Abstract
CD20 was the first B cell differentiation antigen identified, and CD20-specific mAbs are commonly used for the treatment of B cell malignancies and autoantibody-mediated autoimmune diseases. Despite this the role of CD20 in human B cell physiology has remained elusive. We describe here a juvenile patient with CD20 deficiency due to a homozygous mutation in a splice junction of the CD20 gene (also known as MS4A1) that results in “cryptic” splicing and nonfunctional mRNA species. Analysis of this patient has led us to conclude that CD20 has a central role in the generation of T cell–independent (TI) antibody responses. Key evidence to support this conclusion was provided by the observation that although antigen-independent B cells developed normally in the absence of CD20 expression, antibody formation, particularly after vaccination with TI antigens, was strongly impaired in the patient. Consistent with this, TI antipolysaccharide B cell responses were severely impeded in CD20-deficient mice. Our study therefore identifies what we believe to be a novel type of humoral immunodeficiency caused by CD20 deficiency and characterized by normal development of antigen-independent B cells, along with a reduced capacity to mount proper antibody responses.
Introduction
Primary antibody deficiencies are associated with an increased susceptibility to infections by encapsulated bacteria. In recent years, a variety of genetic defects have been identified that explain these disorders (1). Agammaglobulinemia is accompanied by a strong reduction or even complete absence of mature circulating B cells and results from a block of antigen-independent B cell development in the bone marrow. Frequently, this block is caused by mutations in the components of the pre–B cell receptor (pre-BCR) or associated signaling molecules such as Bruton’s tyrosine kinase (2–7). In disorders associated with defects in the responsiveness of mature B cells, normal numbers of circulating mature B cells are found with deficiencies of some but not all immunoglobulin classes. These disorders fall into 2 categories. In the first of these, hyper-IgM syndromes, genetic defects impair immunoglobulin-class switching (e.g., IgM to IgG) and somatic mutation of immunoglobulin genes in response to antigens (8–10). The second and most frequently occurring category is common variable immunodeficiency (CVID), a syndrome involving hypogammaglobulinemia, a lack of antibody production in response to vaccination and sometimes autoimmune phenomena or granulomatous inflammation. Genetic mutations have been found in some CVID patients that can either be B cell intrinsic (CD19) or extrinsic (ICOS, TACI) (11–14). Still, for the majority of patients, the molecular basis for antibody deficiencies have yet to be unraveled.
CD20 was one of the first B cell–specific differentiation antigens identified (15). Nowadays, CD20 immunotherapy using chimeric mAbs is used for the treatment of B cell neoplasia, EBV-associated immunopathology, and a growing list of diseases with presumed autoimmune origin (16, 17). CD20 belongs to the MS4A family of molecules with multiple membrane spanning domains (18, 19) and is expressed on pre-B and mature B cells but is lost upon differentiation into plasma cells (20). CD20 is unlikely to have a natural ligand, but in vitro studies with CD20 mAbs have demonstrated its involvement in the regulation of B cell activation and proliferation (21, 22). Moreover, CD20 is a component of a multimeric cell surface complex that regulates Ca2+ transport across the plasma membrane (23, 24). Although CD20 mAb immunotherapy depletes normal and malignant B cells in vivo by antibody-dependent activation of the innate monocytic network (25), mAb crosslinking of CD20 alters Ca2+ homeostasis, which influences cell cycle progression and can lead to apoptosis of normal and leukemic B cells in vitro (23, 26). The CD20 gene structure and expression pattern is strongly conserved between mouse and human (27–29) and CD20-deficient mice have been independently generated by 2 groups (29, 30). CD20–/– B cells develop and function normally, but spleen B cells exhibit demonstrable alterations in BCR- and CD19-induced Ca2+ responses (29). The importance of CD20 for the generation and function of human B cells has yet to be clarified.
We describe here a patient that completely lacked surface CD20 expression on B cells as a result of a homozygous mutation in the CD20 gene (28). The patient had a persistent hypogammaglobulinemia, normal B cell numbers, and a strong reduction in circulating memory B cells. A decreased frequency of somatic hypermutations in IgG heavy chain genes was found. After repeated vaccinations the patient mounted proper responses to recall antigens but displayed a strongly reduced ability to respond to pneumococcal polysaccharides. In agreement with a conserved role of CD20 in the generation of T cell–independent (TI) antibody responses, we found that CD20-deficient mice have a reduced ability to respond to TI antigens. Thus, CD20 has a nonredundant role in the generation of optimal B cell responses.
Results
Case report.
The patient, a girl of Turkish descent, was the oldest of 2 children of consanguineous parents (second cousins). She was referred to our institute at 4 years of age, with a history of intermittent respiratory infections and recurrent bronchopneumonia, starting at 2 years of age. On physical examination, her heart rate was normal and auscultation of the lung did not reveal abnormalities. A low-titer antinuclear antibody (IgG, 1:40) was found, but there were no indications for autoreactivity. The differential blood count and complement levels were normal. Cultures from sputum were negative. Pulmonary function was normal for age. Upon further investigations, she was given a diagnosis of hypogammaglobulinemia due to CVID. Intravenous immune globulin therapy was initially initiated but stopped after 6 months because of the lack of easy vascular access and discomfort. Instead, antibiotic prophylaxis was provided (co-trimoxazole).
Identification of the CD20 defect.
