Abstract
Tumor hypoxia is a common microenvironmental factor that adversely influences tumor phenotype and treatment response. Cellular adaptation to hypoxia occurs through multiple mechanisms, including activation of the unfolded protein response (UPR). Recent reports have indicated that hypoxia activates a lysosomal degradation pathway known as autophagy, and here we show that the UPR enhances the capacity of hypoxic tumor cells to carry out autophagy, and that this promotes their survival. In several human cancer cell lines, hypoxia increased transcription of the essential autophagy genes microtubule-associated protein 1 light chain 3β (MAP1LC3B) and autophagy-related gene 5 (ATG5) through the transcription factors ATF4 and CHOP, respectively, which are regulated by PKR-like ER kinase (PERK, also known as EIF2AK3). MAP1LC3B and ATG5 are not required for initiation of autophagy but mediate phagophore expansion and autophagosome formation. We observed that transcriptional induction of MAP1LC3B replenished MAP1LC3B protein that was turned over during extensive hypoxia-induced autophagy. Correspondingly, cells deficient in PERK signaling failed to transcriptionally induce MAP1LC3B and became rapidly depleted of MAP1LC3B protein during hypoxia. Consistent with these data, autophagy and MAP1LC3B induction occurred preferentially in hypoxic regions of human tumor xenografts. Furthermore, pharmacological inhibition of autophagy sensitized human tumor cells to hypoxia, reduced the fraction of viable hypoxic tumor cells, and sensitized xenografted human tumors to irradiation. Our data therefore demonstrate that the UPR is an important mediator of the hypoxic tumor microenvironment and that it contributes to resistance to treatment through its ability to facilitate autophagy.
Introduction
Solid tumor microenvironments are characterized by extreme heterogeneities in oxygenation that arise primarily due to a poorly developed and/or poorly functioning vascular network. Gradients of oxygenation exist around individual perfused vessels and range from normal values (~5%) near the vessel wall to complete anoxia in perinecrotic regions. Transient changes in blood flow also lead to strong temporal changes in oxygenation within specific tumor regions (1). The percentage of viable tumor cells within individual tumors with otherwise similar clinical features varies tremendously among patients (2) and is clinically important because high levels of tumor hypoxia correlate with poor prognosis and a more aggressive phenotype (2–6). The source of the variability in hypoxia among different tumors is likely due to the acquisition of changes that drive increased hypoxia tolerance (7). This was first demonstrated by the selective outgrowth of hypoxia-resistant p53-deficient cells (8). Since this time, several hypoxic signaling pathways have been identified that influence hypoxia tolerance and other phenotypes relevant in cancer. This includes activation of the HIF transcription factors (HIF1, HIF2, HIF3), which induce a large number of genes involved in glycolysis, angiogenesis, pH regulation, and cell motility (6, 9). Consequently, HIF1 activity is an important contributor to both cell proliferation under hypoxic conditions (e.g., through glycolysis) as well as overall tumor growth.
A number of HIF-independent signaling pathways are also sensitive to changes in tumor oxygenation (10) including the unfolded protein response (UPR), an evolutionarily conserved pathway that responds to ER stress. The UPR is activated by the coordinate action of 3 separate ER stress sensors that reside in the ER membrane, PKR-like ER kinase (PERK, also known as EIF2AK3), IRE-1, and activating transcription factor 6 (ATF6) (11). These sensors are activated by a common mechanism that involves sequestration of BIP by misfolded ER proteins away from the luminal domains of the sensors. PERK is a kinase that phosphorylates the serine 51 residue of eukaryotic initiation factor 2α (eIF2α) to inhibit the initiation step of mRNA translation. In addition to a general inhibition in protein synthesis, this event enables the selective translation of a subset of transcripts including that encoding the ATF4 transcription factor (12, 13). Phosphorylation of eIF2α and inhibition of mRNA translation is transient because ATF4 induces a second transcription factor, CHOP (also known as GADD153), which in turn regulates expression of the eIF2α phosphatase, GADD34 (also known as MyD116) (14), completing a negative feedback loop to restore protein synthesis. IRE-1 has both kinase activity, which signals through JNK, and an endonuclease activity, which removes an intron in the transcript that codes for the transcription factor XBP1 (15). ATF6 is a transcription factor that is released from the membrane by proteolytic cleavage after ER stress (16). The activation of ATF4, XBP1, and ATF6 transcription factors by each arm of the UPR leads to the induction of a large number of genes involved in increasing folding capacity in the ER and removing misfolded proteins so that they can be degraded in the cytoplasm. Exposure to severe hypoxia (<0.02% O2) leads to rapid activation of PERK, phosphorylation of eIF2α, inhibition of mRNA translation, and activation of ATF4 (17–20). Hypoxic activation of PERK is important because PERK-knockout mouse embryonic fibroblasts (MEFs) or MEFs with a knockin eIF2α mutant allele containing a serine to alanine mutation at position 51 (S51A) show increased cell death during hypoxia and produce slow-growing tumors with reduced regions of viable hypoxia (21). Similarly, IRE-1 is also activated during hypoxia (22) and loss of XBP1 reduces hypoxia tolerance and tumor growth (22, 23).
Recently, several reports have indicated that hypoxia can activate a lysosomal degradation pathway known as autophagy, which mediates both selective and bulk degradation of proteins, cytoplasmic content, and organelles. Nonselective autophagy is termed “macroautophagy” (hereafter referred to as “autophagy”), whereas microautophagy, mitophagy, and chaperone-assisted autophagy refer to more selective forms that are directed by other proteins. Autophagy begins with the formation of a double-membrane structure that grows, capturing cytoplasm and organelles, before closing to form an autophagosome. The outer membrane of the autophagosome then fuses with a lysosome, releasing its contents to the degradative enzymes. Autophagy plays an important role during conditions of starvation or metabolic stress by recycling amino acids and nutrients to maintain energy levels, protein synthesis, and essential metabolic processes (reviewed in ref. 24). Autophagy also plays an increasingly recognized role in quality control during hypoxia by removing mitochondria that might otherwise become cytotoxic (25).
Genetic screens in yeast have identified more than 30 autophagy-related genes (ATGs) (26), many of which have known homologs in mammalian cells. This includes the ATG8 gene and its mammalian homolog microtubule-associated protein 1 (MAP1) light chain 3β (MAP1LC3B). MAP1LC3B is produced in a pro-form (pro-MAP1LC3B), which is cleaved by ATG4 into a cytosolic form referred to as MAP1LC3B-I. During autophagy MAP1LC3B-I is conjugated to the lipid phosphatidylethanolamine (referred to as MAP1LC3B-II), which is then inserted into both inner and outer membranes of the growing autophagic vesicle (27, 28). Consequently, autophagy can be followed in cells by monitoring the cellular distribution and processing of MAP1LC3B. Autophagosome formation and lipidation of MAP1LC3B is also strictly dependent on the essential autophagy gene ATG5. Cells deficient in MAP1LC3B or ATG5 are defective in formation of autophagic vesicles and reveal numerous phenotypes related to the role of autophagy in energy homeostasis and removal of protein aggregates (29, 30).
Here we demonstrate that autophagy is localized primarily to hypoxic regions in tumors and identify a previously unknown link between the UPR and regulation of autophagy that involves PERK-dependent transcriptional induction of MAP1LC3B and ATG5. Regulation of autophagy by the PERK/eIF2α/ATF4 arm of the UPR is an important mediator of hypoxia tolerance, and targeting this pathway through genetic or pharmacological approaches reduces tumor hypoxia and sensitizes tumors to treatment with irradiation.
Results
Hypoxia is the primary site for autophagy in tumors.
