Abstract
Fibroblasts, which are the most numerous cell type in the heart, interact with cardiomyocytes in vitro and affect their function; however, they are considered to play a secondary role in cardiac hypertrophy and failure. Here we have shown that cardiac fibroblasts are essential for the protective and hypertrophic myocardial responses to pressure overload in vivo in mice. Haploinsufficiency of the transcription factor–encoding gene Krüppel-like factor 5 (Klf5) suppressed cardiac fibrosis and hypertrophy elicited by moderate-intensity pressure overload, whereas cardiomyocyte-specific Klf5 deletion did not alter the hypertrophic responses. By contrast, cardiac fibroblast–specific Klf5 deletion ameliorated cardiac hypertrophy and fibrosis, indicating that KLF5 in fibroblasts is important for the response to pressure overload and that cardiac fibroblasts are required for cardiomyocyte hypertrophy. High-intensity pressure overload caused severe heart failure and early death in mice with Klf5-null fibroblasts. KLF5 transactivated Igf1 in cardiac fibroblasts, and IGF-1 subsequently acted in a paracrine fashion to induce hypertrophic responses in cardiomyocytes. Igf1 induction was essential for cardioprotective responses, as administration of a peptide inhibitor of IGF-1 severely exacerbated heart failure induced by high-intensity pressure overload. Thus, cardiac fibroblasts play a pivotal role in the myocardial adaptive response to pressure overload, and this role is partly controlled by KLF5. Modulation of cardiac fibroblast function may provide a novel strategy for treating heart failure, with KLF5 serving as an attractive target.
Introduction
Myocardial hypertrophy is an essential adaptive process through which the heart responds to various mechanophysical, metabolic, and genetic stresses. However, the hypertrophy induced by sustained overload eventually leads to contractile dysfunction and heart failure through mechanisms that remain poorly understood (1). In addition to enlargement of individual cardiomyocytes, the hypertrophied myocardium exhibits complex structural remodeling that involves rearrangement of the muscle fibers, interstitial fibrosis, accumulation of extracellular matrix, and angiogenesis (2, 3), which implies that the non-muscle cells residing in the interstitium likely play important roles in both cardiac hypertrophy and heart failure. In fact, cells other than cardiomyocytes account for approximately 70% of the total cell number in the heart, with the majority being fibroblasts (4, 5). In addition to extracellular matrix proteins (e.g., collagens), cardiac fibroblasts produce a variety of growth factors that likely mediate an interplay between cardiac fibroblasts and cardiomyocytes. For instance, several humoral factors secreted by cardiac fibroblasts, including cardiotrophin-1 (6), endothelin-1 (7), IL-6 (8), periostin (POSTN) (9), and leukemia inhibitory factor (10), have been shown to induce hypertrophic responses in cultured cardiomyocytes. Cardiac fibroblasts also promote proliferation of cardiomyocytes through paracrine interactions in developing hearts (11). And very recently it was shown that inhibition of a fibroblast-selective miRNA ameliorated cardiac fibrosis, hypertrophy, and dysfunction, suggesting that fibroblasts play a detrimental role in cardiac remodeling (12). Still, the precise function of cardiac fibroblasts during adaptive responses of the myocardium remains unclear (2).
Members of the Krüppel-like factor (KLF) family of transcription factors are important regulators of development, cellular differentiation and growth, and the pathogenesis of various diseases, including cancer and cardiovascular disease (13). We previously used Klf5+/– mice to show that KLF5 is required for cardiac hypertrophy and fibrosis in response to continuous infusion of angiotensin II (AII) (14, 15). In primary cultured cardiac fibroblasts, KLF5 directly controls transcription of Pdgfa, encoding platelet-derived growth factor A (PDGF-A) (14), which is known to be involved in tissue remodeling and wound healing (16–19). The precise role played by KLF5 in cardiac hypertrophy and heart failure remains unclear, however.
In the present study, we developed conditional Klf5-knockout mouse lines to examine the cell type–specific functions of KLF5 in cardiac hypertrophy and heart failure. While cardiomyocyte-specific deletion of Klf5 did not alter the hypertrophic responses to pressure overload, cardiac fibroblast–specific deletion of Klf5 ameliorated cardiac hypertrophy in a moderate-intensity pressure overload model, indicating that fibroblasts are essential for hypertrophic responses of the myocardium. Notably, however, cardiac fibroblast–specific Klf5-knockout mice developed severe heart failure when subjected to high-intensity pressure overload, suggesting cardiac fibroblasts have a cardioprotective function. We further demonstrated that KLF5 controls expression of IGF-1, which mediates the interplay between cardiomyocytes and fibroblasts. These data provide compelling evidence that cardiac fibroblasts play a pivotal role in the adaptive response of the myocardium.
Results
KLF5 plays an important role in pressure overload–induced cardiac hypertrophy.
To analyze KLF5’s function in cardiac adaptive responses, we first established models of pressure overload–induced cardiac hypertrophy using transverse aortic constriction (TAC). By applying a low-intensity TAC (LI-TAC) for 2 weeks, we were able to induce cardiac hypertrophy with preserved cardiac systolic function, while application of high-intensity TAC (HI-TAC) induced severe myocardial dysfunction and LV dilation (Supplemental Figures 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI40295DS1). The survival rates in the LI- and HI-TAC groups were 100% and 90%, respectively, after 2 weeks, which suggests that the LI-TAC model induces adaptive hypertrophy, while in the HI-TAC model, the adaptive response fails to protect against the severe pressure overload and enable maintenance of cardiac function.
When we examined the involvement of KLF5 in cardiac hypertrophy using the LI-TAC model, we found that cardiac expression of KLF5 was increased by LI-TAC (Supplemental Figure 3) and that LI-TAC–induced cardiac hypertrophy and fibrosis was diminished in Klf5+/– mice (Figure 1 and Supplemental Figure 4). Moreover, expression of 4 fibrosis-related genes, Col1a1, Fn1, Ctgf, and Spp1, was significantly suppressed in Klf5+/– mice (Supplemental Figure 4). Thus KLF5 appears to play a critical role in pressure overload–induced cardiac hypertrophy and fibrosis.
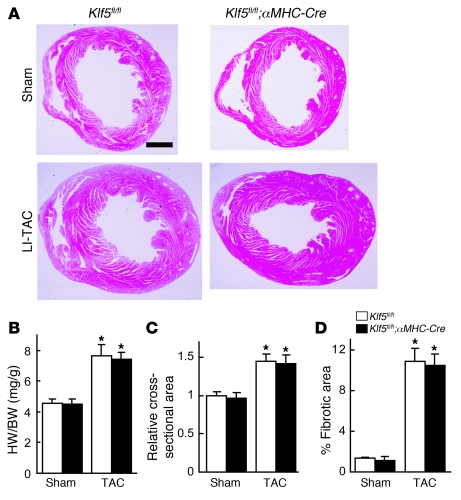
Figure 1. KLF5 is essential for pressure overload–induced hypertrophy.
(A–D) Klf5+/– and wild-type mice were subjected to LI-TAC or sham operation. (A) Representative low-magnification views of H&E-stained heart sections from WT and Klf5+/– mice 2 weeks after the operations. Scale bar: 1 mm. (B and C) Heart weight/body (HW/BW) weight ratios (B) and relative cross-sectional areas of cardiomyocytes (C) from wild-type and Klf5+/– hearts. (D) Fractional areas of fibrosis in cross sections of hearts as determined by elastic picrosirius red staining. *P < 0.01 versus sham control of the same genotype; #P < 0.05 versus wild-type subjected to TAC. n = 7. (E) Expression of KLF5 in normal and hypertrophied hearts 4 days after LI-TAC. Cells were double stained for KLF5 (brown) and a cardiomyocyte marker, αMHC (red); nuclei were counterstained in blue. Scale bar: 20 μm.
Cardiomyocyte-specific deletion of Klf5 does not affect pressure overload–induced cardiac hypertrophy.
