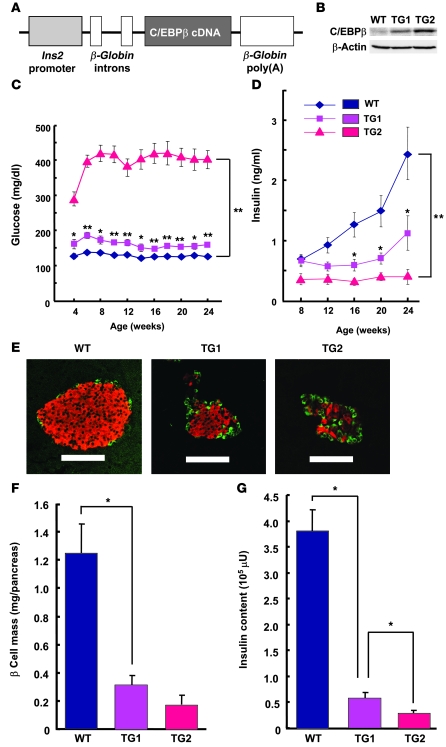
To confirm directly the effects of C/EBPβ accumulation on β cell function and mass, we generated transgenic mice that overexpress C/EBPβ exclusively in β cells under the control of the rat Ins2 promoter (Figure 4A). Two lines of transgenic mice were obtained that differed in islet C/EBPβ expression: transgenic animals with moderate C/EBPβ expression (approximately 3-fold increase relative to the C57BL/6J WT) were designated TG1, and those with high C/EBPβ expression (approximately 10-fold increase) were designated TG2 (Figure 4B). The copy numbers of TG1 and TG2 mice were 3 and 10, respectively, relative to the positive control (Supplemental Figure 4). The expression level of C/EBPβ in TG2 islets was similar to that in Akita islets. The blood glucose concentration in the fed state significantly increased compared with WT in TG2 mice at 4 weeks of age (296 ± 26 versus 126 ± 9 mg/dl, P < 0.01) and was maintained at greater than 400 mg/dl after 6 weeks (Figure 4C). The blood glucose level of TG1 mice was slightly, but significantly, increased and was maintained at greater than 200 mg/dl until at least 24 weeks of age (Figure 4C). The plasma insulin concentration in the fed state was significantly decreased in the transgenic animals in inverse proportion to the level of C/EBPβ expression in islets. TG2 mice thus exhibited low plasma insulin levels at 8 weeks of age and thereafter, whereas TG1 mice showed moderately but significantly decreased plasma insulin levels at 16 weeks and thereafter (Figure 4D). Examination of islet morphology revealed that β cell mass was reduced by about 75% in TG1 mice and about 85% in TG2 mice at 8 weeks of age compared with that in WT animals (Figure 4, E and F). The insulin content of the pancreas also decreased by about 85% in TG1 mice and about 90% in TG2 mice (Figure 4G). Together, these observations suggested that C/EBPβ induces a loss of β cell mass and insulin content in a manner dependent on its expression level, and that these effects subsequently result in lower plasma insulin levels and diabetes onset.
Figure 4. Effects of C/EBPβ overexpression in pancreatic β cells.
(A) Schematic representation of the C/EBPβ transgene construct. The mouse C/EBPβ cDNA was positioned downstream of the rat Ins2 promoter and rat β-globin gene introns as well as upstream of the poly(A)-encoding sequence of the rat β-globin gene. (B) Immunoblot analysis of C/EBPβ in islets isolated from 8-week-old WT, TG1, and TG2 mice. (C and D) Blood glucose (C) and plasma insulin (D) concentrations of WT, TG1, and TG2 mice in the fed state and at the indicated ages. Data are mean ± SEM from at least 12 mice per genotype. *P < 0.05, **P < 0.01 versus WT. (E) Pancreatic sections from 8-week-old WT, TG1, and TG2 mice were stained with antibodies to insulin (red) and to glucagon (green). Scale bars: 100 μm. (F) Quantitation of β cell mass in 8-week-old WT, TG1, and TG2 mice. Data are mean ± SEM from at least 6 mice per genotype. *P < 0.05. (G) Insulin content of the pancreas of 8-week-old WT, TG1, and TG2 mice. Data are mean ± SEM from at least 3 mice per genotype. *P < 0.05.
C/EBPβ regulates ER stress through modulation of GRP78 expression.