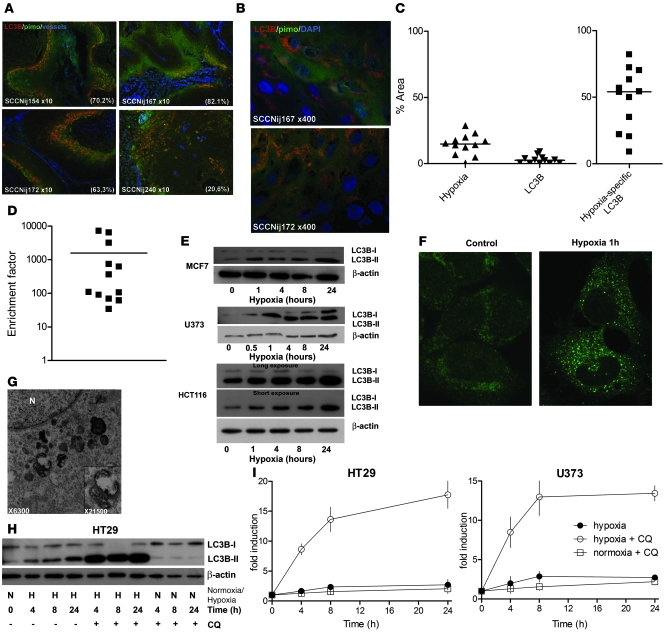
A number of recent studies have demonstrated that human tumor cell lines induce autophagy when exposed in vitro to hypoxia and/or metabolic stress (25, 31–34). To determine whether a similar situation exists in hypoxic tumor regions in vivo, we assessed the prevalence and distribution of autophagy in 12 different human head and neck xenografts established from primary tumors. These xenografts, when maintained by serial passage in vivo, accurately reproduce human tumor microenvironments including regions of tumor hypoxia as assessed using the hypoxia marker pimonidazole (35). Strikingly, we found that autophagy, as evaluated by positive expression of MAP1LC3B, was strongly associated with hypoxic (pimonidazole positive) tumor regions (Figure 1, A and B). Positive areas of MAP1LC3B expression in the 12 tumors ranged from 9% to 82% (median 54%) within the hypoxic regions (Figure 1C), which is dramatically higher than in the whole tumor (median 2.6%). This represents a 33- to 7,096-fold enrichment (median 235-fold) (Figure 1D) in MAP1LC3B expression in the hypoxic regions. It is noteworthy that strong expression of MAP1LC3B staining is observed within the full gradient of hypoxia (the pimonidazole gradient reflects the oxygenation gradient) and is not restricted to the most severe hypoxic (perinecrotic) regions. Strong expression of MAP1LC3B within the hypoxic areas is associated with activation of autophagy as evidenced by a punctate staining pattern characteristic of MAP1LC3B lipidation and insertion into autophagic membranes (Figure 1B).
Figure 1. Autophagy is induced by hypoxia and is primarily present in hypoxic tumor regions.
(A) Immunohistochemical staining of 12 head and neck tumor cell lines (SCCNij) for MAP1LC3B (red), hypoxia (pimonidazole [pimo], green), and vessels (9F1, blue) revealed that MAP1LC3B staining was primarily localized to hypoxic regions. Percentage colocalization of MAP1LC3B and pimonidazole is indicated in the bottom right corner. Representative micrographs are shown at low magnification (×10). (B) Higher magnification (×400) revealed a punctate MAP1LC3B pattern characteristic of autophagosome formation. (C) Quantification of the 12 head and neck tumor cell lines for hypoxia, MAP1LC3B staining, and MAP1LC3B expressed specifically in hypoxic regions showed that MAP1LC3B in tumors is primarily expressed in hypoxic regions of tumors. (D) Enrichment factor (enrichment compared with the statistical probability) on log scale for 12 head and neck tumor cell lines. (E) Immunoblots for MAP1LC3B on MCF7, U373, and HCT116 lysates showed processing and increased expression of MAP1LC3B during hypoxic (<0.02% O2) exposure. (F) Immunohistochemical staining for MAP1LC3B on U373 cells exposed to 1 hour of hypoxia showed redistribution of MAP1LC3B and an increased punctate pattern indicative of induction of autophagy. Original magnification, ×800. (G) Confirmation of autophagy induction during hypoxia by electron microscopy. Numerous autophagosomes were detected after anoxic exposure (8 hours) of U373 cells (original magnification, ×6,300). High magnification (inset, ×21,500) clearly revealed the double membrane, characteristic of autophagosomes. N, nucleus. (H) Immunoblots on HT29 lysates showed rapid activation of MAP1LC3B during hypoxia. CQ addition identified a very high autophagic flux during hypoxia. (I) Flow cytometric assessment of MAP1LC3B levels during hypoxia (filled circles), hypoxia in the presence of CQ (open circles), or normoxic exposure in the presence of CQ (open squares) identified a very high autophagic flux during hypoxia (data are presented as mean ± SEM, n = 3).
Several forms of cell stress expected within tumor microenvironments are capable of inducing autophagy. To test whether hypoxia alone is sufficient to induce autophagy, we monitored the expression and processing of MAP1LC3B in several cancer cell lines of different origin during culture in vitro in complete medium. In HT29, MCF-7, U373, and HCT116 cell lines, hypoxia was sufficient to induce the conversion of MAP1LC3B from its cytosolic form (MAP1LC3B-I) to its lipidated membrane-bound form (MAP1LC3B-II) (Figure 1, E and H). Changes in MAP1LC3B processing and expression were also examined by immunohistochemical staining on U373 cells exposed to hypoxia. In normoxic conditions, MAP1LC3B staining is diffuse with few examples of foci, characteristic of autophagosomes. In contrast, exposure to hypoxia induces a rapid relocalization of MAP1LC3B into a punctate pattern (Figure 1F). Activation of autophagy was confirmed by electron microscopy, in which autophagic vesicles were distinguished from other vesicles by the presence of a double membrane (Figure 1G).
During autophagy, lipidated MAP1LC3B is inserted into both inner and outer membranes of the expanding autophagophores (36). Following fusion with the lysosome, the inner membrane and its contents are exposed to lysosomal degradative enzymes, resulting in partial degradation of MAP1LC3B (29). Surprisingly, despite clear evidence for the activation of autophagy, we found that MAP1LC3B protein levels were maintained or slightly increased during continued hypoxic exposure in each of the 4 cell lines (Figure 1, E and H). To examine changes in MAP1LC3B protein expression more closely, we assessed protein levels during hypoxia in cells incubated with the lysosomotrophic drug chloroquine (CQ). CQ has been shown to inhibit the final step of autophagy (fusion of the lysosome with the autophagosome) (37, 38) and thus prevents MAP1LC3B turnover during conditions that activate autophagy. Under these conditions, the increase in MAP1LC3B, particularly the lipidated MAP1LC3B-II form, was dramatically enhanced during hypoxia but not during normoxic conditions, as assessed by immunoblotting (Figure 1H). This increase was confirmed in both HT29 and U373 by flow cytometry (Figure 1I). Hypoxic exposure leads to a 1.5-fold to 2-fold increase in expression of MAP1LC3B in both lines without CQ, but to an increase of more than 18-fold with CQ. These data not only indicate very high rates of autophagic flux during hypoxia, but also suggest a hypoxia-associated increase in MAP1LC3B protein production.
It is noteworthy that induction of autophagy during hypoxia was observed in both HT29 and HCT116 cells (Figure 1E), as both cell lines are deficient in BNIP3 (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40027DS1) due to promoter methylation (39). BNIP3 is a HIF-1 target gene that has previously been implicated in the induction of autophagy during hypoxia (31). We also found that forced expression of BNIP3 did not induce autophagy in HCT116 (data not shown). Together these data indicate that hypoxia can regulate MAP1LC3B and autophagy through a BNIP3-independent mechanism.
MAP1LC3B and ATG5 mRNA levels increase during hypoxic exposure.
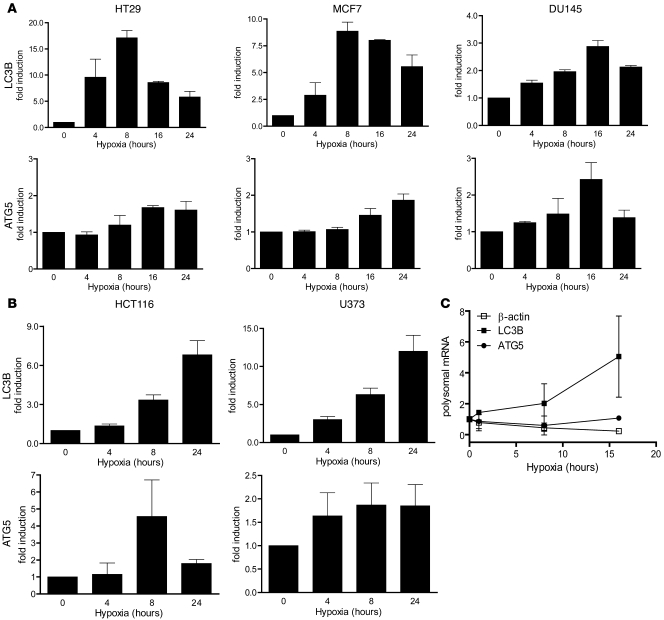
As shown, MAP1LC3B protein synthesis (and degradation) is highly induced during hypoxia. However, relatively little is known regarding the transcriptional regulation. We assessed the expression profiles of MAP1LC3B and other known autophagy genes within a microarray dataset carried out in HT29, DU145, and MCF7 exposed to different durations of hypoxia. Our analysis showed that MAP1LC3B transcript levels rise was significantly induced in each of the 3 cell lines (Figure 2A). In addition, another essential factor during hypoxia, ATG5, was found to be increased (Figure 2A). Analysis of other autophagy genes on the same array revealed no such pattern or were not pursued in this analysis (Supplemental Figure 2). The increases in MAP1LC3B and ATG5 transcript levels during hypoxia were validated using quantitative RT-PCR (Figure 2B) and confirmed in 2 additional cell lines, U373 and HCT116 (Figure 2C), which showed strong protein increases. Although the relative increase in mRNA content of MAP1LC3B and ATG5 differed among the various lines, a similar overall trend was observed with rapid induction during hypoxic exposure. Hypoxia is known to inhibit overall protein translation, yet some mRNA species are known to overcome this inhibition (40). To investigate whether MAP1LC3B and ATG5 can be efficiently translated during hypoxia, we performed polysomal analysis on HELA cells during hypoxia as described previously (19). Although there was a significant reduction in β-actin translation, ATG5 translation remained at levels similar to normoxic conditions. As expected form the protein data, translation of MAP1LC3B was increased during hypoxia (Figure 2C), indicating sufficient levels of translation to ensure increased protein production.