We found that Klf5 is mainly expressed in fibroblasts (Supplemental Figure 3C), which suggests that its function in fibroblasts might contribute to the phenotypes seen in Klf5+/– mice. To test this idea, we generated several conditional Klf5-knockout mouse lines (Supplemental Figure 5). Homozygous Klf5-floxed (Klf5fl/fl) mice appeared normal, and expression of Klf5 was unaltered (data not shown). The Klf5fl/fl mice were then crossed with cardiomyocyte-specific Cre transgenic mice (αMHC-Cre) (20). In cardiomyocytes from adult cardiomyocyte-specific Klf5-knockout (Klf5fl/fl;αMHC-Cre) mice, which were homozygous for both floxed Klf5 and the αMHC-Cre transgene, approximately 70% of the Klf5 gene was deleted (Supplemental Figure 6). When Klf5fl/fl;αMHC-Cre mice and control Klf5fl/fl mice were subjected to LI-TAC, there were no differences in cardiac structure or function, or gene expression, between the 2 groups (Figure 2 and Supplemental Figure 7), which means that the level of Klf5 deletion obtained in Klf5fl/fl;αMHC-Cre mice did not affect the hypertrophic response to TAC.
Figure 2. Cardiomyocyte-specific deletion of Klf5 did not alter pressure overload–induced hypertrophy.
Klf5fl/fl and Klf5fl/fl;αMHC-Cre mice were subjected to LI-TAC or sham operation. (A) Representative low-magnification views of H&E-stained heart sections 2 weeks after LI-TAC. Scale bar: 1 mm. (B and C) Heart weight/body weight ratios (B) and relative cross-sectional areas of cardiomyocytes normalized to those obtained from Klf5fl/fl mice subjected to sham operations (C). (D) Fractional areas of fibrosis. *P < 0.01 versus sham control of the same genotype. n = 7.
Cardiac fibroblast–specific deletion of Klf5 reduces hypertrophic and fibrotic responses.
We then analyzed the function of KLF5 in cardiac fibroblasts using a transgenic mouse line in which Cre recombinase was driven by a 3.9-kb mouse Postn promoter (21, 22), which is restricted to the non-myocyte lineage in the neonatal heart (P. Snider and S.J. Conway, unpublished observations). Periostin, which is encoded by Postn, is not normally expressed in either the normal or pathological cardiomyocyte lineage (23), but is induced in cardiac fibroblasts by TAC (9, 24, 25). The activity of Cre recombinase in the Postn-Cre mice was examined after they were crossed with R26RstoplacZ indicator mice (26). Although only a few β-galactosidase+ cells were found in the heart under basal conditions, LI-TAC induced robust lacZ expression in fibrotic areas in hearts from R26RstoplacZ;Postn-Cre mice (Figure 3A and Supplemental Figure 8). As expected, lacZ expression was not detected in either cardiomyocytes or ECs (Supplemental Figure 8).
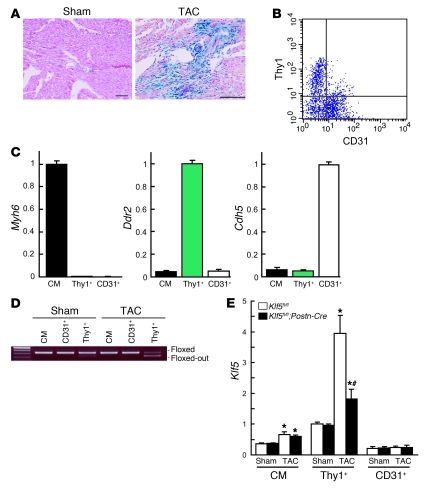
Figure 3. Fibroblast-specific deletion of Klf5 in Klf5fl/fl;Postn-Cre mice.
(A) Fibroblast-specific deletion of the floxed region in Postn-Cre mice was examined using R26RstoplacZ indicator mice. LacZ expression was visualized using X-gal. Scale bars: 100 μm. (B) CD3– cells within non-myocyte-enriched cell populations isolated from adult hearts were analyzed for surface expression of the fibroblast marker Thy1 and the endothelial marker CD31. (C) Relative expression levels of cell-lineage markers in adult cardiomyocytes (CM) isolated using the Langendorff perfusion method, and in Thy1+CD31–CD3– (Thy1+) and Thy1–CD31+CD3– (CD31+) cells sorted from non-myocyte-enriched populations as shown in B. Myh6 (encoding αMHC), Ddr2 (encoding discoidin domain receptor 2), and Cdh5 (encoding VE-cadherin) were used as markers for cardiomyocytes, fibroblasts, and ECs, respectively. The cells were isolated from 8-week-old mice subjected to sham operations. (D) Competitive PCR analysis for quantitation of Cre-mediated recombination of the Klf5 gene region in adult cardiomyocytes, CD31+ ECs, and Thy1+ fibroblasts isolated from Klf5fl/fl and Klf5fl/fl;Postn-Cre mice 2 weeks after either the sham or LI-TAC operation. Competitive PCR was performed as shown in Supplemental Figure 6B. (E) Relative expression levels of Klf5 mRNA in adult cardiomyocytes, Thy1+ fibroblasts, and CD31+ ECs isolated from Klf5fl/fl and Klf5fl/fl;Postn-Cre mice as shown in B 5 days after either sham operation or LI-TAC. Expression levels of Klf5 mRNA were assessed using real-time PCR and normalized to those of 18s rRNA, after which they were further normalized to the levels in Thy1+ cells isolated from Klf5fl/fl mice subjected to the sham operation. *P < 0.01 versus sham control of the same genotype in the same cell lineage group; #P < 0.01 versus Klf5fl/fl mice subjected to LI-TAC in the same cell lineage group.
We then used flow cytometry to further analyze expression of β-galactosidase in populations enriched in either cardiomyocytes or non-myocytes isolated from R26RstoplacZ;Postn-Cre mice subjected to LI-TAC (Supplemental Figure 9). β-Galactosidase+ cells were not found in the cardiomyocyte population. Moreover, β-galactosidase+ cells isolated from the non-myocyte population expressed a fibroblast-specific marker, discoidin domain receptor 2 (Ddr2), but not the cardiomyocyte-specific marker αMHC (Myh6) or the endothelial marker VE-cadherin (Cdh5), which supports the notion that β-galactosidase+ cells are fibroblasts.
Klf5fl/fl mice were then bred with the Postn-Cre mice to generate Klf5fl/fl;Postn-Cre mice.
These animals were born with no apparent abnormalities and were healthy into adulthood. To examine the efficacy of Cre-mediated deletion of Klf5 in each cell type, we isolated cardiomyocytes, cardiac fibroblasts, and ECs from adult mice. Cardiomyocytes were isolated using the Langendorff perfusion method (27). Fibroblasts and ECs were sorted from non-myocyte-enriched cell populations using anti-Thy1 antibody for fibroblasts (11, 28) and anti-CD31 for ECs. Because Thy1 is also expressed in T lymphocytes, CD3– cells were analyzed for surface expression of Thy1 and CD31 (Figure 3B). When the mRNA expression of cell type–specific lineage markers was analyzed, Myh6 was found to be expressed only in cardiomyocytes, while Ddr2 was expressed only in Thy1+CD31–CD3– cells and Cdh5 only in Thy1–CD31+CD3– cells (Figure 3C), which indicates that Thy1+CD31–CD3– cells were fibroblasts. Approximately 72% of the Klf5 gene was deleted in Thy1+CD31–CD3– fibroblasts isolated from Klf5fl/fl;Postn-Cre mice subjected to LI-TAC for 2 weeks, whereas only 4% was deleted in the sham-operated mice (Figure 3D). No Klf5 deletion was observed in cardiomyocytes or ECs. Moreover, Cre mRNA was selectively expressed in Thy1+ fibroblasts, as was endogenous Postn mRNA, which is consistent with fibroblast-specific Cre-mediated deletion by Postn-Cre (Supplemental Figure 10).