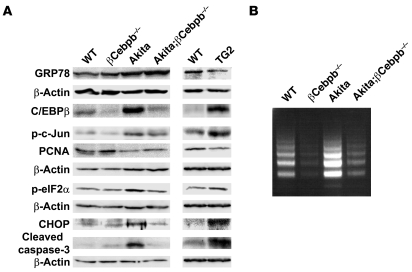
We next examined the mechanism by which C/EBPβ accumulation induces β cell failure. Akita mice manifest increased ER stress in pancreatic β cells caused by accumulation of the unfolded mutant form of insulin. The increased ER stress, in turn, results in activation of the unfolded protein response (UPR). We examined UPR status by immunoblot analysis of GRP78, a molecular chaperone of the ER lumen that stabilizes intermediates in protein folding. The abundance of GRP78 in Akita islets increased compared with that in WT islets (Figure 5A and Supplemental Figure 5). However, GRP78 induction was not sufficient to suppress the augmented ER stress in the islets of Akita mice, as revealed by the increased phosphorylation of c-Jun and eukaryotic translation initiation factor 2α (eIF2α), which are phosphorylated by the ER stress signaling molecules IRE1 and PERK, respectively (Figure 5A and Supplemental Figure 5). The activation of these ER stress signaling molecules was also likely responsible for an increase in the expression of the proapoptotic protein CHOP. The abundance of the cleaved form of caspase-3 and the extent of internucleosomal DNA fragmentation, both markers of apoptosis, increased in Akita islets (Figure 5, A and B). The abundance of proliferating cell nuclear antigen (PCNA) decreased in Akita islets compared with that in WT islets (Figure 5A), which suggests a loss of β cell proliferation in the former.
Figure 5. Effects of C/EBPβ on ER stress and apoptosis in pancreatic β cells.
(A) Islets isolated from 8-week-old mice of the indicated genotypes were subjected to immunoblot analysis with antibodies to the indicated proteins. p-c-Jun and p-eIF2α represent phosphorylated forms of c-Jun and eIF2α, respectively. (B) Islets from 8-week-old mice of the indicated genotypes were incubated in RPMI 1640 medium for 24 hours, after which internucleosomal DNA fragmentation was examined by ligation-mediated PCR analysis.
Deletion of C/EBPβ in pancreatic β cells further increased GRP78 expression in Akita mice (Figure 5A and Supplemental Figure 5). The extent of GRP78 accumulation in Akita;βCebpb–/– mouse islets was sufficient to relieve ER stress, as indicated by the reduced phosphorylation of c-Jun and eIF2α compared with that in Akita islets (Figure 5A and Supplemental Figure 5). Islets of Akita;βCebpb–/– mice also showed a reduced abundance of CHOP as well as a reduced level of apoptosis, as reflected by the amount of the cleaved form of caspase-3 and the extent of internucleosomal DNA fragmentation (Figure 5, A and B). They also showed an increased abundance of PCNA (Figure 5A), possibly reflecting an increased β cell proliferation.
TG2 islets showed reduced GRP78 expression and an increased extent of ER stress, as reflected by increased phosphorylation of c-Jun and eIF2α compared with those of WT islets (Figure 5A and Supplemental Figure 5). Islets of TG2 mice also manifested upregulation of CHOP, increased apoptosis (as revealed by an increased abundance of the cleaved form of caspase-3), and reduced proliferation (as revealed by a reduced abundance of PCNA; Figure 5A). Together, these results suggested that C/EBPβ ablation in β cells promotes the induction of GRP78 expression and thereby confers tolerance to ER stress. Conversely, the accumulation of C/EBPβ likely increases the vulnerability of pancreatic β cells to ER stress.
Amelioration of hyperglycemia in the Lepr–/– mouse by ablation of C/EBPβ.
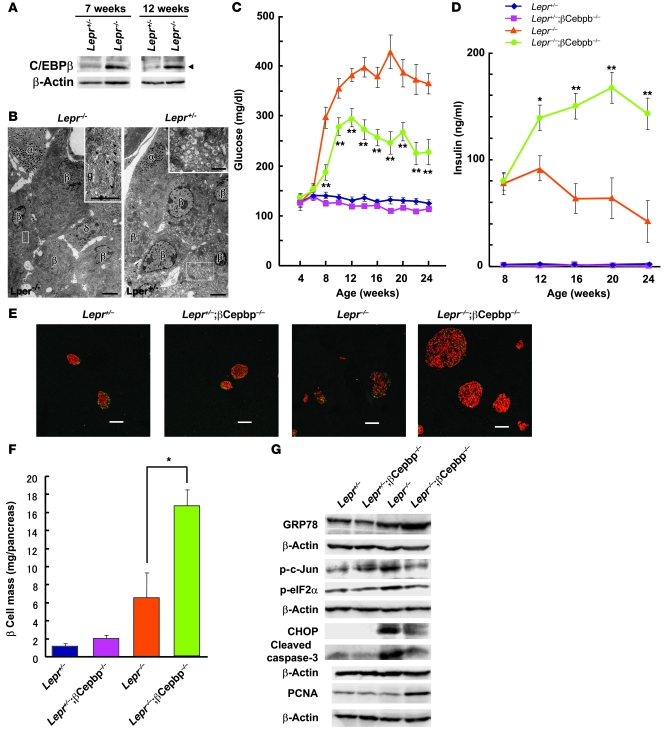
In addition to the Akita mouse, a model of diabetes caused by β cell apoptosis, we examined the role of C/EBPβ in islets of the Lepr–/– mouse, a model of type 2 diabetes associated with obesity (13). Compared with that in Lepr+/– control islets, the amount of C/EBPβ in Lepr–/– islets increased at 12 weeks (Figure 6A), an age at which hyperglycemia was already manifest (Figure 6C). Furthermore, the abundance of C/EBPβ also increased in Lepr–/– islets at 7 weeks (Figure 6A), an age at which the animals exhibited normoglycemia to moderate hyperglycemia together with hyperinsulinemia. Electron microscopy revealed that, although the ER was well organized in the peripheral region, its cisternae were enlarged near the Golgi complex and the number of secretory granules appeared reduced, suggestive of ER stress, in Lepr–/– β cells at 8 weeks of age (Figure 6B). These results thus suggested that C/EBPβ expression might be induced in Lepr–/– islets by ER stress during the stage of insulin resistance before the onset of frank diabetes.