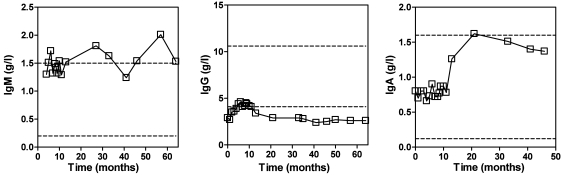
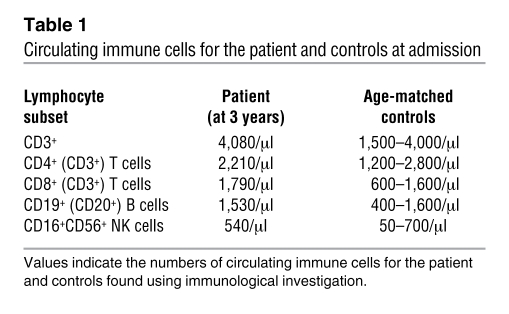
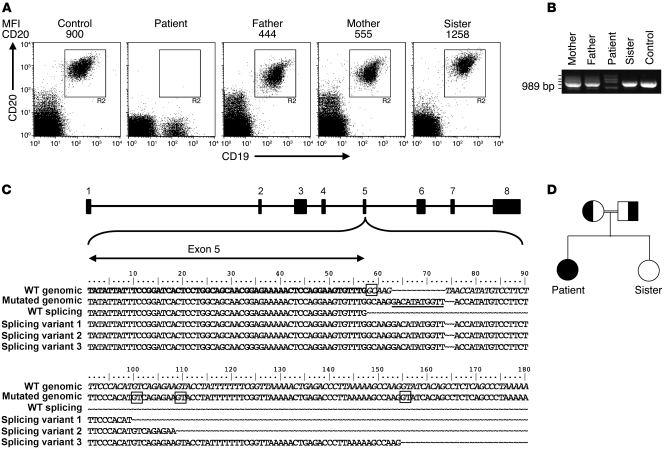
During a follow-up period of 4 years, the patient showed normal IgM and IgA levels but persistently low IgG levels. Intravenous immunoglobulins were administered for several months and induced a moderate rise in IgG levels into the low normal range (months 1–8) (Figure 1). Immunophenotyping revealed that the patient had a normal distribution of circulating immune cells (Table 1). Also, the number of CD19+ B cells was normal, but CD20 expression was conspicuously absent (Figure 2A). Various mAbs against nonoverlapping epitopes on CD20 were tested, and all were found to be negative (data not shown). Both parents expressed CD20, but the expression of the antigen appeared to be 50% of that compared with controls and the patient’s sibling (Figure 2A). Next to the lack of CD20 expression on B cells, it was also observed that a small lymphocyte fraction of CD19negCD20dull cells present in healthy controls was apparently absent in the patient. This CD19negCD20dull population was found to contain T cells, especially those of the CD8+CD45R0+ phenotype, but their specific role in the regulation of immune reaction is as of yet unknown (31).
Figure 1. Serum IgM, IgG, and IgA levels over time in the patient.
A turbidimetric method was used to determine serum immunoglobulin levels in serial samples. The date of first presentation of the patient at the clinic is set as 0. The upper and lower ranges of normal immunoglobulin levels are indicated by a dashed line. Intravenous immune globulin therapy was initially initiated but stopped after 6 months.
Table 1 .
Circulating immune cells for the patient and controls at admission
Figure 2. Analysis of CD20 expression and gene sequences of the patient, parents, and sibling.
(A) FACS analysis of CD20 and CD19 expression in indicated PBMC samples of normal controls and family members. MFI of CD20 staining is indicated above the plots. In a cohort of healthy children (age, 3 months to 10 years; n = 30), mean CD20 expression ranged between 874 and 1,230. (B) Agarose gel analysis of CD20 cDNA fragments of the patient, family members, and a control, revealing aberrant mRNA species in the patient and her parents. Cloning and sequencing of the cDNA fragments showed that the smallest fragment contains a complete deletion of exon 5 (not shown). The 3 fragments with increased MW contained variable parts of intron 5 sequence (see below in C). (C) CD20 gene sequences surrounding the exon 5/intron 5 boundary, as determined from cDNA and genomic fragments. The 11–base pair insertion at the noncanonical splice site is underlined in the mutated genomic sequence. The CD20 intron/exon structure is indicated on top, and the normal and aberrant sequences found in the patient cDNA clones are depicted below. The noncanonical GC splice donor site at intron 5 and the cryptic splice sites used as a result of the mutation are boxed. (D) Family pedigree indicating consanguinity of the parents. Normal and aberrant CD20 alleles are indicated by white and black, respectively.
Analysis of CD20 cDNA fragments showed 4 aberrant mRNA species in the patient (Figure 2B). Sequencing of the CD20 cDNA and genomic DNA of the patient revealed a compound mutation of the noncanonical splice donor sequence of exon 5 of the CD20 gene (homozygous 11-bp insertion as well as a partial deletion). At this site, exon 5 carries a unique donor splice site TTG/GCAAGT, with a C replacing a T in the canonical GT recognition site (28). The alteration at this splice donor site resulted in transcripts with a complete deletion of exon 5 as well as with insertion of intronic sequences due to usage of cryptic splice sites (Figure 2C). The parents each carried only 1 mutated allele as indicated in the family pedigree (Figure 2D) and also expressed, next to the single CD20 transcript found in healthy individuals, the aberrant mRNA species (Figure 2B). A single CD20 transcript at normal levels was found in the RNA isolated from the B cells of the healthy sibling, in accordance with the normal CD20 surface expression on her B cells (Figure 2, A and B).
Functional properties of CD20-deficient B cells.
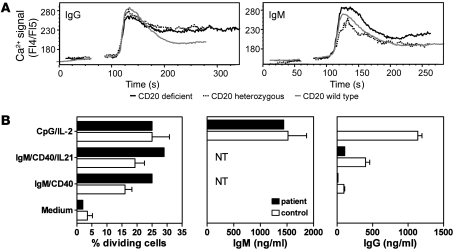
To study the functional properties of CD20-deficient B cells, IgM naive and IgG memory B cell lines were generated by retroviral transduction of patient and control B cells with B cell lymphoma 6 protein (Bcl-6) and the Bcl-2 family member Bcl-xL. Cell growth, in the presence of CD40 ligand (CD40L) and IL-21, of CD20-deficient and CD20-heterozygous lines obtained from the patient and her parents was similar to controls (data not shown). Furthermore, also Ca2+ responses after surface IgM or IgG cross-linking were intact (Figure 3A). The ability of CD20-deficient B cells to respond efficiently to BCR-mediated ligation suggests that the signaling function of the BCR complex is not significantly altered in the absence of CD20 expression.
Figure 3. B cell function in vitro.