We further analyzed expression of Klf5 mRNA in each cell type in mice subjected to either the sham operation or LI-TAC (Figure 3E). In sham-operated hearts, levels of Klf5 expression were higher in Thy1+ fibroblasts than in cardiomyocytes or ECs and did not differ between Klf5fl/fl and Klf5fl/fl;Postn-Cre mice. LI-TAC markedly increased Klf5 expression in fibroblasts (approximately 4-fold) and moderately increased it in cardiomyocytes in Klf5fl/fl mice. While Klf5 expression was clearly reduced in fibroblasts from Klf5fl/fl;Postn-Cre, as compared with Klf5fl/fl mice, it was not altered in cardiomyocytes, which is again consistent with fibroblast-specific deletion of Klf5.
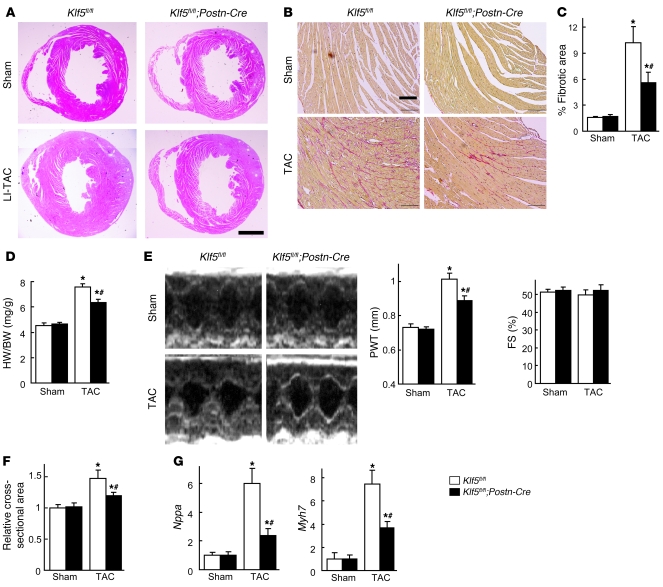
LI-TAC induced less cardiac interstitial fibrosis in Klf5fl/fl;Postn-Cre mice than in control Klf5fl/fl mice, as expected (Figure 4, A–C), and expression of fibrosis-related factors such as Col1a1, Fn1, Ctgf, and Spp1 was reduced (Supplemental Figure 11A). In addition to fibrosis, increases in heart weight/body weight ratios, echocardiographic LV wall thickness, and cardiomyocyte cross-sectional area were smaller in Klf5fl/fl;Postn-Cre mice than Klf5fl/fl mice (Figure 4, D–F). Moreover, expression levels of hypertrophy-related genes such as Nppa, which encodes atrial natriuretic peptide (ANP), and Myh7, which encodes β–myosin heavy chain, were lower in Klf5fl/fl;Postn-Cre than Klf5fl/fl mice (Figure 4G), indicating suppression of hypertrophic responses. These phenotypes clearly demonstrate that not only is KLF5 expressed in cardiac fibroblasts essential for fibrosis, it is also important for mediating subsequent cardiomyocyte hypertrophy.
Figure 4. Fibroblast-specific deletion of Klf5 attenuates cardiac hypertrophy and fibrosis after TAC.
Klf5fl/fl and Klf5fl/fl;Postn-Cre mice were subjected to LI-TAC or sham operation. (A) Representative low-magnification views of H&E-stained heart sections 2 weeks after the operations. Scale bar: 1 mm. The bottom-left panel was composited from 2 photographs of the same section. (B) Representative elastic picrosirius red–stained sections and fibrotic areas. Scale bars: 100 μm. (C) Fibrotic areas. (D) Heart weight/body weight ratios 2 weeks after the operations. (E) Echocardiographic analysis 2 weeks after the operations. (F) Relative cross-sectional areas of cardiomyocytes. (G) Relative expression levels of Nppa and Myh7 mRNA. Expression levels of each gene were normalized to 18s ribosomal RNA levels and then further normalized with respect to those obtained with samples from Klf5fl/fl mice subjected to sham operation. *P < 0.01 versus sham control of the same genotype; #P < 0.01 versus Klf5fl/fl subjected to TAC. n = 7.
IGF-1 controlled by KLF5 mediates hypertrophic responses.
We next investigated the mechanisms by which KLF5 expressed in fibroblasts controls hypertrophy of cardiomyocytes. Because earlier studies have suggested there are paracrine interactions between cardiomyocytes and fibroblasts (2, 3), we hypothesized that KLF5 might directly control the expression of paracrine factors in fibroblasts. To test that idea, we cultured cardiomyocytes in medium conditioned by cardiac fibroblasts transfected with either siRNA against KLF5 (29) or control siRNA (Figure 5A). We found that the medium conditioned by KLF5 knockdown fibroblasts was less able to induce cardiomyocyte hypertrophy and ANP secretion than medium conditioned by control cells (Figure 5, B–D), suggesting that, in fibroblasts, KLF5 does indeed control production of paracrine factors that induce hypertrophic responses in cardiomyocytes. Similarly, Thy1+ cardiac fibroblasts isolated from Klf5fl/fl;Postn-Cre mice subjected to LI-TAC were less able to induce cardiomyocyte hypertrophy than Thy1+ cells from Klf5fl/fl mice (Supplemental Figure 12).
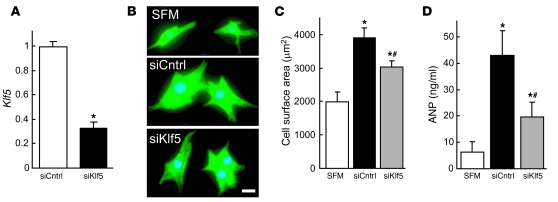
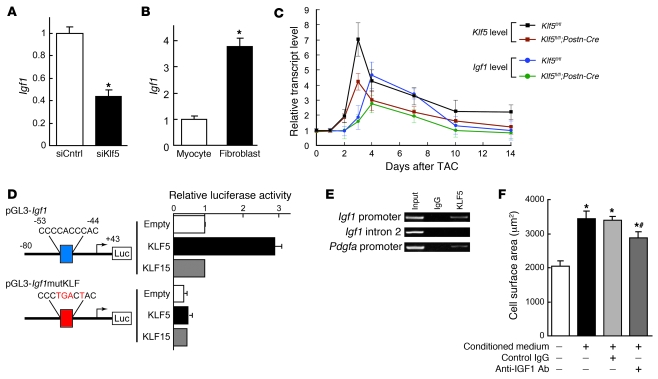
Figure 5. KLF5 controls expression of paracrine factors in cardiac fibroblasts that mediate cardiomyocyte hypertrophy.
(A) siRNA-mediated knockdown of Klf5 in cardiac fibroblasts. Klf5 levels were normalized to those in cells transfected with the control siRNA (siCntrl). *P < 0.01 versus siCntrl. (B–D) Cultured cardiomyocytes were incubated with serum-free medium (SFM) or conditioned medium prepared from cardiac fibroblasts transfected with control or Klf5 siRNA for 48 hours. (B) Representative cardiomyocytes are shown stained for sarcomeric α-actinin (green) and nuclei (Hoechst 33258, blue). Scale bar: 10 μm. (C) Cell surface areas of 100 cells from each group. *P < 0.01 versus cells treated with SFM; #P < 0.05 versus cells treated with medium conditioned by siCntrl transfectants. (D) ANP concentrations in culture medium conditioned by cardiomyocytes. *P < 0.05 versus SFM; #P < 0.05 versus siCntrl.