Figure 6. Effects of C/EBPβ ablation in pancreatic β cells of Lepr–/– mice.
(A) Immunoblot analysis of C/EBPβ (arrowhead) in islets isolated from Lepr+/– and Lepr–/– mice at 7 and 12 weeks of age. (B) Electron microscopy of β cells of Lepr–/– and Lepr+/– mice at 8 weeks and 10 months of age, respectively. Higher-magnification views of the boxed regions are shown in the insets. Secretory granules were fewer in number in β cells, but not in α or δ cells, of Lepr–/– mice. Arrowheads indicate enlarged cisternae of the ER near the Golgi complex (g). Scale bars: 3 μm; 1 μm (insets). (C and D) Blood glucose (C) and plasma insulin (D) concentrations of Lepr+/– (n = 6), Lepr+/–;βCebpb–/– (n = 8), Lepr–/– (n = 7), and Lepr–/–;βCebpb–/– (n = 12) mice in the fed state and at the indicated ages. *P < 0.05, **P < 0.01 versus Lepr–/–. (E) Pancreatic sections from 24-week-old Lepr+/–, Lepr+/–;βCebpb–/–, Lepr–/–, and Lepr–/–;βCebpb–/– mice were stained with antibodies to insulin (red) and to glucagon (green). Scale bars: 100 μm. (F) Quantitation of β cell mass in 24-week-old mice of the indicated genotypes. Data are mean ± SEM from 3 mice per genotype. *P < 0.05. (G) Islets isolated from 16-week-old mice of the indicated genotypes were subjected to immunoblot analysis with antibodies to the indicated proteins.
To examine whether ablation of C/EBPβ in β cells of Lepr–/– mice increases β cell mass and insulin secretion sufficiently to overcome peripheral insulin resistance, we crossed these animals with βCebpb–/– mice to obtain Lepr–/–;βCebpb–/– offspring. Hyperglycemia developed more slowly and was significantly ameliorated in Lepr–/–;βCebpb–/– mice compared with that in Lepr–/– mice (Figure 6C). Ablation of C/EBPβ also significantly increased plasma insulin levels in Lepr–/– mice at 12 weeks of age and thereafter (Figure 6D). Furthermore, the islets of Lepr–/–;βCebpb–/– mice were markedly enlarged compared with those of Lepr–/– mice (Figure 6E). Quantitative analysis revealed that β cell mass substantially increased (about 6-fold) in Lepr–/– mice compared with that in Lepr+/– mice at 24 weeks of age, and that loss of C/EBPβ induced a further increase in this parameter (approximately 15-fold increase compared with Lepr+/– mice; Figure 6F). These results suggested that C/EBPβ might contribute to the onset of β cell failure in obese diabetic mice.
As in Akita islets, the abundance of GRP78 in Lepr–/– islets increased compared with that in Lepr+/– islets (Figure 6G and Supplemental Figure 6). However, the extent of GRP78 induction was not sufficient to overcome ER stress in β cells elicited by peripheral insulin resistance, as indicated by the increased phosphorylation of c-Jun and eIF2α (Figure 6G and Supplemental Figure 6). Ablation of C/EBPβ further increased the abundance of GRP78 in Lepr–/– islets to a level sufficient to reduce ER stress, as revealed by reduced levels of c-Jun and eIF2α phosphorylation. The increased apoptosis in Lepr–/– islets, as suggested by the increased abundance of CHOP and the cleaved form of caspase-3, was also reduced in Lepr–/–;βCebpb–/– islets (Figure 6G). Lepr–/– islets exhibited an increased abundance of PCNA at 8 weeks of age compared with that in Lepr+/– islets (data not shown), indicative of continued β cell proliferation. However, at 16 weeks, the amount of PCNA was no longer increased in the islets of Lepr–/– mice (Figure 6G), consistent with a loss of proliferating β cells at this age. A marked increase in PCNA expression in the islets of Lepr–/–;βCebpb–/– mice was apparent at 16 weeks of age (Figure 6G), consistent with the associated increase in β cell mass at this age (data not shown). Together, these results indicated that C/EBPβ may regulate both proliferation and death by apoptosis of pancreatic β cells, possibly through its modulation of ER stress.
Role of ATF6α in regulation of GRP78 expression by C/EBPβ.