(A) Cells lacking CD20 respond to IgG and IgM BCR stimulation by Ca2+ flux. B cell lines from donors expressing wild-type, intermediate, or no CD20 were cultured for 4 hours without CD40L in the presence of IL-21 before being loaded with Indo-1 AM. Cells were maintained at room temperature and incubated with a control and subsequently with anti-BCR mAbs. Fluorescence ratios of Indo-1 emission at 405/485 nm were measured by flow cytometry. Ca2+ flux curves were adjusted graphically to the same starting level fluorescence 4/fluorescence 5(FL4/FL5) ratio, and the starting point of the stimulus was set at the same time point. In general, all cells responded to BCR stimulation by showing intracellular Ca2+ fluxes. (B) CFSE-labeled PBMCs were stimulated with indicated stimuli for 6 days, and CD19+CD20+ B cells were analyzed by flow cytometry for CFSE dilution. The percentage of dividing B cells (precursor frequencies) was calculated. Supernatants were tested by ELISA for IgM and IgG secretion. NT, not tested due to stimulus. Values represent mean ± SEM and are obtained for 3–4 healthy donors.
Freshly isolated B cells from the patient showed normal proliferation after BCR-dependent and BCR-independent stimuli, but IgG production was low (Figure 3B). Specifically, BCR cross-linking with anti-IgM mAb combined with CD40 mAb in the absence or presence of IL-21 induced cell division, but IgG was not detected in the supernatants of these cultures. Moreover, BCR-independent, TLR9-mediated B cell activation (32) (CpG) induced normal cellular activation of the patient’s B cells, as demonstrated by CD38 upregulation (data not shown), proliferation, and ample IgM secretion. However, using this stimulus, IgG secretion was conspicuously absent (Figure 3B). Thus, in response to either T cell–dependent or TI stimuli, the patient’s B cells displayed adequate proliferation and IgM secretion but very low to absent IgG production in vitro.
Cellular and molecular properties of the circulating B cell compartment.
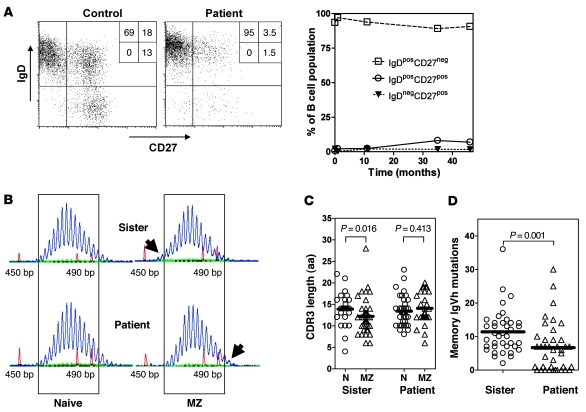
Ig-κ+ B cells and Ig-λ+ B cells were present at normal ratios in blood samples from the patient (data not shown). Moreover, these B cells had normal expression of other membrane-bound molecules, including IgM, IgD, CD22, CD38, CD40, and HLA-DR (data not shown). While normal absolute numbers of CD19+ B cells were present in the peripheral blood, class-switched CD27+ memory B cells were hardly detected (<2%), measured over a 3- to 5-year period, and were decreased when compared with levels of age-matched controls (relative surface immunoglobulin D–CD27+ [sIgD–CD27+] number in children of 3–10 years of age, mean 7.1% ± 2.71%, range 4.8%–15.5%; n = 30; Figure 4A). The number of sIgD+CD27+ marginal zone (MZ) B cells showed a modest increase over a 3-year period, from 2.0% up to 3.5%, in the blood samples from the patient (relative B cell number in healthy children, mean 15.4% ± 8.0%, range 6.2%–17.5%; n = 30; Figure 4A). The numbers of MZ and class-switched sIgD–CD27+ memory B cells in the 2 parents were within the range of the controls (data not shown).
Figure 4. B cell selection.
(A) FACS analysis of B cell subsets. IgD × CD27 dot plot gated on CD19+CD20+ B cells. Percentage of cells in each quadrant are indicated (left). The percentages of the patient’s naive, nonswitched, and switched memory B cells are shown over the course of approximately 4 years since initial presentation (t = 0) (right). (B) Gene scanning analyses of IgM-VH3 transcripts amplified out of IgD+CD27– mature naive and IgD+CD27+ MZ B cells of the patient and her sister show the presence of shorter MZ B cell IgM-VH3 transcripts of the sister and longer IgM-VH3 transcripts of the patient (highlighted by the top and bottom arrows on the right panel, respectively). Numbers on the x axis are lengths in base pairs, as determined using internal length markers. (C) Comparison of IgVH-CDR3 amino acid lengths of IgD+CD27– mature naive (N) and IgD+CD27+ MZ B cells. Of the patient and her sister, 25 and 39 VH3-IgM clones of the naive B cells and 42 and 28 VH3-IgM clones of the MZ B cells were sequenced, respectively. Data for the mother and a normal control were similar to those of the sister (not shown). (D) Numbers of VH3-IgVH mutations of IgG-expressing IgD–CD27+ memory B cells. Of the patient and her sister, 38 and 39 VH3-IgG clones the memory B cells were sequenced, respectively. (C and D) Individual clones are indicated by the separate symbols. Mean CDR3 length and number of mutations is indicated by bars. P values of statistical analyses among groups are shown.
To study B cell selection and affinity maturation in detail, B cells of the patient, her sister, her mother, and of a healthy control were sorted into 3 fractions based on phenotype, i.e., sIgD+CD27– naive B cells, sIgD+CD27+ MZ B cells, and sIgD–CD27+ memory B cells. Global distribution of immunoglobulin variable heavy chain–CDR3 (IgVH-CDR3) length and diversity was assessed by RT-PCR of IgM-VH3 transcripts, using a fluorochrome-labeled Cμ primer and detection by gene scanning. In healthy donors, there is a marked downward shift in the distribution of IgVH-CDR3 lengths of MZ B cells as compared with naive B cells, and this was clearly detectable in the younger sister (Figure 4B, top), the mother, and a control donor (data not shown). Unexpectedly, this shift was not apparent in the patient’s MZ B cells (Figure 4B, bottom). The mean IgVH-CDR3 lengths of individual IgVH clones of the MZ B cells of the sister (Figure 4C), the heterozygous mother, and the control (data not shown) were significantly shorter as compared with those of naive B cells. This selection for shorter CDR3 lengths was not observed in the patient, thereby confirming the gene scanning results (Figure 4C). As expected, counterselection for long IgVH-CDR3 was not observed in the IgG-expressing memory B cells of the different individuals (data not shown) (33).