To identify the paracrine factor genes targeted by KLF5, we compared genome-wide gene expression profiles for the left ventricle in wild-type mice subjected to the sham operation and those subjected LI-TAC; and in Klf5fl/fl and Klf5fl/fl;Postn-Cre mice subjected to LI-TAC (Supplemental Table 1). Thereafter, expression levels were verified by real-time PCR. We screened for genes encoding secreted proteins whose expression levels were significantly increased by LI-TAC in wild-type mice and were lower in Klf5fl/fl;Postn-Cre than Klf5fl/fl mice. Among the 11 genes that met these criteria, 6 were preferentially expressed in cardiac fibroblasts, as compared with cardiomyocytes (Supplemental Table 1), and included 2 growth factor genes, Igf1 and Tgfb3, encoding IGF-1 and TGF-β3, and Postn, encoding periostin. While knocking down KLF5 significantly reduced expression of Igf1 in cultured fibroblasts (Figure 6A), expression levels of Tgfb3 and Postn were not altered (Supplemental Figure 13), suggesting that KLF5 might directly control Igf1 transcription. Moreover, Igf1 expression was highly enriched in fibroblasts (Figure 6B). IGF-1 reportedly promotes cardiac growth and improves cardiac function in patients with LV dysfunction and advanced heart failure (30–32). We therefore further analyzed Igf1 as a likely downstream target of KLF5.
Figure 6. KLF5 transactivates the Igf1 promoter.
(A) KLF5 knockdown reduced Igf1 expression in cardiac fibroblasts. KLF5 was knocked down as shown in Figure 5A. *P < 0.01 versus siCntrl. (B) Fibroblast-selective expression of Igf1. Igf1 mRNA levels in cultured cardiac fibroblasts were normalized to those of 18s rRNA and then further normalized with respect to those in cardiomyocytes. *P < 0.01 versus cardiomyocytes. (C) Cardiac expression of Klf5 and Igf1 mRNA after LI-TAC in Klf5fl/fl and Klf5fl/fl;Postn-Cre mice. Expression levels were normalized to those of 18s rRNA and then further normalized with respect to those in the hearts before TAC. (D) Reporter analysis of KLF5-depedent transactivation of the Igf1 promoter. Luciferase reporter constructs driven by the wild-type Igf1 promoter or a mutant promoter in which the potential KLF-binding site was mutated were cotransfected with either empty vector or a vector harboring Klf5 or Klf15. Data are representative of 3 independent experiments. (E) ChIP assays of KLF5 binding to the Igf1 and Pdgfa promoters. An intronic region of Igf1 that does not contain a KLF-binding motif served as a negative control. (F) Effects of neutralizing IGF-1 on the cardiotrophic activity of fibroblast-conditioned medium. An antibody against IGF-1 (30 μg/ml) or normal IgG was added to the conditioned medium, after which the effect of the medium on cardiomyocyte surface area was analyzed. *P < 0.01 versus cells treated with SFM; #P < 0.01 versus cells treated with fibroblast-conditioned medium.
We found that upregulation of myocardial Klf5 expression after LI-TAC preceded the induction of Igf1 (Figure 6C). Levels of both Klf5 and Igf1 expression were reduced in Klf5fl/fl;Postn-Cre mice, as compared with those in Klf5fl/fl mice, during the 2-week observation period following the LI-TAC operation. The Igf1 promoter contains a KLF-binding motif (CCCCACCCAC) at –53 bp, which, in the rat, is reportedly important for promoter activity and bound by an as-yet-unidentified transcription factor (Figure 6D) (33). Reporter analysis of the Igf1 promoter showed that KLF5 transactivated this promoter, but KLF15, which is expressed in cardiomyocytes and cardiac fibroblasts (34, 35), failed to do so, and mutation within the potential KLF5-binding motif abolished KLF5-dependent transactivation (Figure 6D). ChIP assays confirmed that KLF5 bound to the Igf1 promoter (Figure 6E). As reported (30–32), IGF-1 induced hypertrophy in cultured cardiomyocytes (Supplemental Figure 14A), and inhibition of IGF-1 using a neutralizing antibody significantly suppressed cardiomyocyte hypertrophy induced by the fibroblast-conditioned medium (Figure 6F). Taken together, the results so far demonstrate that KLF5 directly regulates expression of Igf1, which appears to be a major cardiotrophic factor secreted by fibroblasts.
We also found that the numbers of fibroblasts positive for BrdU incorporation following LI-TAC were significantly smaller in Klf5fl/fl;Postn-Cre hearts than Klf5fl/fl hearts (Supplemental Figure 11B), which suggests that KLF5 may also be involved in modulating fibroblast proliferation, either autonomously (36–38) or by regulating autocrine/paracrine factors. Consistent with the latter, we found that IGF-1 induces fibroblast proliferation (Supplemental Figure 14B).
We previously reported that KLF5 also controls Pdgfa, which encodes PDGF-A, in response to angiotensin II (14, 39). At the same concentrations, PDGF-A was less able to induce cardiomyocyte hypertrophy than IGF-1 (Supplemental Figure 14A), though PDGF-A and IGF-1 similarly induced fibroblast proliferation (Supplemental Figure 14B). PDGF-A induced greater migration of fibroblasts in Boyden chamber assays than IGF-1 (Supplemental Figure 14C), suggesting PDGF-A is primarily involved in mediating the migration and proliferation of fibroblasts. Thus, among the paracrine factors controlled by KLF5, it appears to be a change in IGF-1 activity that is primarily responsible for the reduced cardiac hypertrophy observed in LI-TAC Klf5fl/fl;Postn-Cre hearts.
Cardiac fibroblasts are essential for adaptive responses to severe pressure overload.
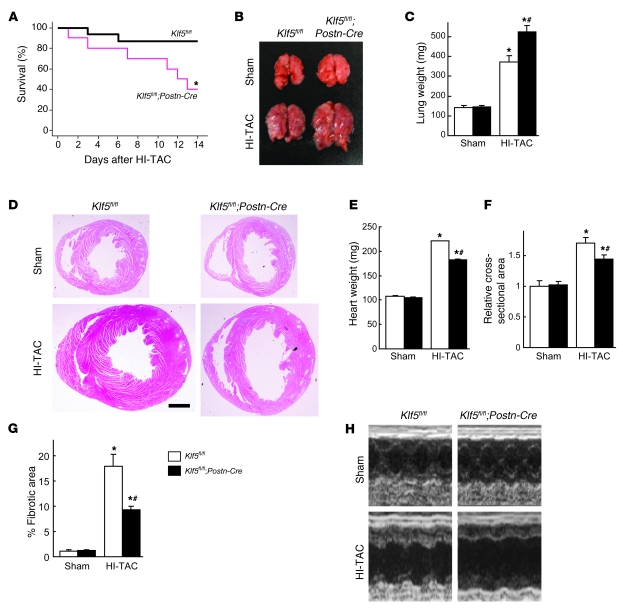
Next we assessed the importance of cardiac fibroblasts in the adaptive responses of the myocardium for maintenance of cardiac function. Klf5fl/fl and Klf5fl/fl;Postn-Cre mice were subjected to HI-TAC, which we previously found to induce cardiac dysfunction within 2 weeks (Supplemental Figures 1 and 2). After 10 days of HI-TAC, Klf5fl/fl;Postn-Cre mice were visibly emaciated, and their body weights were correspondingly diminished (data not shown). In addition, mortality among Klf5fl/fl;Postn-Cre mice was significantly higher than among Klf5fl/fl mice after 14 days of HI-TAC (Figure 7A). Pulmonary edema and/or hemorrhage were noted in most of the dead Klf5fl/fl;Postn-Cre mice, and even in those that survived, lung weights were significantly increased due to pulmonary congestion and alveolar edema (Figure 7, B and C). Many Klf5fl/fl;Postn-Cre mice also exhibited shallow, rapid breathing characteristic of heart failure. Nonetheless, cardiac hypertrophy and fibrosis were markedly attenuated in Klf5fl/fl;Postn-Cre mice, though LV luminal diameters were markedly increased (Figure 7, D–G). Because of the fragility of Klf5fl/fl;Postn-Cre mice, systematic echocardiographic analysis could not be performed, as even a minimum dose of anesthetic was lethal. However, the 1 Klf5fl/fl;Postn-Cre mouse that survived the anesthesia showed marked LV dilation and diminished systolic function (Figure 7H). Apparently, expression of KLF5 by cardiac fibroblasts is necessary for the adaptive and protective activities that take place within the myocardium in response to severe pressure overload.