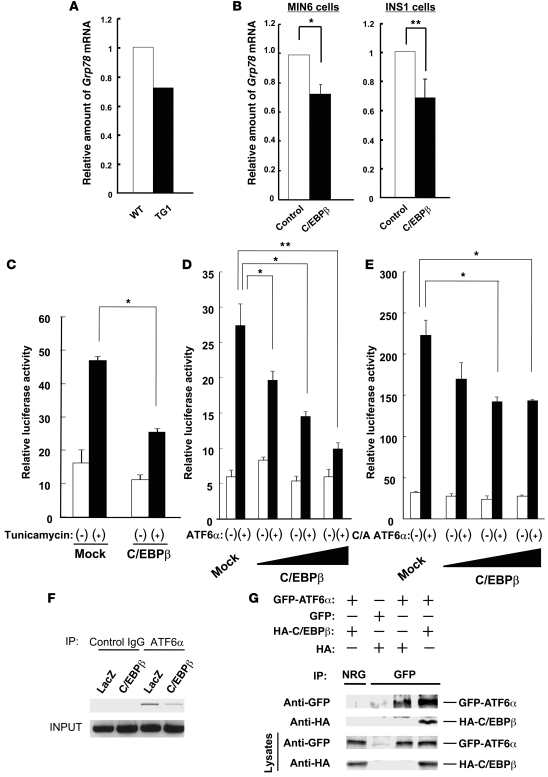
Gene expression profiling with an oligonucleotide microarray showed that the increased expression of C/EBPβ in the islets of TG1 mice resulted in reduced Grp78 mRNA (Figure 7A). Real-time RT-PCR analysis also showed that forced expression of C/EBPβ in MIN6 and INS1 mouse insulinoma cells resulted in downregulated Grp78 mRNA (Figure 7B). These results thus suggested that C/EBPβ might inhibit the expression of GRP78 at the transcriptional level.
Figure 7. Interaction of C/EBPβ with ATF6α and its role in the transcriptional regulation of Grp78.
(A) Relative abundance of Grp78 mRNA in islets isolated from WT and TG1 mice at 8 weeks of age was determined by gene expression profiling with an oligonucleotide microarray. (B) Real-time RT-PCR analysis of Grp78 mRNA in MIN6 and INS1 cells infected with a retrovirus encoding C/EBPβ or with the corresponding empty virus (control). Data are mean ± SEM from 3 independent experiments. *P < 0.05; **P < 0.01. (C) Luciferase assays were performed to examine the effect of C/EBPβ on tunicamycin-induced transcription activation of Grp78 in INS1 cells. Data are mean ± SEM from 3 independent experiments. *P < 0.05. (D and E) Luciferase assays were performed to examine the effect of the various amounts of C/EBPβ on full-length ATF6α–induced (D) or C/A ATF6α–induced (E) transcription activation of Grp78 in INS1 cells. Data are mean ± SEM from 3 independent experiments. *P < 0.05; **P < 0.01. (F) ChIP assay of the Grp78 promoter in INS1 cells infected with a retrovirus encoding either C/EBPβ or β-galactosidase (LacZ) was performed as described in Methods. (G) HEK293 cells were transfected with expression vectors for GFP-tagged ATF6α or for HA epitope–tagged C/EBPβ, as indicated. Cell lysates were subjected to immunoprecipitation with antibodies to GFP or with normal rabbit IgG (NRG), and the resulting precipitates were subjected to immunoblot analysis with antibodies to HA and to GFP.
Among ER stress sensor proteins, ATF6α has been identified as the main inducer of GRP78 (14, 15). ATF6α is proteolytically cleaved and activated in response to ER stress and subsequently translocates to the nucleus. Activated ATF6α contains a bZIP domain at its N terminus, as does C/EBPβ, and it forms a heterodimer with other bZIP proteins. We therefore examined the possible role of interaction between C/EBPβ and ATF6α in GRP78 induction by ER stress. We first performed a luciferase reporter assay in INS1 cells with a reporter construct controlled by a region of the human GRP78 promoter containing 3 ER stress response elements. GRP78 promoter activity increased about 2.5-fold by exposure of the cells to tunicamycin, and this effect was inhibited by forced expression of C/EBPβ (Figure 7C), which supports the notion that C/EBPβ inhibits ER stress–induced transcriptional activation of GRP78.
We next transfected INS1 cells with a vector for full-length ATF6α and confirmed that the ectopic protein increased the activity of the GRP78 promoter. Forced expression of C/EBPβ inhibited the ATF6α-induced activation of the GRP78 promoter in a concentration-dependent manner (Figure 7D). In contrast to C/EBPβ, forced expression of C/EBPα had almost no effect on the ATF6α-induced activation of the GRP78 promoter (Supplemental Figure 7). We also examined the effect of a constitutively active form of ATF6α (C/A ATF6α), which consists of the cytosolic fragment of ATF6α cleaved by the proteases S1P and S2P, and which migrates to the nucleus to activate transcription. The extent of the increase in GRP78 promoter activity induced by C/A ATF6α was markedly greater than that induced by the full-length protein. Forced expression of C/EBPβ also inhibited, in a concentration-dependent manner, the activation of the GRP78 promoter induced by C/A ATF6α (Figure 7E).