Figure 2. Regulation of autophagy genes MAP1LC3B and ATG5 during hypoxic exposure.
(A) Validation of the obtained microarray results by quantitative real-time PCR confirmed MAP1LC3B and ATG5 upregulation during hypoxia (<0.02% O2). n = 3, mean ± SEM. (B) Confirmation of MAP1LC3B and ATG5 gene regulation by quantitative real-time PCR during hypoxia in HCT116 and U373 cells. n = 3, mean ± SEM. (C) mRNA abundance in polysomal fractions indicated translation of MAP1LC3B and ATG5 during hypoxia (<0.02% O2). n = 2, mean ± SD.
Increased MAP1LC3B and ATG5 expression during hypoxia is regulated by PERK-dependent activation of ATF4 and CHOP.
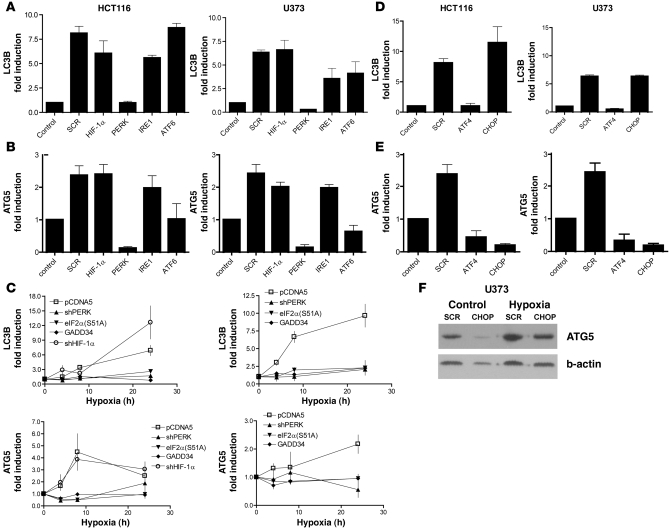
To explore the mechanisms behind MAP1LC3B and ATG5 regulation we used siRNA to interfere with known hypoxia-responsive signaling pathways, including HIF-1α or each of the 3 known sensors of ER stress that activate different arms of the UPR: ATF-6, IRE-1, and PERK. Knockdown efficiency varied between 60% and 90% (Supplemental Figure 3A) but was effective in causing a significant reduction in downstream signaling through each of these pathways (Supplemental Figure 3, B and C). Knockdown of HIF-1α had no effect on either MAP1LC3B or ATG5 induction. However, knockdown of the UPR sensor PERK completely inhibited hypoxic induction of both MAP1LC3B and ATG5 (P < 0.05; Figure 3, A and B). Interestingly, loss of PERK also significantly reduced basal expression of ATG5. Knockdown of ATF6 also prevented hypoxic induction of ATG5 (P < 0.05) but did not affect its basal expression or the hypoxic regulation of MAP1LC3B. Knockdown of the other UPR sensor, IRE-1, did not affect expression of either MAP1LC3B or ATG5 in these cell lines.
Figure 3. Induction of MAP1LC3B and ATG5 mRNA is dependent on PERK signaling.
(A and B) Knockdown of HIF-1α, PERK, IRE-1, or ATF6 in HCT116 and U373 cells showed that hypoxia-dependent induction (<0.02% O2 for 24 hours) of MAP1LC3B (A) and ATG5 (B) is dependent on PERK signaling. Samples were normalized to normoxic exposure (control) and compared with scrambled (SCR) siRNA knockdown. n = 3 mean ± SEM. (C) MAP1LC3B and ATG5 dependency on UPR signaling was further confirmed in HCT116 and U373 cells hampered in UPR signaling. n = 3, mean ± SEM. (D) Knockdown of ATF4 prevented upregulation of MAP1LC3B mRNA during hypoxia. Control was normoxic exposure. n = 3, mean ± SEM. (E) Knockdown of ATF4 and its downstream transcription factor CHOP prevented ATG5 mRNA upregulation during hypoxia. n = 3, mean ± SEM. (F) CHOP knockdown reduced ATG5 protein expression during normoxia and hypoxia as detected by immunoblotting.
These data suggest that the PERK arm of the UPR is required for hypoxia-mediated induction of MAP1LC3B and ATG5. To test whether activation of this pathway is also sufficient for MAP1LC3B expression, we treated the same cell lines with well-described UPR-activating agents (thapsigargin, tunicamycin, or dithiotreitol). Each of these treatments led to substantial increases in both MAP1LC3B and ATG5 (Supplemental Figure 4).
To further investigate the dependence of MAP1LC3B and ATG5 mRNA upregulation on PERK signaling during hypoxia, we used isogenic HCT116 and U373 cells engineered with various tetracycline/doxycycline constructs that interfere with the ability of PERK to phosphorylate eIF2α. Isogenic cell lines (18) defective in PERK signaling were created in 3 different ways including inducible expression of a shRNA against PERK, overexpression of the non-phosphorylatable eIF2α allele [eIF2α(S51A)], or overexpression of the constitutively active mutant ΔGADD34 that promotes dephosphorylation of eIF2α (41). Each of these 3 approaches effectively blocked the ability of hypoxic cells to signal through the PERK arm of the UPR (Supplemental Figure 5) after 24 hours of tetracycline pretreatment. Consistent with the transient siRNA experiments, hypoxic induction of MAP1LC3B was not observed in any of the cell lines with defects in PERK signaling (P < 0.05 in HCT116 at 24 hours compared with pCDNA5, P < 0.05 in U373 at 4, 8, and 24 hours compared with pCDNA5; Figure 3C). ATG5 displayed a similar UPR dependency (P < 0.05 in HCT116 at 4 and 8 hours, P < 0.05 in U373 at 24 hours). Furthermore, as observed with transient knockdown, stable knockdown of HIF-1α (shHIF-1α) (Supplemental Figure 6B) in these same cell lines had no effect on MAP1LC3B or ATG5 mRNA levels.
The transcription factors ATF4 and CHOP are activated following PERK phosphorylation of eIF2α and are strongly induced by hypoxia (12, 13). ATF4 expression is induced due to its selective translation during hypoxia, which is mediated by upstream ORFs in its 5′UTR, whereas CHOP is at least partially transcriptionally induced by ATF4 (42). In both cell lines, transient knockdown of ATF4 (Supplemental Figure 3A) prevented MAP1LC3B induction during hypoxia (P < 0.05; Figure 3D), but MAP1LC3B induction was not prevented by CHOP knockdown. These data are in agreement with the recent finding that proteasomal stabilization of ATF4 upregulates MAP1LC3B (43). In contrast, both basal and hypoxia-induced expression of ATG5 are significantly reduced following knockdown of either ATF4 or CHOP (P < 0.05; Figure 3E). Since knockdown of ATF4 also leads to a reduction in CHOP in these cells (Supplemental Figure 3B), these data suggest that CHOP, rather than ATF4, may regulate ATG5. Loss of ATG5 mRNA following knockdown of CHOP also led to a reduction in ATG5 protein (Figure 3F).