Figure 7. Cardiac fibroblasts are essential for the protective response elicited by severe pressure overload.
(A–H) Klf5fl/fl and Klf5fl/fl;Postn-Cre mice were subjected to HI-TAC or sham operation. (A) Kaplan-Meier survival analysis of Klf5fl/fl (n = 16) and Klf5fl/fl;Postn-Cre (n = 10) mice after HI-TAC. *P < 0.05 versus Klf5fl/fl. (B) Representative pictures of lungs 2 weeks after the operations. Note the severe lung edema in Klf5fl/fl;Postn-Cre mice subjected to HI-TAC. (C) Lung weights in Klf5fl/fl (n = 5) and Klf5fl/fl;Postn-Cre (n = 3) mice 2 weeks after the operations. (D) Representative low-magnification views of H&E-stained heart sections 2 weeks after the operations. Scale bar: 1 mm. (E–G) Heart weight/body weight ratios (E), relative cross-sectional areas of cardiomyocytes (F), and fibrotic areas (G) in Klf5fl/fl (n = 5) and Klf5fl/fl;Postn-Cre (n = 3) mice 2 weeks after the HI-TAC operation. *P < 0.01 versus sham control of the same genotype; #P < 0.05 versus Klf5fl/fl mice subjected to HI-TAC. (H) M-mode echocardiographic tracings obtained 2 weeks after the operations.
Finally, we tested whether IGF-1 is required for the cardioprotective responses to pressure overload by using JB1, a peptide IGF-1 receptor antagonist, to inhibit IGF-1 signaling in vivo (40). JB1 did not affect survival, cardiac function, heart weights, or cardiac histology in sham-operated control mice (data not shown). In mice subjected to HI-TAC, by contrast, JB1 administration led to the emaciation and shallow, rapid breathing characteristic of heart failure as well as early death (Figure 8A). In JB1-treated mice subjected to HI-TAC, the lung weights were significantly increased (Figure 8, B and C), cardiac hypertrophy was attenuated (Figure 8, D–F), and the left ventricle was significantly enlarged (Figure 8, D and F). Moreover, systolic function was impaired more than in vehicle-treated mice (Figure 8F), indicating that JB1 exacerbated the heart failure. IGF-1 secreted from cardiac fibroblasts thus appears to be a crucial mediator of the cardioprotective response to severe pressure overload.
Figure 8. An IGF-1 receptor antagonist aggravates heart failure induced by severe pressure overload.
(A) Kaplan-Meier survival analysis of wild-type mice treated with vehicle or JB1, a peptide IGF-1 receptor antagonist, after HI-TAC. n = 10 in each group. *P < 0.001 versus vehicle. (B) Representative photographs of lungs taken 1 week after the operations. Note the severe lung edema in JB1-treated mice subjected to HI-TAC. (C) Lung weights in vehicle-treated and JB1-treated groups 1 week after the operations. n = 5 in each group. (D) Representative low-magnification views of H&E-stained heart sections 1 week after the operations. Scale bars: 1 mm. (E) Heart weights. (F) Echocardiographic analysis carried out 1 week after the operation. *P < 0.05 versus sham controls in the same treatment group; #P < 0.05 versus vehicle controls subjected to HI-TAC.
Discussion
The results of the present study clearly demonstrate that cardiac fibroblasts play essential roles in cardioprotection and cardiomyocyte hypertrophy, at least in part by producing paracrine factors, including IGF-1. Based on the observation that in addition to extracellular matrix proteins, cardiac fibroblasts produce a variety of paracrine factors — some of which can induce hypertrophic responses in vitro — it has been postulated that an interplay between cardiomyocytes and fibroblasts is involved in cardiac hypertrophy and pathology (2, 3). However, the potential requirement for fibroblasts in the cardiac response to pathological stress in vivo has not been fully appreciated. Our study provides clear evidence that cardiac fibroblasts functionally contribute to the adaptive response to pressure overload in vivo. Particularly noteworthy is our finding that cardiac fibroblasts are absolutely required for protection of cardiac function in severe pressure overload. This means that cardiac fibroblasts are not mere bystanders acting only in fibrosis, but are crucial mediators of myocardial hypertrophy and adaptive responses in the heart. It was recently suggested that inappropriate angiogenesis plays an important role in heart failure (41). That finding, together with those summarized here, highlights the importance of the activities of the non-muscle cells residing in the myocardial interstitium. In addition to contributing to pathological remodeling, it is likely that these cells mediate homeostatic responses to physiological stress.
Our findings also indicate that KLF5 expressed in cardiac fibroblasts is a key regulator controlling the stress response in the myocardium. Moreover, the results obtained with cardiac fibroblast–specific Klf5-knockout mice suggest that IGF-1 produced by fibroblasts is also important for protective responses. These findings are consistent with the results of earlier studies of the effects of transgenic overexpression of IGF-1 and IGF-1 receptor in cardiomyocytes, which were also suggestive of IGF-1’s role in physiological hypertrophy and cardiac protection (42–46). Thus IGF-1 produced locally by fibroblasts appears to be a key mediator of cardiac hypertrophy and myocardial protection against pressure overload.
The present study identified Igf1 as the target of KLF5 in fibroblasts involved in the cardiac adaptive response. However, KLF5 also likely controls the expression of genes other than Igf1 and Pdgfa, including those encoding paracrine factors involved in regulating fibroblast function (Supplemental Table 1). We would therefore expect future studies of the gene networks controlled by KLF5 in cardiac fibroblasts to shed additional light on their homeostatic and pathological functions. In that regard, it will also be important to analyze the possible interplay between KLF5 and other members of the KLF family. Previous studies have demonstrated the functional roles played by several KLFs in the heart (34, 35, 47, 48). For instance, KLF15 negatively regulates cardiac fibrosis (34). It is therefore conceivable that networks of KLFs contribute to the myocardial responses to stress. Finally, our results suggest that therapeutic modulation of cardiac fibroblast function may represent a novel approach to the prevention and/or treatment of heart failure.
Methods
For experimental procedures not described herein, see Supplemental Methods.
Animals.
Mice were housed in temperature-controlled rooms with a 12-hour light/12-hour dark cycle. All experiments were approved by the University of Tokyo Ethics Committee for Animal Experiments and strictly adhered to the guidelines for animal experiments of the University of Tokyo.
Generation of Klf5-floxed mice.
A 12-kb Klf5 fragment containing exons 1–3 was used to construct the targeting vector. The scheme for construction of the targeting vectors is shown in Supplemental Figure 5. The targeting construct was introduced into ES cells by electroporation, and G418-resistant clones were then examined for homologous recombination using Southern blot analysis with appropriate 3′ probes. Six ES clones that contained the correctly targeted Klf5 locus were obtained, and 2 were injected into 129/Sv blastocysts to obtain chimeric mice. Male chimeras were bred with female transgenic mice expressing the enhanced site-specific recombinase FLP to remove the FRT-flanked neomycin cassette to generate heterozygous floxed Klf5 (Klf5fl/+) mice. Klf5fl/+ mice were then backcrossed with C57BL/6 mice using the marker-assisted speed congenic method (49). The mice were genotyped by Southern blot analysis or PCR. Southern blot analysis was performed after HindIII digestion using a 396-bp PCR-amplified probe for hybridization (Supplemental Figure 5). The probe for intron 1 was made by PCR using primers 5′-TGTCGTGGTGCTTTGAGAAG-3′ and 5′-TATCTTCCAGGCCCTGATTG-3′. PCR for genotyping was performed with primers A (5′-GCATCAGGAGGGTTTCATGT-3′) and B (5′-GTCTCGGCCTCATTGCTAAG-3′), which yielded 164-bp and 331-bp products for wild-type and floxed Klf5 alleles, respectively.