To assess somatic mutations of the IgVH of different B cell subsets, IgM-VH3 PCR products of the naive and MZ B cells and IgG-VH3 PCR products of the memory B cell were cloned and sequenced. We observed no difference in the mean number of somatic mutations (i.e., 8 mutations per VH3-IgVH) of the MZ B cells when isolated from the patient or from her 3-year-old sister. This number is slightly lower than those found in the MZ B cells of the patient’s heterozygous mother and of a healthy adult control, which was a mean of 13 and 11 mutations per VH3-IgVH, respectively (data not shown). The IgG memory B cells of the patient contained a mean of 7 mutations per VH3-IgVH, which is significantly lower than those of her 3-year-old sister (11 mutations per VH3-IgVH), her mother (14 mutations per VH3-IgVH), and the healthy adult control (19 mutations per VH3-IgVH) (Figure 4D and data not shown).
Specific antibody responses after vaccination.
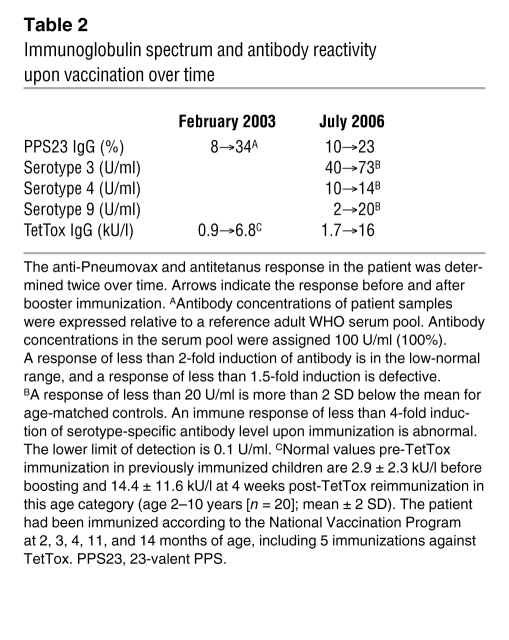
To test whether the altered selection of B cells with a MZ phenotype influenced TI antibody responses, the patient was vaccinated twice in a 3-year period with TI pneumococcal polysaccharide vaccine. The humoral response against the overall pool of 23 capsular serotypes was reduced, albeit detectable. Moreover, selective responses to serotypes 3, 4, and 9 were also abnormal when separately determined (Table 2). Remarkably, the ability of the patient to generate antitetanus toxoid IgG after repeated booster immunization was normal (Table 2). Moreover, the affinity of the tetanus-reactive antibodies was within the normal range (34) (data not shown).
Table 2 .
Immunoglobulin spectrum and antibody reactivity upon vaccination over time
TI antibody responses in CD20-deficient mice.
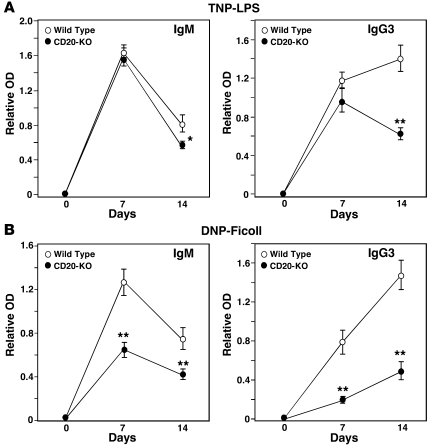
To test whether CD20 also plays a role in the generation of TI antibody responses in mice, humoral immune responses were compared in Cd20–/– and wild-type mice immunized with trinitrophenyl-LPS (TNP-LPS) (a TI-1 antigen) or dinitrophenol-Ficoll (DNP-Ficoll) (a TI-2 antigen). Following TNP-LPS immunizations, hapten-specific IgM and IgG3 antibody levels were similar in both CD20 and wild-type littermates at day 7, but were significantly decreased in Cd20–/– mice by day 14 (Figure 5A). By contrast, both hapten-specific IgM and IgG3 responses were significantly reduced in Cd20–/– mice following DNP-Ficoll immunizations (Figure 5B). Although TI antigens primarily induce IgM and IgG3 antibodies (35), similar results were obtained for all IgG isotypes (data not shown). Thus, CD20 deficiency had significant inhibitory effects on both TI-1 and TI-2 antibody responses, which contrasts with the lack of effect of CD20 deficiency on T cell–dependent antibody responses in mice (29).
Figure 5. CD20 deficiency impairs TI antibody responses in mice.
Wild-type (n = 9/antigen) and Cd20–/– (n = 9–10/antigen) mice were immunized with either (A) TNP-LPS or (B) DNP-Ficoll on day 0, with sera collected at the indicated times. TNP- and DNP-specific IgM and IgG3 levels in all sera were assessed simultaneously by ELISA. Values represent mean ± SEM relative antigen-specific antibody levels, with 9 or more mice in each group. Significant differences between mean IgM and IgG3 levels are indicated: *P < 0.05, **P < 0.01.
Discussion
We describe a patient with a homozygous mutation in the CD20 gene, affecting the splice donor site at the 5′ site of exon 5. By consequence, exon 5 is deleted or various abnormal transcripts appear, all without translation into cell surface–expressed protein. In more than 200 children tested, who were either healthy or suspected of minor or major immunodeficiencies, CD20 surface expression on B cells was always normally present, underlining the unique nature of this mutation.
CD20, high-affinity IgE receptor β chain (FcεRIβ), HTm4, and a growing number of proteins are structurally related cell surface proteins, expressed by distinct hematopoietic cells. These homologous proteins have at least 4 potential membrane-spanning regions, with N- and C-terminal cytoplasmic domains encoded by distinct exons. This family of proteins was therefore named the membrane-spanning 4-domains, subfamily A, with at least 12 subgroups (MS4A1, which is identical to CD20, through MS4A12), currently representing more than 20 distinct human and mouse proteins. Each family member has unique patterns of expression among hematopoietic cells and nonlymphoid tissues. CD20, along with FCER1B, HTM4, and several but not all human MS4A genes identified map to chromosome 11q12-q13.1 (18).