We confirmed the binding of ATF6α to the promoter region of GRP78 by ChIP analysis and further showed that this binding was inhibited by overexpression of C/EBPβ (Figure 7F and Supplemental Figure 8). We also found by ChIP analysis that C/EBPβ bound to the promoter region of Grp78 (Supplemental Figure 9). Others have previously shown that mutation of C-terminal cysteine 296 adjacent to the leucine zipper domain of C/EBPβ eliminates DNA-binding activity (16). Thus, we generated mutated CebpbC296S and found that the ATF6α-induced activation of the Grp78 promoter was not suppressed by the CebpbC296S mutant (Supplemental Figure 10). Finally, we examined the potential association of ATF6α and C/EBPβ in HEK293 cells by immunoprecipitation analysis. HA epitope–tagged C/EBPβ was coimmunoprecipitated from cell lysates together with GFP-tagged ATF6α (Figure 7G). Furthermore, we detected association of C/EBPβ with ATF6α in liver cells expressing abundant amounts of C/EBPβ physiologically, such as HepG2 cells (Supplemental Figure 11).
Discussion
We found that C/EBPβ was expressed in mouse pancreatic β cells, and the insulin content of the pancreas significantly increased and plasma insulin concentrations tended to be higher, whereas peripheral insulin sensitivity was not altered, in mice that lack C/EBPβ specifically in β cells. Together with the previous observation that C/EBPβ contributed to inhibition of insulin gene transcription in β cell lines exposed to supraphysiological concentrations of glucose (8), our present results suggest that C/EBPβ regulates insulin expression, likely at the transcriptional level, under physiological conditions. We observed that C/EBPβ expression increased in pancreatic β cells before the onset of frank diabetes in mouse models of this condition. In addition, we found that prevention of C/EBPβ accumulation in the pancreatic β cells of such mice normalized β cell mass and plasma insulin levels and improved glucose tolerance. Such accumulation of C/EBPβ in pancreatic β cells of diabetic mice implicates this protein in β cell failure.
In vitro studies of HepG2 and C6 cells have shown that C/EBPβ expression is upregulated by stimuli that induce ER stress, such as glucose or amino acid deprivation or exposure to drugs such as tunicamycin and thapsigargin (17–19). We have now shown that the expression of C/EBPβ in MIN6 mouse insulinoma cells was also induced by tunicamycin. In addition, we found that C/EBPβ was upregulated in the pancreatic islets of Akita mice, in which expression of a mutant insulin protein induces ER stress in pancreatic β cells. Furthermore, we detected increased ER stress and C/EBPβ accumulation in the pancreatic β cells of Lepr–/– mice, a model of obesity-associated diabetes. C/EBPβ expression in MIN6 cells was also found to be increased upon exposure of the cells to cytokines or free fatty acids (data not shown). Thus, in addition to the ER stress attributable to excessive insulin production and secretion triggered to compensate for insulin resistance, increases in the levels of cytokines and free fatty acids in Lepr–/– mouse plasma might also contribute directly to C/EBPβ accumulation in the pancreatic β cells of these animals.
Our results suggest that excessive accumulation of C/EBPβ blocks ATF6-mediated Grp78 transcription in pancreatic β cells and thereby prevents the induction of GRP78 expression. Under normal conditions, GRP78 inhibits the UPR by binding to the luminal domains of ER stress sensors such as IRE1α, PERK, and ATF6 within the ER and by maintaining these proteins in an inactive state. Unfolded protein accumulation results in the recruitment of GRP78 to facilitate their processing and in the consequent activation of IRE1α, PERK, and ATF6 and induction of the UPR (20–23). Dissociation of GRP78 from ATF6 results in translocation of the latter protein to the Golgi apparatus, where its transmembrane domain is cleaved by the proteases S1P and S2P, leading to the generation of the active form (p50) of the transcription factor and a consequent increase in GRP78 transcription in the nucleus. ATF6 exists as α and β paralogs. Experiments with mouse embryonic fibroblasts deficient in ATF6α or ATF6β revealed that ATF6α is important for transcriptional induction of ER chaperones (14, 15). Furthermore, ATF6α overexpression has been shown to increase GRP78 transcription in a manner dependent on the ER stress response element sequences in the gene promoter, and independent of the presence or absence of ER stress (24). In the present study, we found that binding of ATF6α to the promoter region of Grp78 was inhibited by C/EBPβ overexpression (Figure 7F). Furthermore, C/EBPβ bound to the promoter region of Grp78 (Supplemental Figure 9), and the C/EBPβ mutation, which eliminates its DNA-binding activity, did not have any effect on the ATF6α-induced activation of the Grp78 promoter (Supplemental Figure 10). These results suggest that DNA-binding activity is necessary for C/EBPβ to inhibit the ATF6α-induced activation of the Grp78 promoter. It is likely that accumulated C/EBPβ binds to the promoter region of Grp78 and occupies the ATF6α-binding sites. ATF6 is a member of the family of proteins with a bZIP domain, which form heterodimers with other bZIP transcription factors. The C/EBP family of proteins is also composed of bZIP transcription factors that have been shown to bind to ATF family members through the bZIP domain (25–27). Thus, we cannot completely exclude the possibility that accumulated C/EBPβ inhibits the binding of ATF6α to the promoter region of Grp78 as a result of complex formation between C/EBPβ and ATF6α.