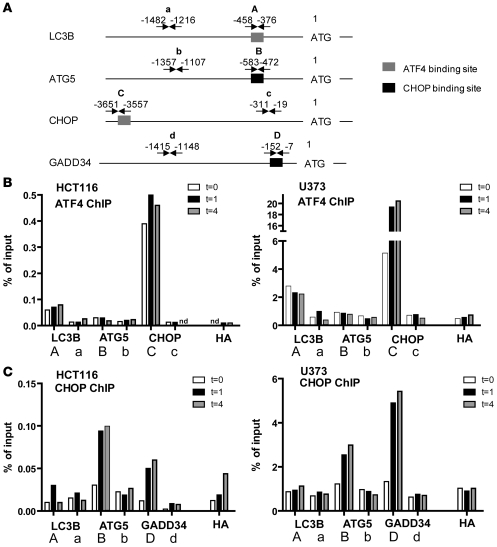
To investigate whether ATF4 and CHOP are capable of directly transactivating MAP1LC3B and ATG5, respectively, we performed ChIP assays followed by promoter-specific quantitative real-time PCR during normal oxygen concentration (0 hours), or hypoxia (< 0.02% O2) after 1 and 4 hours. The ATF4 ChIP demonstrated enrichment of MAP1LC3B (–458 to –376 from start codon) and CHOP (–3,651 to –3,557, positive control) in both HCT116 and U373 cells, but not of ATG5 or different regions of the respective promoters (Figure 4A). Enrichment was observed under both basal and hypoxic conditions. Transcription factor binding analysis using Genomatix software indicated an ATF4-binding consensus (GTGACGCG) in the analyzed part of the promoter. Furthermore, we also found a strong enrichment of CHOP binding to the ATG5 promoter (region –583 to –472) following exposure to 1 or 4 hours of hypoxia in both cell lines (Figure 4B), similar to that for the known CHOP target gene GADD34 (region –152 to –7). Using ChIP with hypoxia-regulated transcription factors ATF6 and HIF-1α did not show enrichment of MAP1LC3B or ATG5 promoter above background (data not shown). These data identify MAP1LC3B and ATG5 as direct targets of the PERK-dependent transcription factors ATF4 and CHOP, respectively.
Figure 4. The UPR transcription factors ATF4 and CHOP bind the MAP1LC3B and the ATG5 promoters, respectively.
(A) Locations of primer pairs used for quantitative real-time PCR analysis following ChIP. (B) ATF4 ChIP analysis revealed enrichment of putative ATF4 transcription factor binding sites in MAP1LC3B (A primer) but not at a region further upstream (a primer) in both HCT116 and U373 cell lines. Enrichment of the CHOP promoter was used as a positive control for ATF4. (C) CHOP ChIP enriched the promoters of ATG5 (B primer) but not at a region further upstream (b primer) in both lines. Enrichment of GADD34 was used as a positive control for CHOP. HA denotes the signal obtained after HA-ChIP followed by PCR for the respective MAP1LC3B promoter or the ATG5 promoter. t = 0, 1, or 4 indicates the amount of time (in hours) exposed to hypoxia (<0.02% O2).
PERK induction of ATF4 during hypoxia is required for maintenance of autophagy.
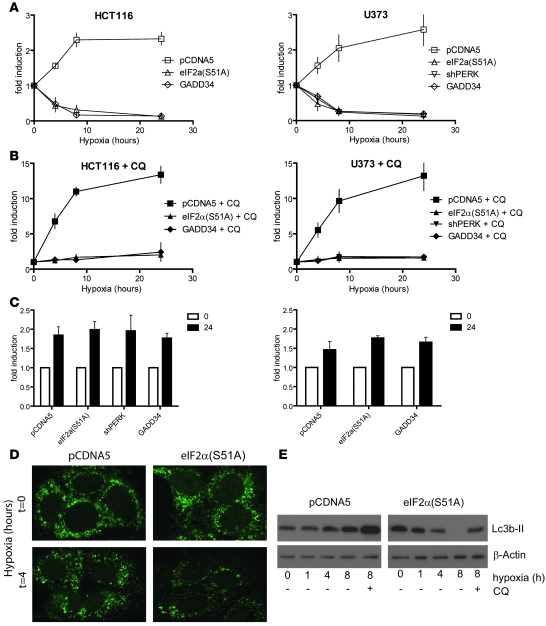
High MAP1LC3B turnover during hypoxia (Figure 1, H and I) indicates a high autophagic flux. We hypothesized that PERK-mediated induction of MAP1LC3B transcription during hypoxia functions as a source of MAP1LC3B replenishment during extended hypoxic exposures. To test this idea, we used flow cytometry to quantify MAP1LC3B expression in individual cells as a function of time during hypoxic exposure. Similar to what was observed in immunoblots and by flow cytometry (Figure 1, E and H), we found that MAP1LC3B protein increased during hypoxia (Figure 5A). However, cells deficient in eIF2α (shPERK, GADD34, eIF2α) signaling displayed a steady loss of MAP1LC3B protein during hypoxia. In addition, in U373 and MCF7 cells transfected with siRNA targeting PERK, we were unable to detect any remaining MAP1LC3B protein after 24 hours of hypoxia (data not shown). As shown previously, addition of CQ prevented MAP1LC3B turnover (degradation in the lysosome) and led to robust accumulation (up to 15-fold compared with normoxic control) of MAP1LC3B in U373 and HCT116 cells (Figure 5, B and C). Addition of CQ to cells deficient in eIF2α signaling displayed no such accumulation, yet the observed decrease (Figure 5A) was prevented. These data suggest that autophagy is activated by hypoxia in both WT and eIF2α-deficient cells, leading to an increase in the rate of MAP1LC3B degradation. In eIF2α-deficient cells, this led to loss of the MAP1LC3B protein due to the lack of a compensatory induction at the transcript level. Indeed, we found that cells deficient in eIF2α(S51A) showed no defect in their ability to form autophagosomes during short exposures to hypoxia (Figure 5D), although overall protein levels were reduced by 4 hours. In addition, immunoblotting showed that addition of CQ to cells expressing eIF2α(S51A) prevented MAP1LC3B loss during hypoxia, indicating an increased autophagic flux in these cells (Figure 5E). Thus, ATF4-mediated induction of MAP1LC3B appears to function to replenish MAP1LC3B levels during extended hypoxic exposures characterized by high autophagic flux but is not required for basal MAP1LC3B expression or activation of autophagy.
Figure 5. UPR signaling is required for maintenance of autophagy during hypoxia.
(A) Inhibition of UPR signaling caused depletion of MAP1LC3B protein during hypoxic exposure as measured by flow cytometry. (B) Addition of CQ led to the accumulation of MAP1LC3B and prevented depletion of MAP1LC3B in UPR-deficient cells during hypoxia. (C) Addition of CQ to cells exposed to normoxia led to a 1.5- to 2-fold accumulation of MAP1LC3B. All data are n = 3, mean ± SEM. (D) Immunohistochemical staining for MAP1LC3B of HCT116-pCDNA5 and –pCDNA5-eIF2α(S51A) cells under normal conditions or after 4 hours of hypoxia (<0.02% O2). Original magnification, ×600. (E) Immunoblots for MAP1LC3B on HCT116-pCDNA5 and –pCDNA5-eIF2α(S51A) lysates during hypoxia (<0.02% O2). CQ was added as a control for autophagic flux.
The PERK/eIF2a/ATF4 arm of the UPR is required for autophagy in hypoxic regions of tumors.
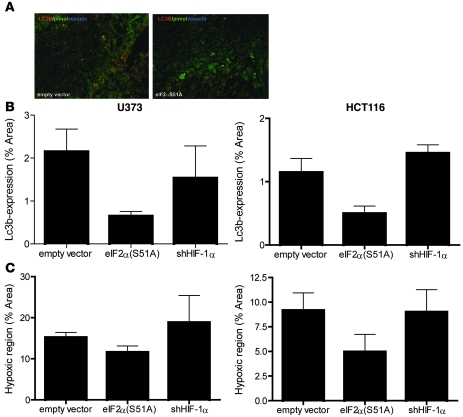
These in vitro data suggest an essential role for UPR signaling to maintain MAP1LC3B levels and autophagy during hypoxia. To investigate this in a more relevant tumor setting, we assessed MAP1LC3B and autophagy in tumors derived from cell lines with defects in either PERK or HIF hypoxia response pathways. Disruption of PERK signaling [eIF2α(S51A)] was confirmed by diminished CHOP and MAP1LC3B levels during hypoxia, and HIF-1α knockdown was confirmed by quantitative RT-PCR (Supplemental Figure 6). HCT116 and U373 xenografts derived from eIF2α(S51A) cells produced tumors with decreased MAP1LC3B expression (Figure 6, A and B). In comparison, inhibition of HIF-1 signaling had no effect on MAP1LC3B expression in hypoxic regions (Figure 6B). Interestingly, eIF2α(S51A) tumors also contained reduced regions of viable hypoxic tumor compared with WT controls (Figure 6C). This decrease was not due to differences in angiogenesis, as PERK-deficient and control tumors revealed similar vessel densities (9F1 immunohistochemistry; data not shown). These data support a role for PERK-dependent autophagy in mediating hypoxia tolerance.