Similar to FcεRIβ and the human MS4A family members, CD20 is likely to be a component of an oligomeric cell surface complex involved in signal transduction. In B cells, engagement of CD20 molecules initiates a signal transduction cascade that involves tyrosine kinases involved in cell adhesion, proliferation, and survival (21, 23, 24, 36, 37). In knockout mice, CD20 loss had no detectable effect on mitogen-induced proliferation, which is in agreement with the data obtained with B cells of the CD20-deficient patient. Normal levels of all immunoglobulin isotypes were found in sera from Cd20–/– mice, and they also generated primary and secondary antibody responses following immunization with a T cell–dependent antigen, with affinities of primary and secondary IgG1 antigen-specific antibody responses similar to those generated in their wild-type littermates. Therefore, CD20 function was suggested to be not required for T–B cell interactions, isotype switching, or affinity maturation during the generation of humoral immune responses (30). Despite normal B cell development in Cd20–/– mice (splenic B220+), B cells from Cd20–/– mice generated mildly reduced [Ca2+]i responses following IgM ligation and CD19 cross-linking (29, 30).
In human CD20 deficiency, the loss of CD20 did not disturb the differentiation of precursor B cells in the bone marrow, because B cell count and differentiation status was normal, as determined by a variety of other surface markers, in accordance with the CD20-knockout mice (29, 30). The severe reduction of class-switched sIgD–CD27+ memory B cells might reflect suboptimal germinal center formation and is likely to cause the profound hypogammaglobulinemia. Although IgG antibodies and circulating memory B cells are not defective in the CD20-knockout mice (29, 30), the selection of naive B cells into the MZ B cell compartment seems disturbed in the patient. In healthy donors the transition of naive B cells into MZ B cells is accompanied by a reduction of the mean IgVH-CDR3 lengths and acquisition of IgV mutations (38, 39). Concomitantly, a counterselection against poly- and self-reactive antibodies and enrichment for antibodies reactive with specific bacterial polysaccharides occurs (38). This selection process is potentially driven by tonic BCR signals that may be assumed to be different between BCRs with longer and shorter IgVH-CDR3 lengths. Alternatively, particular self- or commensal-related antigens, recognized with low avidity, may provide positive BCR signals, favoring the selection of B cells that have short IgVH-CDR3 lengths in the MZ B cell compartment. In the MZ B cells of the patient, no counterselection for long IgVH-CDR3 lengths was observed, which is in accordance with a diminished response against polysaccharide vaccination. The notion of a disturbed counterselection against poly- and self-reactive antibodies is strengthened by the observation that, in the absence of any clinical signs of autoimmunity, antinuclear antibodies in the serum of the patient were found repeatedly (data not shown). It is noteworthy that the mean IgVH mutation frequencies of the MZ B cells are comparable to those of her 3-year-old healthy sister (i.e., 8 mutations per VH3-IgVH) and to those reported for young adults (~10 mutations per IgVH; range, 4–18 mutations). This finding contrasted with the mean mutation number found in the IgG-VH3–expressing memory B cells of 7 mutations per VH3-IgVH, which was significantly lower in the patient as compared with her 3-year-old sister (Figure 4D). The mutation numbers reported for VH5-IgG transcripts in young adults varies significantly, ranging between 12 and 23 (with a mean of 17.3 mutations) per VH5-IgVH (40). The remarkable finding of normal mutation rates in the patient’s MZ B cells seems contradictory but may possibly be explained by BCR- and GC-independent, intrinsic immunoglobulin gene diversification program, as has been described in sheep and birds and for which also evidence has been obtained in humans (41, 42).
Human CD20 deficiency results in decreased IgG antibody levels and relatively increased IgM levels with weak responses against polysaccharides after vaccination. Importantly, we found that CD20 deficiency in mice also results in an impaired ability to make TI antibody responses. Thus, although the phenotypes of CD20-deficient humans and mice are subtly different, the current data imply that CD20 signals are essential to enable B cells to optimally respond to antigenic stimuli.
Methods
Blood samples.
Heparinized venous blood was collected from healthy donors and from the patient and her family members, after obtaining informed consent. The study was approved by the AMC institutional medical ethical committee, in accordance with the standards laid down in the 1964 Declaration of Helsinki.
Sequence analysis of the CD20 gene and mRNA.
DNA was isolated from granulocytes purified by Ficoll gradient centrifugation. cDNA was prepared from mRNA from PBMCs. CD20 cDNA was amplified by PCR and analyzed on 0.8% agarose gel. Normal and aberrant PCR fragments were purified and cloned in T vector (Promega) and sequenced (ABI Prism 377; Applied Biosystems). Fragments of CD20 genomic DNA were amplified and sequenced to verify the mutations found in the patient’s cDNA. Primers for cDNA PCR were 5′-GAGACTCAGGAGTTTTGAGAGC-3′ (forward) and 5′-GCTTCCAAGAGACATGCTGACT-3′ (reverse); primers for genomic PCR were 5′-TTACAGTATATTATTTCCGGATCACTCCTG-3′ (forward intron 4/exon 5 boundary) and 5′-CACACAGCATTTAACGTTCTTAAGACTGT-3′ (reverse, intron 5).
Immunofluorescence and flow cytometry.
For determination of absolute numbers of T, B, and NK-lymphocytes, the 6-color Multitest system was used, and samples were measured on a FacsCanto II with FacsCanto software (all from BD Biosciences). For flow cytometric immunophenotyping of B cells, fresh PBMCs (or viable cryopreserved PBMCs after thawing) were resuspended in PBS containing 0.5% (w/v) BSA, and 200,000 PBMCs were incubated with fluorescent label–conjugated mAbs using saturating concentrations. APC-conjugated CD20, PerCP Cy 5.5–conjugated CD19, and PE-conjugated IgD were from BD Biosciences and FITC-conjugated CD27 was from Sanquin.