Quantification of Cre-mediated recombination.
Competitive PCR was performed to calculate the relative deletion frequency using primers A, B, and C (5′-TGACCCATTACCGAATCTACTG-3′), which produced 331-bp and 250-bp bands for the floxed and floxed-out Klf5 alleles, respectively. The respective abundances of the floxed and floxed-out Klf5 alleles were analyzed using real-time PCR with the same primer sets. To calculate absolute numbers of alleles in a given cell sample, we used external standards containing known numbers of DNA fragments derived from Klf5-floxed and floxed-out alleles.
TAC model.
TAC was performed as described previously (50). Briefly, mice (8–10 weeks old, 21–24 g body weight) were anesthetized by intraperitoneal injection of a mixture of xylazine (5 mg/kg) and ketamine (100 mg/kg). The animals were then placed in a supine position, an endotracheal tube was inserted, and the animals were ventilated using a volume-cycled rodent ventilator with a tidal volume of 0.4 ml room air and a respiratory rate of 110 breaths/minute. The chest cavity was exposed by cutting open the proximal portion of the sternum. After the aortic arch between the innominate and left common carotid arteries was isolated, it was constricted with a 7-0 nylon suture tied firmly 3 times against a 25- or 27-gauge blunted needle for LI- and HI-TAC, respectively. Sham-operated mice underwent the identical surgical procedure, including isolation of the aorta, but without placement of the suture.
Administration of JB1, a peptide IGF-1 receptor antagonist.
C57BL/6 mice (8–10 weeks old, 21–24 g body weight) were anesthetized by intraperitoneal injection of pentobarbital sodium (50 mg/kg). An incision was made in the midscapular region under sterile conditions, and an osmotic minipump (Alzet) containing either JB1 (BACHEM) dissolved in 0.15 mol/l NaCl and 1 mmol/l acetic acid or vehicle only was implanted. The delivery rate was 1 mg/kg/d for 14 days.
Echocardiography.
Animals were lightly anesthetized with 2,2,2-tribromoethanol (200 mg/kg) and set in a supine position. Two dimensional (2D) guided M-mode echocardiography was performed using an echocardiogram (model 4500, Sonos) equipped with a 15 to 6 L MHz transducer (Philips). The heart at the level of the LV papillary muscle was imaged in the 2D mode in the parasternal short-axis view with a depth setting of 2 cm. LV diastolic posterior wall thickness (PWT), LV diastolic dimensions (LVDd), and LV end-systolic dimensions (LVDs) were measured. LV fractional shortening (%FS) was calculated as (LVDd – LVDs)/LVDd × 100.
Histological analysis.
Heart sections were prepared as described previously (14) and stained with H&E for overall morphology. Immunohistochemical staining of KLF5 was performed using an anti-KLF5 monoclonal antibody (KM1784). A biotin-conjugated goat anti-rat antibody, streptavidin-conjugated horseradish peroxidase (Dako), and 3,3′-diaminobenzidine (DAB) were used to visualize labeling. For double staining of KLF5 and αMHC, we also used anti-αMHC monoclonal antibody CMA19 (51). Simple Stain AP(M) (Nichirei) and an Alkaline Phosphatase Substrate Kit I (Vector) were used to visualize labeling. To quantify cardiac interstitial fibrosis, we stained sections with elastic picrosirius red, after which images were captured with a digital camera and analyzed using Photoshop (Adobe) and Scion Image.
β-Galactosidase staining.
Heart tissues were fixed for 12 hours at 4°C in PBS containing 0.4% glutaraldehyde, 0.01% Na deoxycholate, 0.1% NP40, 0.1 M MgCl2, and 5 mM EGTA; rinsed 3 times for 30 minutes in PBS; and then incubated for 24 hours at 37°C in a staining solution [1 mM MgCl2, 20 mM K3Fe(CN)6, 20 mM K4Fe(CN)6, 1 mg/ml X-gal in PBS]. LacZ-stained sections were counterstained with nuclear fast red for nuclei, biotin-conjugated isolectin B4 (Vector) for ECs, and elastic picrosirius red for fibrosis.
Cardiomyocyte cross-sectional area.
Heart sections were deparaffinized, rehydrated, and incubated for 1 hour at room temperature with FITC-labeled wheat germ agglutinin (Sigma-Aldrich) to visualize myocyte membranes. Regions that included the circular shapes of capillaries were selected from the epicardial side of the LV free walls. The mean cross-sectional area of cardiomyocytes was determined for each mouse from 60 to 80 cells.
RNA extraction and real-time PCR.
Heart tissues were stored in RNAlater RNA stabilization reagent (QIAGEN) at 4°C. Total RNA was extracted using an RNeasy Fibrous Tissue Mini Kit (QIAGEN). First-strand cDNA synthesis was performed with 1 μg of total RNA, random hexamers, and SuperScript III Reverse Transcriptase (Invitrogen). Real-time PCR was performed using a QuantiTect SYBR Green PCR kit (QIAGEN) in a LightCycler (Roche). The expression level of each gene was normalized to that of 18s rRNA, which served as an endogenous internal control. The sequences of the PCR primers are shown in Supplemental Table 2.
Isolation of neonatal and adult cardiomyocytes and non-myocytes.
Neonatal ventricles from 1-day-old ICR mice were separated and minced in ice-cold balanced salt solution, as described previously with minor modifications (52). To isolate cardiac cells, the tissues were incubated in a balanced salt solution containing 0.2% collagenase type 2 (Worthington Biochemical) for 6 minutes at 37°C with agitation. The digestion buffer was replaced 7 times, at which point the tissues were completely digested. The dispersed cells were incubated in 10-cm culture dishes for 80 minutes to remove non-myocytes. The unattached viable cells, which were rich in cardiomyocytes, were cultured on gelatin-coated dishes at 37°C in DMEM supplemented with 10% FBS and 10 μM cytosine 1-β-d-arabinofuranoside (Ara C) to inhibit fibroblast proliferation. Using this protocol, we consistently obtained cell populations containing at least 90%–95% cardiomyocytes. Non-myocyte cells that attached to the dishes were cultured in DMEM supplemented with 10% FBS and allowed to grow to confluence, then they were trypsinized and passaged at1:3. This procedure yielded cell cultures that were almost exclusively fibroblasts by the first passage. Experiments were carried out after 2 or 3 passages.
Adult cardiomyocytes were isolated using the Langendorff perfusion method as previously described (27). For isolation of non-myocyte-enriched cells, hearts were dissected free of vessels and atria, washed in ice-cold modified Krebs-Henseleit bicarbonate (KHB) buffer (pH 7.2) (Sigma-Aldrich), and quickly cut into pieces. The heart pieces were incubated in 5 ml of digesting solution (0.25 mg/ml Liberase TH [Roche] and 10 mM HEPES in balanced salt solution containing calcium and magnesium) for 8 minutes at 37°C with vigorous stirring. The supernatant was then added to 10 ml of ice-cold KHB. Five milliliters of fresh digesting solution was added to the remaining tissue fragments, and the digestion and sampling steps were repeated until all the tissue was dissolved. The collected cells were filtered through 35-μm nylon mesh (BD Falcon) and then used for flow cytometry. Levels of Myh6 mRNA expression were much lower in the isolated cell populations than in normal whole hearts, indicating that the cell populations obtained using the method described here were enriched in non-myocytes (Supplemental Figure 15). Flow cytometric analysis (Figure 3) showed that the cell populations contained large fractions of fibroblasts and ECs.
Flow cytometric analysis and sorting.
Single cells were isolated from adult hearts and incubated with PE-conjugated anti-Thy1 antibody (eBioscience), FITC-conjugated anti-CD31 antibody (BD Biosciences), and allophycocyanin-conjugated CD3e antibody (eBioscience), after which they were analyzed and sorted using a FACSAria II (BD Biosciences) and FlowJo software.