Figure 6. UPR signaling is required for MAP1LC3B expression in hypoxic areas of tumor xenografts.
(A) Representative images of control (pCDNA3) of UPR-inhibited pCDNA3-eIF2α(S51A) xenografts. MAP1LC3B is displayed in red, hypoxia (pimonidazole) in green, and vessels (9F1) in blue. Original magnification, ×10. (B) Quantification of MAP1LC3B expression and hypoxia showed decreased expression of MAP1LC3B in pCDNA3-eIF2α(S51A) xenografts. Knockdown of HIF-1α had no effect on MAP1LC3B expression in hypoxic regions. n = 4, mean ± SEM. (C) Expression of pCDNA3-eIF2α(S51A) decreased the hypoxic regions in xenografts. Knocking down HIF-1α had no effect. n = 4, mean ± SEM.
Inhibition of autophagy sensitizes cells to hypoxia.
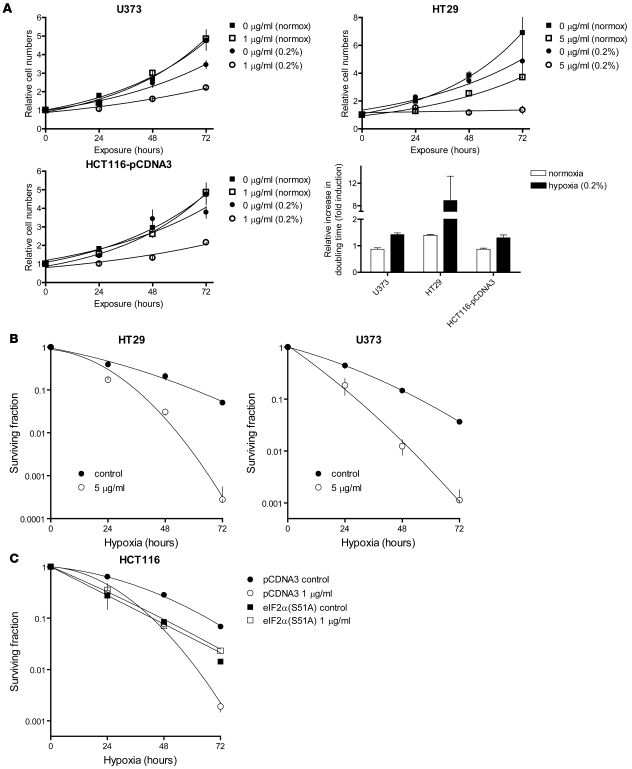
Several previous studies have demonstrated that PERK-, eIF2α-, and ATF4-defective cells are sensitive to hypoxia-induced cell death (reviewed in ref. 10). Although our data are consistent with a role for MAP1LC3B and autophagy in this effect, PERK regulates a large number of other proteins through both translational and transcriptional mechanisms that could also contribute to hypoxia tolerance. To more directly examine the role of autophagy in maintaining cell viability during hypoxia, we monitored both proliferation and cell survival in the presence of pharmacological inhibitors of autophagy. For proliferation studies, we cultured cells under conditions of moderate hypoxia that support continued proliferation (0.2% O2). As expected, moderate hypoxia alone led to decreased proliferation in the cell lines tested (Figure 7A). Addition of CQ caused a further decrease in proliferation during hypoxia but had little or no effect on normoxic cells (Figure 7A). Similarly, autophagy inhibition with 3-methyladenine (3-Ma) also caused a hypoxia-specific decrease in proliferation (Supplemental Figure 7). For cell survival studies, we cultured cells under condition of severe hypoxia (<0.02% O2) sufficient to directly cause cell death (44). Exposure to CQ (Figure 7B) or 3-Ma (Supplemental Figure 8) at concentrations 10-fold lower than those required to affect the survival of normoxic cells (data not shown) caused a significant increase in cell death during hypoxia. Importantly, CQ exposure did not lead to further hypoxic sensitization of eIF2α(S51A) cells, which were already defective in MAP1LC3B induction during hypoxia (Figure 7C). This suggests that the enhanced hypoxic sensitivity of cells deficient in PERK/eIF2α/ATF4 signaling is directly related to defects in autophagy.
Figure 7. Inhibition of autophagy by CQ addition sensitizes cells to hypoxia.
(A) Growth curves under normoxic and moderately hypoxic (0.2% O2) conditions showed that CQ addition selectively inhibited cell proliferation in U373, HT29, and HCT116 cells under hypoxic conditions (0.2% O2). Lower right: Quantification of the CQ-mediated inhibition of cell proliferation during normoxic and hypoxic conditions. n = 3, mean ± SEM. (B and C) Clonogenic survival after hypoxic (<0.02% O2) exposure with or without the addition of CQ in HT29 and U373 cells (B) or HCT116-pCDNA3 and HCT116–pCDNA3-eIF2α(S51A) cells (C). n = 3, mean ± SEM.
Blockade of hypoxia-induced autophagy sensitizes cells to hypoxia and tumors to treatment.
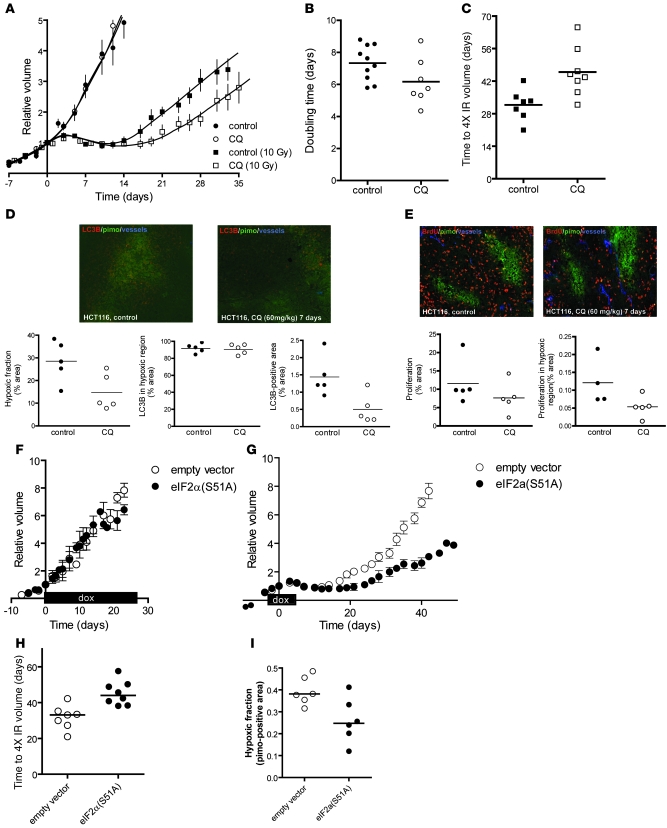
Since tumor hypoxia is known to cause treatment resistance with radiation, we tested whether inhibition of autophagy with CQ would reduce the number of viable radioresistant hypoxic cells and thus sensitize tumors to treatment. We found that treatment of established HCT116 xenografts with CQ (60 mg/kg i.p.) caused no measurable effect on tumor growth as assessed by changes in tumor size (Figure 8, A and B). This was not unexpected, as hypoxic cells represent a small minority of the tumor, proliferate slowly, and thus do not substantially affect the rates of overall tumor growth. However, small fractions of hypoxic cells are sufficient to cause substantial resistance to treatment (45) and thus are expected to influence overall tumor response to treatment. Indeed, administration of CQ 4 days prior to and 3 days after irradiation significantly sensitized HCT116 tumors to a single subcurative irradiation dose (P = 0.0093; Figure 8, A and C). The increased tumor radiosensitivity was not due to a change in intrinsic cellular radiosensitivity, as CQ did not influence radiosensitivity of HCT116 cells in vitro (Supplemental Figure 9A). Instead, the enhanced tumor radiosensitivity was consistent with a reduction in the fraction of viable radiation-resistant hypoxic cells in the CQ-treated tumors. Pimonidazole staining demonstrated that CQ treatment for only 7 days reduced the fraction of viable hypoxic cells by approximately half (28% to 14%, P = 0.0352; Figure 8D). Although CQ causes an accumulation of MAP1LC3B protein, total levels of positive MAP1LC3B decreased in the treated tumors. This is due to the fact that approximately 90% of MAP1LC3B expression was localized to hypoxic regions, which were substantially decreased in the treated tumors at the time points analyzed.