Generation of B cell lines.
B cell lines were created using retroviral transduction of B cells with Bcl-6 and Bcl-xL (43, 44) as described previously (45, 46). Briefly, CD19+ B cells from PBMCs were isolated by FACS sorting (FACSAria; BD Biosciences) and cultured on γ-irradiated (50 Gy) mouse L cell fibroblasts, stably expressing CD40L (CD40L cells, 105 cells/ml) and recombinant mouse IL-21 (25 ng/ml; R&D systems) in culture medium containing Iscove’s modified Dulbecco’s medium (IMDM) (Gibco), 8% FBS (HyClone), and penicillin/streptomycin (Roche). After 36 hours, cells were incubated for 6 hours with a retrovirus encoding Bcl-6, Bcl-xL, and GFP. Transduced cells were cultured on CD40L cells in the presence of IL-21 and subsequently cloned at a single cell level. Surface IgM- or IgG-expressing clones were identified by flow cytometry. The cell lines from the patient expressed no CD20, whereas cell lines from the parents had intermediate CD20 levels compared with controls (data not shown).
Measurement of Ca2+ mobilization.
To measure Ca2+ mobilization in the B cell lines after BCR stimulation, 5 × 106 cells were incubated with 2 μM Indo-1 AM (Sigma-Aldrich) in loading buffer (HBSS, 1 mM Ca2+, 1 mM Mg2+, 1% FBS) for 30 minutes at 37°C. Then, cells were washed and resuspended at 3 × 106/ml. Fluorescence ratios of Indo-1 emission at 405/485 nm were measured by flow cytometry on a FACSVantage SE (BD). Cells were incubated at 37°C, and data were first acquired without stimulation. Before activation was induced by adding either 5 μg/ml F(Ab′)2 goat anti-human IgG (Jackson ImmunoResearch Laboratories Inc.) or mouse anti-human IgM mAb (MH15; Sanquin) cells were incubated with a nonbinding control antibody (5 μg/ml). Data were analyzed using FlowJo software (Tree Star). Ionomycin (Molecular Probes) was used to control for intracellular loading of Indo-1.
B cell activation in vitro.
PBMCs were resuspended in PBS at a concentration of 5–10 × 106 cells/ml and labeled with 0.5 μM CFSE (Molecular Probes) in PBS for 10 minutes at 37°C under constant agitation. Cells were washed and subsequently resuspended in IMDM supplemented with 10% fetal calf serum (BioWhittaker), antibiotics, and 3.57 × 10–4% (v/v) β-mercapto-ethanol (Merck).
Labeled PBMCs containing a fixed number of B cells (2 × 105 per well) were cultured in 48-well flat-bottomed plates for 6 days at 37°C and stimulated with saturating amounts of anti-IgM mAb (clone MH15; Sanquin), anti-CD40 mAb (clone 14G7; Sanquin), 20 ng/ml IL-21 (Invitrogen), or 200 μg/ml CpG oligodeoxynucleotide 2006 (32) (Invivogen), with 50 U/ml IL-2 (R&D Systems). Proliferation of the B cells was assessed by measuring CFSE dilution. The precursor frequency (percentage of cells in the initial population that underwent one or more divisions after culture) was calculated as follows: [Σn≥1(Pn/2n)]/[Σn≥0(Pn/2n)], where n is the division number that cells have gone through and Pn is the number of cells in division n. The mean number of divisions of the divided cells was calculated as follows: [Σn≥1(n Pn/2n)]/[ Σn≥1(Pn/2n)].
Supernatants were tested for secreted IgM and IgG by ELISA using polyclonal rabbit anti-human IgG and IgM reagents and a serum protein calibrator all from Dako.
Immunoglobulin sequence analyses.
Total RNA isolated using the Pico Pure RNA isolation kit (Arcturus) was converted to cDNA using pd(N)6 primer (Pharmacia Biotech) and standard procedures. From cDNA of naive and MZ B cells, IgM-VH3 transcripts were amplified using a VH3 family–specific leader primer in combination with a Cμ primer, and from memory B cells, IgG-VH3 transcripts were amplified using a VH3 family–specific leader primer in combination with a Cγ primer (47). In gene scanning experiments, a fluorochrome-labeled Cμ was used to enable automatic detection of PCR product lengths (48). Each time RT-PCR was run, water controls were included, which were negative in all cases. IgM-VH3 and IgG-VH3 RT-PCR products were cloned into pTOPO-TA vectors, transformed into TOP10 bacteria (Invitrogen), and sequenced on both strands using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). To determine the germline genes used in the IgVH-DH-JH rearrangements, the somatic IgVH mutations therein, and the IgVH-CDR3 amino acid sequence lengths, the sequences were compared with published germline IgVH genes using the Vbase database (49) and DNAplot online (MRC Centre for Protein Engineering; http://www.mrc-cpe.cam.ac.uk).
Determination of whole-vaccine and type-specific polysaccharide antibodies.
The 23-valent pneumococcal capsular polysaccharide (PPS) vaccine (Pneumovax; Merck, Sharp & Dohme), containing 25 μg of purified type-specific capsular polysaccharides of 23 pneumococcal serotypes (i.e., 1, 2, 3, 4, 5, 6B, 7F, 8, 9N, 9V, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 20, 22F, 23F, 33F), was used to assess the patient’s capacity to respond to polysaccharide antigens. Serum was drawn before and 4 weeks after each vaccination and stored at –80°C until use.
Detection of anti-PPS vaccine-specific IgG antibodies against the complete pool of 23 polysaccharide antigens has been validated in the past (50). A standard serum from a normal nonvaccinated adult was included in every ELISA as a control. Antibody concentrations of patient samples were expressed relative to a reference adult WHO serum pool. Antibody concentrations in this serum pool were assigned 100% (100 U/ml).
In addition, IgG antibodies against 3 different immunogenic PPSs of the well-defined serotypes 3, 4, and 9 were determined as a separate confirmation of the anti-polysaccharide response (51) and expressed in U/ml. The lower limit of detection was 0.1 U/ml.