For analysis of β-galactosidase expression and the sorting of LacZ+ cells, a FluoReporter LacZ Flow Cytometry Kit (Molecular Probes, Invitrogen) was used according to the manufacturer’s recommendations. Cells were stained with the fluorogenic β-galactosidase substrate fluorescein di-β-d-galactopyranoside (FDG).
Production of medium conditioned by cardiac fibroblasts.
Cardiac fibroblasts were grown to subconfluency in 10-cm dishes. The medium was then replaced with fresh serum-free DMEM, and the cells were incubated for additional 48 hours. The medium was then collected as conditioned medium.
siRNA.
siRNA for Klf5 and control siRNA were purchased from Dharmacon. Using Lipofectamine RNAiMax (Invitrogen), the siRNA at a final concentration of 20 nM in 10 ml of culture medium was transfected into mouse cardiac fibroblasts plated in 10-cm culture dishes. siRNA-mediated knockdown of Klf5 did not affect mRNA expression levels in KLF family members that reportedly function in the cardiovascular system (e.g., Klf2, Klf4, Klf10, Klf13, and Klf15) (Supplemental Figure 16).
Morphometric studies of cells.
Neonatal cardiomyocytes cultured in 3.5-cm dishes were maintained in serum-free DMEM for 24 hours, after which the culture medium was replaced with either fresh serum-free DMEM or medium conditioned by cardiac fibroblasts. To analyze the effects of neutralizing IGF-1, either anti–IGF-1 neutralizing antibody (Millipore) or control IgG (Sigma-Aldrich) was then added, and the cells were cultured for an additional 24 hours. To analyze the effects of growth factors, cells were first cultured for 24 hours in serum-free DMEM. The medium was then replaced with serum-free DMEM containing vehicle, IGF-1 (Wako), or PDGF-A (R&D Systems), and the cells were cultured for an additional 24 hours. The cells were then fixed in 2% paraformaldehyde and permeabilized for 10 minutes with 0.5% Triton X-100 (Sigma-Aldrich) in PBS, after which they were incubated in PBS containing 1% bovine serum albumin for 10 minutes to block nonspecific staining. They were then incubated with anti–sarcomeric α-actinin antibody (Sigma-Aldrich), followed by treatment with an Alexa-conjugated secondary antibody. Cardiomyocyte size was determined by measuring the surface area of sarcomeric α-actinin–positive cells.
Enzyme-linked immunoassay.
To analyze secretion of ANP, cardiomyocytes were cultured in the serum-free DMEM for 24 hours and then stimulated with fresh serum-DMEM or medium conditioned by cardiac fibroblasts for 48 hours. The concentration of ANP protein in the culture medium was measured using an enzyme-linked immunoassay kit (Phoenix Pharmaceuticals).
Analysis of expression of alternatively spliced Igf1 mRNA.
IGF-1 is controlled by 2 distinct promoters associated with untranslated exons 1 and 2, which generate two types of mRNA, containing either exon 1 (class 1 mRNA) or exon 2 (class 2 mRNA) plus a common block of translated exons (53–55). Expression of the two transcripts is differentially regulated in different tissues and species and during development. We therefore determined which mRNA was the major transcript in mouse heart. The respective abundances of the two transcripts were analyzed using real-time PCR with primer sets that specifically amplified either the class 1 or class 2 mRNA. To calculate absolute numbers of transcripts in a given amount of total RNA, we used external standards containing known numbers of class 1 or 2 cDNA fragments. We found that class 2 transcripts were much more abundant in both cardiac fibroblasts and cardiomyocytes than class 1 transcripts, and also in hearts after LI-TAC (Supplemental Figure 17). We therefore analyzed expression of class 2 mRNA as the major Igf1 transcript in the heart and cardiac fibroblasts (Figure 6, A–C). Igf1 class 1 and class 2 cDNA fragments were amplified using a forward primer specific for each leader exon (exon 1, 5′-ATGGGGAAAATCAGCAGTC-3′; exon 2, 5′-CTGCCTGTGTAAACGACCCGG-3′) and a reverse primer (exon 3-4 junction, 5′-GGCTGCTTTTGTAGGCTTCAGTGG-3′) (56).
Igf1 promoter–luciferase constructs.
Because the class 2 Igf1 mRNA is much more abundantly expressed than class 1 mRNA in the heart, we analyzed the promoter associated with exon 2 (33). A genomic fragment of the 5′-flanking region and a part of exon 2 (–80 to +43 bp) was obtained by PCR using mouse genomic DNA. The promoter fragment was then subcloned into a pGL3 basic luciferase reporter vector (Promega) to generate pGL3-Igf1. A mutation within the potential KLF-binding motif was introduced by PCR to generate pGL3-igf1mutKLF. NIH-3T3 cells grown to 60%–80% confluence were then transfected with the vectors, after which luciferase activities were measured and normalized to β-galactosidase activity. The Klf5 expression vector was described previously (57). The Klf15 expression vector was obtained by inserting the Klf15 cDNA into pCAGMS (14).
ChIP.
ChIP assays were performed as previously described (29, 58). Mouse cardiac fibroblasts were formalin fixed, and then chromatin samples were immunoprecipitated using anti-KLF5 mouse monoclonal antibody (KM3918) or control IgG antibody. PCR was performed with the following region-specific primers: for the mouse Igf1 KLF5 site within the exon 2–associated promoter, 5′-ACCCAGGCTCAGAGCATACC-3′ and 5′-GGGTCGTTTACACAGCAGGT-3′; for intron 2 (an intronic region +750 bp from the translation initiation site that does not contain KLF-binding motifs; negative control), 5′-CCTTCACCGCTCTCTGAAAC-3′ and 5′-CATCAGGGCTTCATGGTTCT-3′; and for the Pdgfa KLF5 site, 5′-ATGTAGCTGCCTGCGTGAG-3′ and 5′-CGACAGGGAGGGGGTTATAG-3′.
Statistics.
Data are shown as mean ± SEM. Paired data were evaluated using Student’s t test. Comparisons between multiple groups were made using 1-way ANOVA followed by a post-hoc Bonferroni test. Survival rates among mice were analyzed using long-rank test. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank N. Yamanaka, Y. Xiao, A. Ono, M. Hayashi, and E. Magoshi for their excellent technical assistance. This study was supported by Grants-in-Aid for Scientific Research (to N. Takeda, I. Manabe, R. Nagai) and a grant for Translational Systems Biology and Medicine Initiative from the Ministry of Education, Culture, Sports, Science and Technology of Japan; a research grant from the National Institute of Biomedical Innovation (to R. Nagai); and a research grant from the Japan Science and Technology Institute (to I. Manabe). P. Snider is supported by NIH T32 HL079995 Training Grant in Vascular Biology and Medicine, and S.J. Conway is partially supported by the Riley Children’s Foundation and the NIH.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 120:254–265 (2010). doi:10.1172/JCI40295.