Figure 8. CQ treatment of tumors decreases the hypoxic fraction and sensitizes to radiation.
(A) Growth curves of HCT116 xenografts: untreated (filled circles; saline, n = 10), treated from day –4 to +3 with CQ (60 mg/kg) (open circles; CQ, n = 7), irradiated at day 0 with a single, tumor-specific dose of 10 Gy (filled squares; saline, 10 Gy, n = 7), and treated from day –4 to +3 with CQ and irradiated with 10 Gy (open squares; CQ, 10 Gy, n = 8). (B) Doubling times of unirradiated tumors treated or untreated with CQ. (C) Time to reach 4-fold irradiated tumor volume. (D) Top: Micrographs of LC3/pimonidazole/vessel staining after 7 days of CQ or saline treatment. Original magnification, ×10. Bottom: Hypoxic fraction, MAP1LC3B/pimonidazole colocalization, and total MAP1LC3B staining in the sections (n = 5). (E) Top: Proliferation (BrdU) and pimonidazole staining of tumor sections after 7 days of CQ or saline treatment (n = 5). Bottom: Quantification of overall proliferation and proliferation in hypoxic regions. (F) HCT116 isogenic xenografts harboring doxycycline-inducible genes [empty vector or eIF2α(S51A)] were established in nude mice. When tumors reached a size of 150 mm3, mice were fed doxycycline (2 g/l) continuously and tumor growth was assessed [empty vector, n = 6; eIF2α(S51A), n = 5; mean ± SEM]. (G) HCT116 isogenic xenografts [empty vector, n = 7; eIF2α(S51A), n = 8] were established in nude mice and irradiated with 10 Gy when they reached 150 mm3 (day 0). Doxycycline (2 g/l) was administered for 7 days (day –4 to +3) to induce the transgene, and tumor size was assessed as a function of time (mean ± SEM). (H) Time for tumors in G to reach 4 times the irradiated volume. (I) The hypoxic fraction in individual isogenic xenografts was determined after 7 days of doxycycline treatment using pimonidazole immunohistochemistry.
In addition to inhibiting autophagy, CQ has been reported to activate ATM and p53 (46–48). Consequently, the increase in radiosensitivity observed in the HCT116 tumors, which have WT p53, might be related to this effect. However, CQ did not affect the radiosensitivity of either HCT116 WT or HCT116 p53–/– isogenic cells in vitro at concentrations that were sufficient to sensitize these same cells to hypoxia-induced cell death (Supplemental Figure 9, A and B). More importantly, we repeated the in vivo experiments to test the effects of CQ treatment on the radiosensitivity of xenografts established from p53 mutant U373 cells. Consistent with the results with p53 WT HCT116 tumors, U373 tumors were significantly more sensitive to irradiation when treated with CQ (Supplemental Figure 9, D and E). These data demonstrate that the ability of CQ to sensitize tumors is unrelated to changes in cellular radiosensitivity and is not dependent on p53 status.
Our in vitro experiments indicated that CQ treatment also reduced proliferation of cells exposed to moderate hypoxia (Figure 7). We therefore assessed proliferation in hypoxic tumor regions by BrdU and pimonidazole double staining. CQ treatment resulted in an overall decrease in proliferation, which was much more pronounced in hypoxic cells with a trend toward significance (P = 0.063; Figure 8E). Tumor cell apoptosis occurred primarily (73%–93%) within hypoxic regions of the tumor (data not shown), but administration of CQ had no effect on overall apoptosis in the tumor or within hypoxic regions (Supplemental Figure 10). Thus, the increased sensitivity of hypoxic tumor cells to CQ treatment is consistent with an effect on autophagy rather than apoptosis.
Finally, we tested whether genetic inhibition of PERK/eIF2α in the HCT116 tumors would recapitulate the effects observed in the same tumors treated with CQ to block autophagy. To do this we used HCT116 isogenic cells, which express a doxycycline-regulated eIF2α(S51A) allele in which we could temporally regulate transgene expression. This allowed us to establish tumors with an intact UPR pathway and then transiently inhibit signaling with doxycycline, thereby mimicking the treatment scenario with CQ. No difference was observed between the empty vector and eIF2α(S51A)-expressing tumors (Figure 8F) when established tumors were treated with doxycycline alone, consistent with what we observed when the same tumors were treated with CQ alone (Figure 8, A and B). In contrast, inhibition of eIF2α phosphorylation in established tumors caused a significant sensitization to irradiation (Figure 8, G and H) and reduced the hypoxic fraction (Figure 8I). The magnitude of the effects observed with transient genetic inhibition of UPR are entirely consistent with that observed with CQ treatment. These data support our observations that UPR regulation of autophagy is an important determinant of hypoxic fraction and treatment response in vivo.
Discussion
Hypoxia is a common feature of solid tumors and a major limiting factor in successful cancer treatment (7). Here we reveal what we believe is a novel and important mode of autophagy regulation by hypoxia, mediated by transcriptional activation of MAP1LC3B and ATG5. We also demonstrate that MAP1LC3B and ATG5 are direct transcriptional targets of the PERK-dependent transcription factors ATF4 and CHOP. Cells with defective PERK signaling failed to induce MAP1LC3B during hypoxia and lost MAP1LC3B protein during hypoxic exposure. More importantly, PERK/ATF4 regulation of MAP1LC3B was a critical regulator of autophagy and hypoxic cell survival in tumors. We found that in 2 different cell line xenografts and 12 head and neck tumor xenografts, MAP1LC3B and autophagy was strongly activated in hypoxic tumor areas. Blockade of UPR signaling or autophagy through genetic and pharmacological methods reduced hypoxia tolerance in vitro, reduced the levels of viable hypoxic cells in tumor xenografts, and sensitized tumors to irradiation. Treatment with inhibitors of autophagy also selectively reduced cell proliferation during mild hypoxia both in vitro and in tumor xenografts. Taken together, our findings indicate that activation of PERK increases hypoxia tolerance and the resistance of tumors to treatment with irradiation by increasing the capacity of hypoxic cells to carry out autophagy. Consequently, targeting PERK signaling or autophagy may be an effective way to eliminate treatment-resistant, hypoxic tumor cells. Interestingly, previous published data have already demonstrated the potential beneficial effects of targeting autophagy in combination with other standard treatments (49).
It is not yet clear how autophagy exerts a protective effect on cell proliferation and survival during hypoxia. During conditions of stress and starvation, cellular organelles and cytoplasmic content are degraded, thereby enabling the cells to recycle amino acids and nutrients to maintain protein synthesis and ATP generation (24, 50). Since energy homeostasis is compromised during hypoxia, autophagy may function as an important source of metabolic requirements to sustain cell survival. Survival of cells after metabolic stress induced by glucose or amino acid deprivation are similarly dependent on autophagy (51, 52). However, autophagy also functions as a cytoplasmic quality control mechanism to eliminate protein aggregates and damaged organelles (24, 53). Defects in autophagy cause accumulation of cytoplasmic inclusion bodies and protein aggregates in the cytoplasm, causing toxicity (54–57). During carcinogenesis autophagy suppresses tumor progression through stabilization of the genome (58, 59), perhaps related to the inability to clear the cell of damaged organelles or toxic protein aggregates. Our results, which link PERK signaling to autophagy through regulation of MAP1LC3B and ATG5, are interesting in this regard since accumulation of unfolded or misfolded proteins in the ER acts as the initial signal for UPR activation. Thus, PERK may stimulate autophagy during hypoxia to help clear the ER of toxic protein aggregates that are responsible for its activation. Several ER stress–inducing agents have previously been shown to induce autophagy in both yeast and mammalian cells. Additionally, both the PERK/eIF2α (60–62) and IRE-1 (63, 64) arms of the UPR have been implicated in autophagy regulation. In yeast, which lacks both PERK and ATF6, IRE-1 promotes an ER-selective form of autophagy that mediates removal of the ER and misfolded protein aggregates. In mammalian cells, PERK signaling is required for autophagy following expression of the expanded polyglutamine 72 repeat (60) and following exposure to sorafenib and vorinostat (61). Similar to what we report for hypoxia, PERK-dependent stimulation of autophagy is protective in these examples.