In both tests pre- and postvaccination serum samples were analyzed simultaneously. All serum samples were preincubated overnight with excess free common cell wall polysaccharide (CPS), a procedure that neutralized anti-CPS antibodies. Peroxidase-conjugated rabbit anti-human IgG (Dako) was used as detecting antibody. Antibody levels are expressed as geometric means. Fold increase in antibody titers was calculated by dividing individual post- and preimmunization titers.
Mouse immunizations and antigen-specific ELISAs.
Cd20–/– mice were generated as described (29) and backcrossed with C57BL/6 mice (The Jackson Laboratory) for more than 8 generations. Two independent groups of 3-month-old mice (n = 4–5 mice/group/antigen) were immunized i.p. with TNP-LPS (50 μg; catalog no. T-5065-1; Biosearch Technologies) or DNP-Ficoll (25 μg; catalog no. F-1200; Biosearch Technologies) in 200 μl PBS. Wild-type C57BL/6 control mice in one immunization group (n = 4/antigen) were from The Jackson Laboratory, while the second group of wild-type mice (n = 5) were from the National Cancer Institute-Frederick Laboratory. Serum was obtained before and after immunizations as indicated.
DNP- and TNP-specific antibody levels in individual serum samples were determined in duplicate using antibody isotype-specific ELISAs as described (52). ELISA plates were coated with DNP-BSA (Calbiochem-Novabiochem) or TNP-BSA (Biosearch Technologies) overnight at 4°C. The plates were washed and blocked with 1% gelatin/2% BSA for 90 minutes at 37°C. Sera were diluted in Tris-buffered saline containing 1% BSA (TNP-LPS, diluted 1:400; DNP-Ficoll, diluted 1:1000) and incubated in duplicate wells at room temperature for 90 minutes. The plates were washed, incubated with alkaline phosphatase–conjugated polyclonal goat anti-mouse IgM and IgG isotype-specific antibodies (Southern Biotechnology Associates) for 1 hour at room temperature, and developed using p-nitrophenyl phosphate substrate (Sigma-Aldrich). OD405 values were measured when the OD405 of any well approached 2.0. Mean background OD405 levels (≤0.16) from wells that did not contain serum were subtracted from all values. Sera from wild-type and Cd20–/– mice from both groups gave similar results, which were therefore pooled.
Statistics.
For analysis of immunoglobulin gene sequences, data were statistically evaluated using the Kruskal-Wallis rank-sum test, followed by the Dunn procedure for subsequent comparison of specific groups. P values of less than 0.05 were considered statistically significant. Differences between murine IgM and IgG3 levels were calculated by 2-sided, 2-tailed Student’s t test.
Acknowledgments
We thank Maarten van Tol for determining the avidity maturation of antitetanus toxoid antibodies in the patient’s serum samples over time after (re)vaccination, Jan van Aten for expert assistance with the statistical analyses of immunoglobulin sequences, Thera Wormhout for technical support, and Guglielmo Venturi for mouse immunizations and analysis. This work was supported by grants from the National Institutes of Health (CA105001 and AI56363 to T.F. Tedder). We thank Dörte Hamann (Sanquin, Amsterdam) for her help with the determination of the antinuclear antibodies.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:214–222 (2010). doi:10.1172/JCI40231.
References
- 1.Shearer WT, Fischer A. The last 80 years in primary immunodeficiency: how far have we come, how far need we go? J Allergy Clin Immunol. 2006;117(4):748–752. doi: 10.1016/j.jaci.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Conley ME, Rohrer J, Rapalus L, Boylin EC, Minegishi Y. Defects in early B-cell development: comparing the consequences of abnormalities in pre-BCR signaling in the human and the mouse. Immunol Rev. 2000;178:75–90. doi: 10.1034/j.1600-065X.2000.17809.x. [DOI] [PubMed] [Google Scholar]
- 3.Minegishi Y, et al. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187(1):71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minegishi Y, et al. Mutations in Igalpha (CD79a) result in a complete block in B-cell development. . J Clin Invest. 1999;104(8):1115–1121. doi: 10.1172/JCI7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukada S, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72(2):279–290. doi: 10.1016/0092-8674(93)90667-F. [DOI] [PubMed] [Google Scholar]
- 6.Vetrie D, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361(6409):226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 7.Yel L, et al. Mutations in the mu heavy-chain gene in patients with agammaglobulinemia. N Engl J Med. 1996;335(20):1486–1493. doi: 10.1056/NEJM199611143352003. [DOI] [PubMed] [Google Scholar]
- 8.Durandy A, Peron S, Fischer A. Hyper-IgM syndromes. Curr Opin Rheumatol. 2006;18(4):369–376. doi: 10.1097/01.bor.0000231905.12172.b5. [DOI] [PubMed] [Google Scholar]
- 9.Imai K, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4(10):1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 10.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102(5):565–575. doi: 10.1016/S0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 11.van Zelm MC, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354(18):1901–1912. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 12.Castigli E, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37(8):829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 13.Grimbacher B, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4(3):261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 14.Salzer U, et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113(9):1967–1976. doi: 10.1182/blood-2008-02-141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stashenko P, Nadler LM, Hardy R, Schlossman SF. Characterization of a human B lymphocyte-specific antigen. J Immunol. 1980;125(4):1678–1685. [PubMed] [Google Scholar]
- 16.Arnold DM, et al. Systematic review: efficacy and safety of rituximab for adults with idiopathic thrombocytopenic purpura. Ann Intern Med. 2007;146(1):25–33. doi: 10.7326/0003-4819-146-1-200701020-00006. [DOI] [PubMed] [Google Scholar]
- 17.Dorner T, Lipsky PE. B-cell targeting: a novel approach to immune intervention today and tomorrow. Expert Opin Biol Ther. 2007;7(9):1287–1299. doi: 10.1517/14712598.7.9.1287. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Buckley TR, Tu L, Langdon SD, Tedder TF. Structural organization of the human MS4A gene cluster on Chromosome 11q12. Immunogenetics. 2001;53(5):357–368. doi: 10.1007/s002510100339. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Tedder TF. Identification of a CD20-, FcepsilonRIbeta-, and HTm4-related gene family: sixteen new MS4A family members expressed in human and mouse. Genomics. 2001;72(2):119–127. doi: 10.1006/geno.2000.6472. [DOI] [PubMed] [Google Scholar]