C/EBPβ accumulates in the liver of mice fed a diet deficient in methionine and choline, resulting in the development of nonalcoholic steatohepatitis through its effect on the expression of genes involved in lipid synthesis, inflammation, and possibly ER stress (28). Deletion of Cebpb in diabetic mice reduces blood glucose levels by inhibiting gluconeogenesis in the liver and lipolysis in white adipocytes (29, 30). Such ablation of C/EBPβ also increases IRS1 expression in skeletal muscle (31). It is thus possible that systemic insulin resistance might be ameliorated by reducing the expression level of C/EBPβ in the target tissues of insulin.
In summary, we have shown that C/EBPβ not only inhibits the biosynthesis of insulin in pancreatic β cells, but also controls ER function in these cells by inhibiting the transcriptional induction of the molecular chaperone GRP78. C/EBPβ therefore plays a key role in pancreatic β cell failure. Taking into account the amelioration of peripheral insulin resistance induced by C/EBPβ ablation in addition to the role of this protein in pancreatic β cells, limiting C/EBPβ induction in individuals in the early stages of diabetes or in those with metabolic syndrome might be a promising approach to the prevention or treatment of type 2 diabetes.
Methods
Mice.
A pBlueScript vector containing the promoter of rat Ins2 was provided by P.L. Herrera (University of Geneva Medical School, Geneva, Switzerland; ref. 10). We isolated a C/EBPβ cDNA from mouse embryonic fibroblasts by PCR amplification and inserted it into the ClaI and EcoRI sites of the vector downstream of the insulin gene promoter. The resulting construct was linearized and microinjected into pronuclear oocytes of C57BL/6J mice in the Laboratory for Animal Resources and Genetic Engineering at Riken. The resulting offspring were screened for transgene transmission by PCR analysis and Southern hybridization.
A targeting vector based on pKSTKNEOLOXP, which contains a loxP-flanked (floxed) neomycin resistance gene (pGK Neo), was constructed to flank the coding exon of the mouse Cebpb gene with 2 loxP sites (32) (Supplemental Figure 1). The thymidine kinase gene of herpes simplex virus (HSV TK) was included for negative selection of clones with random integration of the targeting vector. E14-1 ES cells were transfected with the linearized targeting vector by electroporation. After double selection with G418 and ganciclovir, 4 homologous recombinants were obtained out of 768 double-resistant clones. Of these 4 homologous recombinants, 2 contained a loxP site upstream of the C/EBPβ exon. The neomycin resistance gene was removed by Cre-mediated recombination from the ES clones with a targeted allele to generate the floxed Cebpb allele. ES cell clones with a heterozygous floxed Cebpb allele were injected into C57BL/6 blastocysts to generate chimeric mice. The floxed Cebpb allele was transmitted through the germline, and Cebpbfl/fl mice were generated by intercrossing Cebpb+/fl mice.
Ins-Cre mice were generated as described previously (10).
βCebpb–/– mice were generated by breeding Cebpbfl/fl mice with Ins-Cre mice and were maintained on the C57BL/6J background. Akita mice on the C57BL/6J background were obtained from Japan SLC and were crossed with βCebpb–/– mice to obtain Akita;βCebpb+/– offspring. We also obtained Lepr+/– mice on the C57BL/KsJ background from Clea Japan and crossed them with βCebpb–/– mice on the C57BL/6J background to obtain Lepr+/–;βCebpb+/– offspring on a mixed C57BL/6J × C57BL/KsJ background.
Mice were maintained on a 12-hour light, 12-hour dark cycle and fed normal chow from the time of weaning (3 weeks old), as described previously (33, 34). Blood glucose and plasma insulin concentrations were determined as described previously (33, 34). All experiments were performed with male mice. The Animal Ethics Committee of Kobe University Graduate School of Medicine reviewed and approved the present studies.
Real-time RT-PCR analysis.
Real-time RT-PCR analysis was performed as described previously (35). Primers were as follows: mouse C/EBPβ, sense, 5′-ACCGGGTTTCGGGACTTGA-3′; antisense, 5′-GTTGCGTAGTCCCGTGTCCA-3′; mouse GRP78, sense, 5′-TGGAGTTCCCCAGATTGAAG-3′; antisense, 5′-CCTGACCCACCTTTTTCTCA-3′.
Immunoblot analysis.
Lysates of isolated islets were prepared as described previously (34, 36) and probed with antibodies to C/EBPβ, GRP78, CHOP, and GFP (Santa Cruz Biotechnology Inc.); phosphorylated c-Jun, phosphorylated eIF2α, and cleaved caspase-3 (Cell Signaling); PCNA (Dako); HA (Roche); and β-actin (Sigma-Aldrich).
Immunostaining and morphometric analysis.
The pancreas was immersed in Bouin’s solution, embedded in paraffin, and sectioned at a thickness of 4–5 μm. Sections were stained with antibodies to insulin and to glucagon (Dako). Immune complexes were detected with secondary antibodies conjugated with either Cy3 or FITC (Jackson ImmunoResearch Laboratories). Quantitation of β cell mass was described previously (34, 36).
Analysis of internucleosomal DNA fragmentation.