Although previous studies have implicated UPR signaling in autophagy, no direct links between the UPR transcriptional program and autophagy have been reported during hypoxia. Recently it was shown that lack of ATF4 decreased MAP1LC3B expression in bortezemib-treated cells (43).
To our knowledge, our study provides the first direct link implicating the PERK-dependent transcription factors ATF4 and CHOP in the transcriptional activation of MAP1LC3B and ATG5 during hypoxia. MAP1LC3B and ATG5 each play a central role in the 2 ubiquitin-like conjugation systems involved in the formation of autophagosomes (65). During autophagy, the fraction of MAP1LC3B at the inner membrane of the autophagosome is subject to degradation upon fusion with a lysosome, which must be replenished to allow the cell to maintain autophagy (29, 30). Our data suggest that this role is fulfilled by the PERK/eIF2α/ATF4 arm of the UPR during hypoxia, and we speculate that this allows cells to survive during prolonged periods of hypoxia. The only other known transcriptional regulator of MAP1LC3B is FoxO3. In skeletal muscles, FoxO3 has been reported to play a similar role in replenishing MAP1LC3B and protecting against muscle atrophy (66, 67). Interestingly, FoxO3 activity is increased after hypoxic exposure (68) and thus is an additional potential mediator of autophagy. However, this was not responsible for increased MAP1LC3B during hypoxia, since cells lacking FoxO3 showed normal induction of MAP1LC3B (Supplemental Figure 11).
ATG5 becomes modified on an internal lysine with the ubiquitin-like protein ATG12 as part of the second conjugation system that promotes autophagy. The ATG12-ATG5 conjugate is involved in the initial steps of autophagosome formation (69), and loss of ATG5 effectively inhibits autophagy. Interestingly, the ATG12-ATG5 complex is also important for processing and lipidation of ATG8/MAP1LC3B, which suggests that the 2 conjugation systems work in synergy to promote the development of the autophagosome (70). The consequences of hypoxic induction of ATG5 during hypoxia are not clear but may promote activation of autophagy or support an increase in autophagic flux. Understanding the role of ATG5 is further complicated by the fact that it also can promote apoptosis (71). Overexpression of ATG5 sensitizes to apoptosis upon stimulation with death signals through physically interacting with Bcl-xL and the Fas-associated death domain (FADD) (reviewed in ref. 71). The involvement of ATG5 in both pro-apoptotic and autophagy pathways suggests it may be instrumental in deciding between autophagy and apoptosis. Interestingly, cells derived from ATG5-deficient MEFs are somewhat more resistant to hypoxia-induced cell death and produce tumors that grow more quickly than their WT counterparts (33). It is not clear whether these differences arise due to the effects of ATG5 on apoptosis or autophagy, or whether they are MEF dependent. In agreement with our data, Bellot et al. (72) showed that disruption of autophagy sensitized cells to hypoxia. Here we show that ATG5 is a direct target of CHOP, which, besides autophagy (as shown here), has also been shown to regulate apoptosis (73, 74). It is therefore tempting to speculate that the connection between CHOP and apoptosis is related to the ability of CHOP to influence the balance between autophagy and apoptosis, but this requires further research.
Our results demonstrate a new mechanism linking the PERK arm of the UPR to hypoxia-induced autophagy. They expand on previous reports demonstrating activation of autophagy by hypoxia through mechanisms involving AMPK (33) and the HIF1 target gene, BNIP3 (25, 31). In the conditions used in our study, hypoxia-induced autophagy occurred at least in part independently of BNIP3, since BNIP3 was not expressed in either HT29 or HCT116 cells. Recent evidence showed that removal of BNIP3 is insufficient to block autophagy, but simultaneous removal of BNIP3 and BNIP3L did block autophagy (72). Additionally, BNIP3 may regulate a more specific form of “mitochondrial autophagy which is important for reducing mitochondrial mass, respiration to prevent formation of toxic reactive oxygen species” (25). We suggest that PERK- and ATF4-dependent regulation of MAP1LC3B acts to support autophagy by replenishing MAP1LC3B protein levels and thus the capacity to carry out autophagy during extended periods of hypoxic stress. Consequently, this pathway may act in concert with these other mechanisms that activate or initiate the autophagic process during hypoxia (Figure 9). Importantly, this supportive pathway appears to mediate hypoxia tolerance and levels of viable hypoxia in vivo. Targeting this pathway is able to reduce the hypoxic fraction and sensitize tumors to treatment and is thus an attractive therapeutic option to pursue clinically.
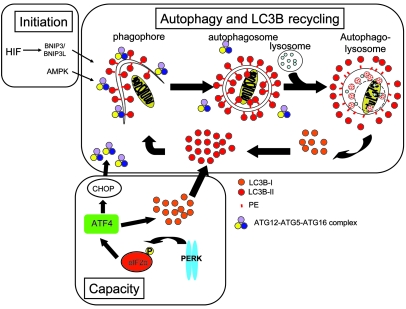
Figure 9. Model for recycling and regeneration of MAP1LC3B during hypoxia.
During the process of autophagy, the extending membrane (phagophore) requires coating with MAP1LC3B to form an autophagosome. The ATG5-ATG12-ATG16 complex redistributes and recruits MAP1LC3B to the membrane. Before degradation of the autolysosomal content, the ATG5-ATG12-ATG16 complex and the MAP1LC3B at the outer membrane is released to be recycled and the portion of MAP1LC3B inside the autolysosome is degraded. Hypoxia activates autophagy through BNIP3/BNIP3L signaling or AMPK (initiation). Activation of UPR/PERK signaling increases the capacity to maintain autophagy as it replenishes the overturned MAP1LC3B, but this activation also increases ATG5 expression to form ATG5-ATG12-ATG16 complexes.
Methods
Cell models.
Our studies included the human cell lines HT29 (colorectal adenocarcinoma), MCF7 (mammary adenocarcinoma derived from pleural effusion), DU145 (prostate carcinoma derived from brain metastasis), HCT116 (colorectal adenocarcinoma), and U373 MG (glioblastoma-astrocytoma) and were obtained from, and maintained as instructed by, the ATCC. Lipofectamine 2000 (Invitrogen) was used for plasmid transfections, and siRNA (Ambion and Applied Biosystems; see Supplemental Table 1 for siRNA IDs) was transfected with oligofectamine (Invitrogen). HIF-1α knockdown using shRNA was derived from pRetrosuper with the targeting sequence of 5′-GGACAAGTCACCACAGGAC-3′. U373 MG and HCT116 cells expressing the constitutively active C-terminal part of GADD34, eIF2α(S51A), and the shRNA against PERK (targeting sequence 5′-AGCGCGGCAGGTCATTAGTAA-3′) were generated as described previously (18). For hypoxia exposure, cells were transferred to a hypoxic culture chamber (MACS VA500 microaerophilic workstation; Don Whitley Scientific).
Tumor models.
Tumor cell lines (denoted as SSCNij) derived from squamous cell carcinoma of the head and neck were maintained in the flanks of nude mice (Balb/c nu/nu mice) as described previously (35). For CQ treatment experiments, colorectal HCT116 carcinoma cells were injected s.c. into NMRI-nu (nu/nu) female mice (28–32 g) under anesthesia. When tumor volume reached 200 mm3, animals were treated daily with CQ (60 mg/kg i.p.; Sigma-Aldrich) or saline for 7 days (49). When applicable, tumors were irradiated (10 Gy, 16 MeV) at day 4 of CQ or saline treatment. For immunohistochemical analysis, mice were killed after 7 days of treatment. Animals were injected with pimonidazole (60 mg/kg i.p. hypoxyprobe-1; Chemicon) and BrdU (30 mg/kg i.p.; Sigma-Aldrich) 60 minutes prior to killing. All animal experiments were conducted in accordance with national guidelines and approved by the animal ethics committees of KU Leuven and Maastricht University.
Affymetrix microarray.
Hypoxic exposure, RNA isolation, labeling, hybridization, and detection were performed as described previously (40).
Quantitative real-time PCR analysis.
RNA was reverse-transcribed using I-script (Bio-Rad). Real-time PCR was performed in ABI 7500 (Applied Biosystems). Gene abundance was detected using power SYBR Green I (Applied Biosystems) or, for the probe, using Taqman Gene expression Assays (Applied Biosystems). The abundance of every gene was normalized to the 18S signal. For a list of primers and probes on cDNA, see Supplemental Table 2.
Immunohistochemistry and image processing.