- 20.Tedder TF, Engel P. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol Today. 1994;15(9):450–454. doi: 10.1016/0167-5699(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 21.Tedder TF, et al. The B cell surface molecule B1 is functionally linked with B cell activation and differentiation. J Immunol. 1985;135(2):973–979. [PubMed] [Google Scholar]
- 22.Tedder TF, et al. Antibodies reactive with the B1 molecule inhibit cell cycle progression but not activation of human B lymphocytes. Eur J Immunol. 1986;16(8):881–887. doi: 10.1002/eji.1830160802. [DOI] [PubMed] [Google Scholar]
- 23.Bubien JK, Zhou LJ, Bell PD, Frizzell RA, Tedder TF. Transfection of the CD20 cell surface molecule into ectopic cell types generates a Ca2+ conductance found constitutively in B lymphocytes. J Cell Biol. 1993;121(5):1121–1132. doi: 10.1083/jcb.121.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanzaki M, Lindorfer MA, Garrison JC, Kojima I. Activation of the calcium-permeable cation channel CD20 by alpha subunits of the Gi protein. J Biol Chem. 1997;272(23):14733–14739. doi: 10.1074/jbc.272.23.14733. [DOI] [PubMed] [Google Scholar]
- 25.Uchida J, et al. The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. J Exp Med. 2004;199(12):1659–1669. doi: 10.1084/jem.20040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Kolk LE, et al. CD20-induced B cell death can bypass mitochondria and caspase activation. Leukemia. 2002;16(9):1735–1744. doi: 10.1038/sj.leu.2402559. [DOI] [PubMed] [Google Scholar]
- 27.Tedder TF, et al. Cloning of a complementary DNA encoding a new mouse B lymphocyte differentiation antigen, homologous to the human B1 (CD20) antigen, and localization of the gene to chromosome 19. J Immunol. 1988;141(12):4388–4394. [PubMed] [Google Scholar]
- 28.Tedder TF, Klejman G, Schlossman SF, Saito H. Structure of the gene encoding the human B lymphocyte differentiation antigen CD20 (B1). J Immunol. 1989;142(7):2560–2568. [PubMed] [Google Scholar]
- 29.Uchida J, et al. Mouse CD20 expression and function. Int Immunol. 2004;16(1):119–129. doi: 10.1093/intimm/dxh009. [DOI] [PubMed] [Google Scholar]
- 30.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Mice carrying a CD20 gene disruption. Immunogenetics. 1998;48(2):125–132. doi: 10.1007/s002510050412. [DOI] [PubMed] [Google Scholar]
- 31.Hultin LE, Hausner MA, Hultin PM, Giorgi JV. CD20 (pan-B cell) antigen is expressed at a low level on a subpopulation of human T lymphocytes. Cytometry. 1993;14(2):196–204. doi: 10.1002/cyto.990140212. [DOI] [PubMed] [Google Scholar]
- 32.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298(5601):2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 33.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26(2):205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroon FP, et al. Immunoglobulin G (IgG) subclass distribution and IgG1 avidity of antibodies in human immunodeficiency virus-infected individuals after revaccination with tetanus toxoid. . Clin Diagn Lab Immunol. 1999;6(3):352–355. doi: 10.1128/cdli.6.3.352-355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23(1):7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Deans JP, et al. Association of 75/80-kDa phosphoproteins and the tyrosine kinases Lyn, Fyn, and Lck with the B cell molecule CD20. Evidence against involvement of the cytoplasmic regions of CD20. J Biol Chem. 1995;270(38):22632–22638. doi: 10.1074/jbc.270.38.22632. [DOI] [PubMed] [Google Scholar]
- 37.Kansas GS, Tedder TF. Transmembrane signals generated through MHC class II, CD19, CD20, CD39, and CD40 antigens induce LFA-1-dependent and independent adhesion in human B cells through a tyrosine kinase-dependent pathway. . J Immunol. 1991;147(12):4094–4102. [PubMed] [Google Scholar]
- 38.Tsuiji M, et al. A checkpoint for autoreactivity in human IgM+ memory B cell development. J Exp Med. 2006;203(2):393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller S, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104(12):3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong Y, et al. Age-related accumulation of Ig V(H) gene somatic mutations in peripheral B cells from aged humans. Clin Exp Immunol. 2003;133(1):59–66. doi: 10.1046/j.1365-2249.2003.02185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weller S, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98(3):1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weller S, et al. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J Exp Med. 2008;205(6):1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheeren FA, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6(3):303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 44.Shvarts A, et al. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 2002;16(6):681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diehl SA, et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180(7):4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing BCR+ human memory B cells by genetic programming. . Nat Med. doi: 10.1038/nm.2071. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aarts WM, et al. VH gene analysis of primary cutaneous B-cell lymphomas: evidence for ongoing somatic hypermutation and isotype switching. Blood. 1998;92(10):3857–3864. [PubMed] [Google Scholar]
- 48.van Dongen JJ, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 49.Cook GP, Tomlinson IM. The human immunoglobulin VH repertoire. Immunol Today. 1995;16(5):237–242. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- 50.Ruths S, Driedijk PC, Weening RS, Out TA. ELISA procedures for the measurement of IgG subclass antibodies to bacterial antigens. J Immunol Methods. 1991;140(1):67–78. doi: 10.1016/0022-1759(91)90127-2. [DOI] [PubMed] [Google Scholar]
- 51.Sanders LA, et al. Immunoglobulin isotype-specific antibody responses to pneumococcal polysaccharide vaccine in patients with recurrent bacterial respiratory tract infections. Pediatr Res. 1995;37(6):812–819. doi: 10.1203/00006450-199506000-00023. [DOI] [PubMed] [Google Scholar]
- 52.Engel P, et al. Abnormal B lymphocyte development, activation, and differentiation in mice that lack or overexpress the CD19 signal transduction molecule. Immunity. 1995;3(1):39–50. doi: 10.1016/1074-7613(95)90157-4. [DOI] [PubMed] [Google Scholar]