Groups of 50 islets isolated from mice at 8 weeks of age were cultured in RPMI 1640 medium for 24 hours, after which DNA fragmentation was examined by ligation-mediated PCR, as described previously (37).
Electron microscopy.
Two Lepr+/– and Lepr–/– mice were anesthetized with pentobarbital (25 mg/kg i.p.) and subjected to cardiac perfusion with 0.1 M sodium phosphate buffer (pH 7.2) containing 2% glutaraldehyde and 2% paraformaldehyde. The pancreas was excised from each mouse, cut into small pieces, and immersed overnight in the same fixative. The tissue was then exposed to 2% osmium tetroxide, stained with 2% uranyl acetate, dehydrated with ethanol, and embedded in Epon812 (TAAB). For light microscopy, 1-μm-thick sections were cut and stained with toluidine blue. For electron microscopy, thin sections were stained with uranyl acetate and lead citrate before examination with a Hitachi 7100 electron microscope (Hitachi).
DNA microarray analysis.
Total cellular RNA was isolated from WT and TG1 mice with the use of an RNeasy kit (Qiagen). The RNA was then subjected to transcriptome/DNA microarray analysis using an oligo-DNA chip (AceGene Mouse Oligo Chip 30 K; HitachiSoft). All the experiments were done according to the manufacturer’s instructions.
Infection with a retrovirus encoding C/EBPβ.
Platinum-E (PLAT-E) ecotropic packaging cells were transfected with the retroviral vector pMX-C/EBPβ (encoding mouse C/EBPβ) with the use of the LipofectAMINE PLUS reagent (Invitrogen). Recombinant retroviruses released into the culture medium were harvested 48 hours after transfection and added to MIN6 or INS1 cells cultured in DMEM supplemented with 15% FBS or in RPMI 1640, respectively. After culture for 8 hours, the infected cells were subjected to selection in medium containing puromycin.
Construction of plasmids.
The plasmid pcDNA-ATF6 (encoding full-length human ATF6α) was provided by K. Mori (Kyoto University, Kyoto, Japan; ref. 38). A cDNA encoding C/A ATF6α was constructed by PCR-mediated amplification of the region corresponding to amino acids 1–1,177 of ATF6α together with a stop codon and was inserted into the XmaI-XhoI sites of the pMX vector.
Luciferase assay.
Based on examination of the published sequence of human GRP78, we amplified by PCR a 311-bp fragment of the GRP78 promoter (nucleotides –304 to +7 relative to the transcription start site) from genomic DNA of HeLa cells and cloned it into the KpnI-XhoI sites of the pGL3-Basic vector (Promega), which contains the coding sequence for firefly luciferase but lacks a eukaryotic promoter or enhancer elements.
The GRP78-luciferase reporter plasmid, as well as vectors for C/EBPβ, ATF6α, and C/A ATF6α, were introduced into INS1 cells. Cells were exposed to tunicamycin or assayed for luciferase activity at 48 hours after transfection.
ChIP analysis.
INS1 cells were fixed with 1% formaldehyde for 30 minutes at room temperature and then subjected to ultrasonic disruption in a solution containing a protease inhibitor cocktail. The lysates were centrifuged to remove debris, diluted 1:10 with a solution containing 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), and 150 mM NaCl, and incubated for 2 hours at 4°C with salmon sperm DNA conjugated to protein A–Sepharose. The Sepharose beads were removed by centrifugation, and the lysates were subjected to immunoprecipitation by incubation at 4°C, first overnight with antibodies to ATF6 or normal mouse IgG (Santa Cruz Biotechnology Inc.), and then for 1 hour with salmon sperm DNA conjugated to protein A–Sepharose. The precipitates were washed and then subjected to extraction for 4 hours at 65°C with 1% SDS in 100 mM NaHCO3, after which proteins were digested with proteinase K and the remaining DNA was purified with the use of a QIAquick PCR purification kit (Qiagen). The DNA was finally subjected to PCR with the GRP78-specific primers 5′-GGCGGCCGTTAAGAATGACCAGTAG-3′ (sense) and 5′-GACGCTTACCTCTCAAACCCGCAAA-3′ (antisense).
Statistics.
Data are presented as mean ± SEM and were compared by ANOVA followed by 2-tailed Student’s t tests. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank K. Mori for helpful discussion and S. Hirahara, M. Nagano, and M. Oya for technical assistance. This work was supported by a grant for the 21st Century COE Program “Center of Excellence for Signal Transduction Disease: Diabetes Mellitus as Model” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) to M. Kasuga; a grant for the Cooperative Link of Unique Science and Technology for Economy Revitalization (CLUSTER) from MEXT to M. Kasuga; and a Grant-in-Aid for Creative Scientific Research from MEXT to M. Kasuga.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:115–126 (2010). doi:10.1172/JCI39721.
Tsuneyasu Kaisho’s and Takashi Tanaka’s present address is: RIKEN Research Center for Allergy and Immunology, Yokohama, Japan.
Masato Koike’s and Yasuo Uchiyama’s present address is: Department of Cell Biology and Neurosciences, Juntendo University Graduate School of Medicine, Tokyo, Japan.