Frozen, acetone-fixed sections were stained using anti-cleaved MAP1LC3B (Abgent), anti-pimonidazole (Chemicon), 9F1 (rat monoclonal antibody to mouse endothelium; Department of Pathology, Radboud University Nijmegen Medical Center), mouse anti-BrdU (Sigma-Aldrich), or rabbit anti-cleaved caspase-3 (Cell Signaling Technology). For quantitative analysis, the slides were scanned by a computerized digital image processing system using a high-resolution intensified solid-state camera on a fluorescence microscope (Zeiss Axioskop) with a computer-controlled motorized stepping stage as described previously (75). Necrotic regions were assessed based on morphology and were excluded from analysis. MAP1LC3B was expressed as the percentage localized in hypoxic regions. MAP1LC3B enrichment factors in hypoxic regions were calculated as the fraction expressed in hypoxic regions divided by random colocalization (MAP1LC3B in hypoxia/[MAP1LC3B total × hypoxia total]). For detection of autophagy after hypoxia exposure, cells were seeded on glass coverslips, exposed to hypoxia, and fixed in 4% PBS-buffered paraformaldehyde.
Western blotting.
Cells were lysed in 50 mM Hepes-KOH pH 7.5, 150 mM KCl, 1 mM EDTA, 2 mM DTT, and 0.2% Tween-20 with protease inhibitors (1 “complete” pill per 25 ml; Roche), sonicated 3 times for 10 seconds each time, at 10 MHz, and resolved by SDS-PAGE. Protein transfers were probed with antibodies directed against MAP1LC3B (PD014; MBL) or β-actin (Sigma-Aldrich). Bound antibodies were visualized using HRP-linked secondary antibodies (anti-rabbit [Cell Signaling Technology] and anti-mouse [Sigma-Aldrich]) and ECL (Pierce Biotechnology).
ChIP.
ChIP was performed as described elsewhere (76). For precipitation the following antibodies were used: ATF4 (CREB2 [C-20]; Santa Cruz Biotechnologies Inc.), CHOP (GaDD153 [R-20]; Santa Cruz Biotechnologies Inc.), ATF6 (ab22341; Abcam,), HIF-1α (NB 100-105; Novus Biologicals). ChIP was performed as described with quantitative PCR (see Supplemental Table 3 for primers), and results were normalized to the chromatin input of the IP.
Flow cytometry.
After anoxia exposure, cells were fixed in 4% PBS-buffered paraformaldehyde, scraped, and permeabilized using 0.5% Tween-20 in PBS. Cells were probed with anti-cleaved MAP1LC3B (Abgent) followed by anti-rabbit Alexa Fluor 488 (DAKO) and analyzed by flow cytometry.
Statistics.
Data were analyzed with GraphPad Prism. Student’s t test was used for single comparisons. Multiple testing was done using a repeated-measures ANOVA with a Bonferroni post-hoc test. Data were considered statistically significant when P was less than 0.05.
Supplementary Material
Acknowledgments
The authors would like to thank Hans Peters and Hans Duimel for their excellent technical support. This work was financially supported by the Dutch Science Organization (ZonMW VENI grants 916.76.158 to K.M.A. Rouschop and 016.056.015 to M. Koritzinsky; ZonMW-NWO Top grants 912-03-047 to B.G. Wouters and 908-02-040 and 016.046.362 to J.W. Voncken), the Dutch Cancer Society (KWF grant UM 2003-2821 to B.G. Wouters), and the European Union Sixth Framework Programme (Euroxy program to B.G. Wouters).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:127–141 (2010). doi:10.1172/JCI40027.
References
- 1.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8(6):425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordsmark M, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol. 2005;77(1):18–24. doi: 10.1016/j.radonc.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Brizel DM, Sibley GS, Prosnitz LR, Scher RL, Dewhirst MW. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 1997;38(2):285–289. doi: 10.1016/s0360-3016(97)00101-6. [DOI] [PubMed] [Google Scholar]
- 4.Hockel M, Schlenger K, Aral B, Mitze M, Schaffer U, Vaupel P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56(19):4509–4515. [PubMed] [Google Scholar]
- 5.Young SD, Marshall RS, Hill RP. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc Natl Acad Sci U S A. 1988;85(24):9533–9537. doi: 10.1073/pnas.85.24.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 7.Wouters BG, van den Beucken T, Magagnin MG, Lambin P, Koumenis C. Targeting hypoxia tolerance in cancer. Drug Resist Updat. 2004;7(1):25–40. doi: 10.1016/j.drup.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Graeber TG, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379(6560):88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 9.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5(5):343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 10.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8(11):851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 11.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 12.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(31):11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167(1):27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima E, et al. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 2003;17(11):1573–1575. doi: 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 17.Koumenis C, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22(21):7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koritzinsky M, et al. Phosphorylation of eIF2alpha is required for mRNA translation inhibition and survival during moderate hypoxia. Radiother Oncol. 2007;83(3):353–361. doi: 10.1016/j.radonc.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Koritzinsky M, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25(5):1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blais JD, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24(17):7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24(19):3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero-Ramirez L, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64(17):5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 23.Lin JH, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36(12):2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27(6):421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 29.Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata A, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005;102(37):13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27(17):6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. Am J Pathol. 1980;98(2):425–444. [PMC free article] [PubMed] [Google Scholar]
- 33.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15(10):1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 34.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4(5):563–566. [PMC free article] [PubMed] [Google Scholar]
- 35.Wijffels KI, et al. Patterns of proliferation related to vasculature in human head-and-neck carcinomas before and after transplantation in nude mice. Int J Radiat Oncol Biol Phys. 2001;51(5):1346–1353. doi: 10.1016/s0360-3016(01)02605-0. [DOI] [PubMed] [Google Scholar]
- 36.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23(1):33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 39.Bacon AL, Fox S, Turley H, Harris AL. Selective silencing of the hypoxia-inducible factor 1 target gene BNIP3 by histone deacetylation and methylation in colorectal cancer. Oncogene. 2007;26(1):132–141. doi: 10.1038/sj.onc.1209761. [DOI] [PubMed] [Google Scholar]
- 40.Koritzinsky M, et al. The hypoxic proteome is influenced by gene-specific changes in mRNA translation. Radiother Oncol. 2005;76(2):177–186. doi: 10.1016/j.radonc.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 41.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153(5):1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 43.Milani M, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with bortezomib. Cancer Res. 2009;69(10):4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 44.Papandreou I, Krishna C, Kaper F, Cai D, Giaccia AJ, Denko NC. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 2005;65(8):3171–3178. doi: 10.1158/0008-5472.CAN-04-3395. [DOI] [PubMed] [Google Scholar]
- 45.Powers WE, Tolmach LJ. Demonstration of an anoxic component in a mouse tumor-cell population by in vivo assay of survival following irradiation. Radiology. 1964;83:328–336. doi: 10.1148/83.2.328. [DOI] [PubMed] [Google Scholar]
- 46.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118(1):79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Manzano C, et al. Adenovirus-mediated transfer of the p53 gene produces rapid and generalized death of human glioma cells via apoptosis. Cancer Res. 1996;56(4):694–699. [PubMed] [Google Scholar]
- 49.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117(2):326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 51.Marx J. Autophagy: is it cancer’s friend or foe? Science. 2006;312(5777):1160–1161. doi: 10.1126/science.312.5777.1160. [DOI] [PubMed] [Google Scholar]
- 52.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3(1):28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 55.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 56.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21(13):1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kouroku Y, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 61.Park MA, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7(10):1648–1662. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talloczy Z, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99(1):190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4(12):e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Bakker WJ, Harris IS, Mak TW. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell. 2007;28(6):941–953. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 69.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152(4):657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yousefi S, Simon HU. Apoptosis regulation by autophagy gene 5. Crit Rev Oncol Hematol. 2007;63(3):241–244. doi: 10.1016/j.critrevonc.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Bellot G, et al. Hypoxia-induced autophagy is mediated through HIF-induction of BNIP3 and BNIP3L via their BH3-domains. Mol Cell Biol. 2009;29(10)):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tajiri S, et al. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ. 2004;11(4):403–415. doi: 10.1038/sj.cdd.4401365. [DOI] [PubMed] [Google Scholar]
- 74.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 75.Rijken PF, Peters JP, Van der Kogel AJ. Quantitative analysis of varying profiles of hypoxia in relation to functional vessels in different human glioma xenograft lines. Radiat Res. 2002;157(6):626–632. doi: 10.1667/0033-7587(2002)157[0626:QAOVPO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 76.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.