References
- 1.Leahy JL. Natural history of beta-cell dysfunction in NIDDM. Diabetes Care. 1990;13(9):992–1010. doi: 10.2337/diacare.13.9.992. [DOI] [PubMed] [Google Scholar]
- 2.Weyer C, Bogardus C, Mott DM, Pratley RE. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Invest. 1999;104(6):787–794. doi: 10.1172/JCI7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes CJ. Type 2 diabetes-a matter of beta-cell life and death? Science. 2005;307(5708):380–384. doi: 10.1126/science.1104345. [DOI] [PubMed] [Google Scholar]
- 4.Kasuga M. Insulin resistance and pancreatic beta cell failure. J Clin Invest. 2006;116(7):1756–1760. doi: 10.1172/JCI29189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51(suppl 3):S455–S461. doi: 10.2337/diabetes.51.2007.S455. [DOI] [PubMed] [Google Scholar]
- 6.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(pt 3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273(44):28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 8.Lu M, Seufert J, Habener JF. Pancreatic beta-cell-specific repression of insulin gene transcription by CCAAT/enhancer-binding protein beta. Inhibitory interactions with basic helix-loop-helix transcription factor E47. J Biol Chem. 1997;272(45):28349–28359. doi: 10.1074/jbc.272.45.28349. [DOI] [PubMed] [Google Scholar]
- 9.Seufert J, Weir GC, Habener JF. Differential expression of the insulin gene transcriptional repressor CCAAT/enhancer-binding protein beta and transactivator islet duodenum homeobox-1 in rat pancreatic beta cells during the development of diabetes mellitus. J Clin Invest. 1998;101(11):2528–2539. doi: 10.1172/JCI2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshioka M, Kayo T, Ikeda T, Koizumi A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes. 1997;46(5):887–894. doi: 10.2337/diabetes.46.5.887. [DOI] [PubMed] [Google Scholar]
- 13.Coleman DL. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14(3):141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Kim JW, Tang QQ, Li X, Lane MD. Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPbeta. Proc Natl Acad Sci U S A. 2007;104(6):1800–1804. doi: 10.1073/pnas.0611137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Dudenhausen EE, Pan YX, Zhong C, Kilberg MS. Human CCAAT/enhancer-binding protein beta gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J Biol Chem. 2004;279(27):27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, et al. Amino-acid limitation induces transcription from the human C/EBPbeta gene via an enhancer activity located downstream of the protein coding sequence. Biochem J. 2005;391(pt 3):649–658. doi: 10.1042/BJ20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. Differential control of the CCAAT/enhancer-binding protein beta (C/EBPbeta) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J Biol Chem. 2008;283(33):22443–22456. doi: 10.1074/jbc.M801046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277(21):18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 21.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu CY, Xu Z, Kaufman RJ. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1alpha. J Biol Chem. 2003;278(20):17680–17687. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- 23.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273(50):33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo M, Ron D, Miller CP, Habener JF. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad Sci U S A. 1993;90(10):4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez AB, et al. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem J. 2007;402(1):163–173. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283(50):35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman SM, et al. CCAAT/enhancing binding protein beta deletion in mice attenuates inflammation, endoplasmic reticulum stress, and lipid accumulation in diet-induced nonalcoholic steatohepatitis. Hepatology. 2007;45(5):1108–1117. doi: 10.1002/hep.21614. [DOI] [PubMed] [Google Scholar]
- 29.Arizmendi C, Liu S, Croniger C, Poli V, Friedman JE. The transcription factor CCAAT/enhancer-binding protein beta regulates gluconeogenesis and phosphoenolpyruvate carboxykinase (GTP) gene transcription during diabetes. J Biol Chem. 1999;274(19):13033–13040. doi: 10.1074/jbc.274.19.13033. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, et al. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPbeta gene. J Clin Invest. 1999;103(2):207–213. doi: 10.1172/JCI4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, et al. Increased insulin receptor substrate-1 and enhanced skeletal muscle insulin sensitivity in mice lacking CCAAT/enhancer-binding protein beta. J Biol Chem. 2000;275(19):14173–14181. doi: 10.1074/jbc.M000764200. [DOI] [PubMed] [Google Scholar]
- 32.Takeda K, et al. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161(9):4652–4660. [PubMed] [Google Scholar]
- 33.Kido Y, et al. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J Clin Invest. 2000;105(2):199–205. doi: 10.1172/JCI7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto N, et al. PKClambda regulates glucose-induced insulin secretion through modulation of gene expression in pancreatic beta cells. J Clin Invest. 2005;115(1):138–145. doi: 10.1172/JCI22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto M, et al. PKClambda in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest. 2003;112(6):935–944. doi: 10.1172/JCI18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchida T, et al. Deletion of Cdkn1b ameliorates hyperglycemia by maintaining compensatory hyperinsulinemia in diabetic mice. Nat Med. 2005;11(2):175–182. doi: 10.1038/nm1187. [DOI] [PubMed] [Google Scholar]
- 37.Ishihara H, et al. Disruption of the WFS1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13(11):1159–1170. doi: 10.1093/hmg/ddh125. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20(18):6755–6767. doi: 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.