References
- 1.Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 2.Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291(3):H1015–H1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 3.Manabe I, Shindo T, Nagai R. Gene expression in fibroblasts and fibrosis: involvement in cardiac hypertrophy. Circ Res. 2002;91(12):1103–1113. doi: 10.1161/01.RES.0000046452.67724.B8. [DOI] [PubMed] [Google Scholar]
- 4.Eghbali M, et al. Collagen chain mRNAs in isolated heart cells from young and adult rats. J Mol Cell Cardiol. 1988;20(3):267–276. doi: 10.1016/S0022-2828(88)80059-2. [DOI] [PubMed] [Google Scholar]
- 5.Zak R. Cell proliferation during cardiac growth. Am J Cardiol. 1973;31(2):211–219. doi: 10.1016/0002-9149(73)91034-5. [DOI] [PubMed] [Google Scholar]
- 6.Kuwahara K, et al. Involvement of cardiotrophin-1 in cardiac myocyte-nonmyocyte interactions during hypertrophy of rat cardiac myocytes in vitro. Circulation. 1999;100(10):1116–1124. doi: 10.1161/01.cir.100.10.1116. [DOI] [PubMed] [Google Scholar]
- 7.Harada M, et al. Significance of ventricular myocytes and nonmyocytes interaction during cardiocyte hypertrophy: evidence for endothelin-1 as a paracrine hypertrophic factor from cardiac nonmyocytes. Circulation. 1997;96(10):3737–3744. doi: 10.1161/01.cir.96.10.3737. [DOI] [PubMed] [Google Scholar]
- 8.Sano M, et al. Interleukin-6 family of cytokines mediate angiotensin II-induced cardiac hypertrophy in rodent cardiomyocytes. J Biol Chem. 2000;275(38):29717–29723. doi: 10.1074/jbc.M003128200. [DOI] [PubMed] [Google Scholar]
- 9.Oka T, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101(3):313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King KL, et al. Phenylephrine, endothelin, prostaglandin F2alpha’ and leukemia inhibitory factor induce different cardiac hypertrophy phenotypes in vitro. Endocrine. 1998;9(1):45–55. doi: 10.1385/ENDO:9:1:45. [DOI] [PubMed] [Google Scholar]
- 11.Ieda M, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16(2):233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 13.Haldar SM, Ibrahim OA, Jain MK. Kruppel-like Factors (KLFs) in muscle biology. J Mol Cell Cardiol. 2007;43(1):1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shindo T, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8(8):856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 15.Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3(8):1569–1576. doi: 10.1111/j.1538-7836.2005.01366.x. [DOI] [PubMed] [Google Scholar]
- 16.Forsberg K, Valyi-Nagy I, Heldin CH, Herlyn M, Westermark B. Platelet-derived growth factor (PDGF) in oncogenesis: development of a vascular connective tissue stroma in xenotransplanted human melanoma producing PDGF-BB. Proc Natl Acad Sci U S A. 1993;90(2):393–397. doi: 10.1073/pnas.90.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277(5323):242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 18.Raines EW. PDGF and cardiovascular disease. Cytokine Growth Factor Rev. 2004;15(4):237–254. doi: 10.1016/j.cytogfr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100(1):169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsley A, et al. Identification and characterization of a novel Schwann and outflow tract endocardial cushion lineage-restricted periostin enhancer. Dev Biol. 2007;307(2):340–355. doi: 10.1016/j.ydbio.2007.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph NM, et al. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell. 2008;13(2):129–140. doi: 10.1016/j.ccr.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway SJ, Molkentin JD. Periostin as a Heterofunctional Regulator of Cardiac Development and Disease. Current Genomics. 2008;9(8):548–555. doi: 10.2174/138920208786847917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snider P, et al. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102(7):752–760. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.N Engl J Med. 2007;357(15):1552–1554. doi: 10.1056/NEJMcibr074816. [DOI] [PubMed] [Google Scholar]
- 26.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 27.Shioya T. A simple technique for isolating healthy heart cells from mouse models. J Physiol Sci. 2007;57(6):327–335. doi: 10.2170/physiolsci.RP010107. [DOI] [PubMed] [Google Scholar]
- 28.Hudon-David F, Bouzeghrane F, Couture P, Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. J Mol Cell Cardiol. 2007;42(5):991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Oishi Y, et al. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat Med. 2008;14(6):656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 30.Serose A, Salmon A, Fiszman MY, Fromes Y. Short-term treatment using insulin-like growth factor-1 (IGF-1) improves life expectancy of the delta-sarcoglycan deficient hamster. J Gene Med. 2006;8(8):1048–1055. doi: 10.1002/jgm.934. [DOI] [PubMed] [Google Scholar]
- 31.Abbas A, Grant PJ, Kearney MT. Role of IGF-1 in glucose regulation and cardiovascular disease. Expert Rev Cardiovasc Ther. 2008;6(8):1135–1149. doi: 10.1586/14779072.6.8.1135. [DOI] [PubMed] [Google Scholar]
- 32.McMullen JR. Role of insulin-like growth factor 1 and phosphoinositide 3-kinase in a setting of heart disease. Clin Exp Pharmacol Physiol. 2008;35(3):349–354. doi: 10.1111/j.1440-1681.2007.04873.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Talamantez JL, Adamo ML. A CACCC box in the proximal exon 2 promoter of the rat insulin-like growth factor I gene is required for basal promoter activity. Endocrinology. 1998;139(3):1054–1066. doi: 10.1210/en.139.3.1054. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, et al. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J Mol Cell Cardiol. 2008;45(2):193–197. doi: 10.1016/j.yjmcc.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisch S, et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104(17):7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ema M, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3(5):555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Parisi S, et al. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121(Pt 16):2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 38.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 39.Aizawa K, et al. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J Biol Chem. 2004;279(1):70–76. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 40.Pietrzkowski Z, Wernicke D, Porcu P, Jameson BA, Baserga R. Inhibition of cellular proliferation by peptide analogues of insulin-like growth factor 1. Cancer Res. 1992;52(23):6447–6451. [PubMed] [Google Scholar]
- 41.Sano M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446(7134):444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 42.Welch S, et al. Cardiac-specific IGF-1 expression attenuates dilated cardiomyopathy in tropomodulin-overexpressing transgenic mice. Circ Res. 2002;90(6):641–648. doi: 10.1161/01.RES.0000013780.77774.75. [DOI] [PubMed] [Google Scholar]
- 43.Li B, et al. Insulin-like growth factor-1 attenuates the detrimental impact of nonocclusive coronary artery constriction on the heart. Circ Res. 1999;84(9):1007–1019. doi: 10.1161/01.res.84.9.1007. [DOI] [PubMed] [Google Scholar]
- 44.Kajstura J, et al. IGF-1 overexpression inhibits the development of diabetic cardiomyopathy and angiotensin II-mediated oxidative stress. Diabetes. 2001;50(6):1414–1424. doi: 10.2337/diabetes.50.6.1414. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, et al. Overexpression of insulin-like growth factor-1 in mice protects from myocyte death after infarction, attenuating ventricular dilation, wall stress, and cardiac hypertrophy. J Clin Invest. 1997;100(8):1991–1999. doi: 10.1172/JCI119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMullen JR, et al. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. . J Biol Chem. 2004;279(6):4782–4793. doi: 10.1074/jbc.M310405200. [DOI] [PubMed] [Google Scholar]
- 47.Lavallee G, et al. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006;25(21):5201–5213. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramaniam M, Hawse JR, Johnsen SA, Spelsberg TC. Role of TIEG1 in biological processes and disease states. J Cell Biochem. 2007;102(3):539–548. doi: 10.1002/jcb.21492. [DOI] [PubMed] [Google Scholar]
- 49.Wakeland E, Morel L, Achey K, Yui M, Longmate J. Speed congenics: a classic technique in the fast lane (relatively speaking). Immunol Today. 1997;18(10):472–477. doi: 10.1016/S0167-5699(97)01126-2. [DOI] [PubMed] [Google Scholar]
- 50.Rockman HA, et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komuro I, et al. Isolation and characterization of two isozymes of myosin heavy chain from canine atrium. J Biol Chem. 1986;261(10):4504–4509. [PubMed] [Google Scholar]
- 52.Komuro I, et al. Stretching cardiac myocytes stimulates protooncogene expression. J Biol Chem. 1990;265(7):3595–3598. [PubMed] [Google Scholar]
- 53.Lin WW, Oberbauer AM. Alternative splicing of insulin-like growth factor I mRNA is developmentally regulated in the rat and mouse with preferential exon 2 usage in the mouse. Growth Horm IGF Res. 1998;8(3):225–233. doi: 10.1016/S1096-6374(98)80115-9. [DOI] [PubMed] [Google Scholar]
- 54.Shemer J, Adamo ML, Roberts CT, Jr, LeRoith D. Tissue-specific transcription start site usage in the leader exons of the rat insulin-like growth factor-I gene: evidence for differential regulation in the developing kidney. Endocrinology. 1992;131(6):2793–2799. doi: 10.1210/endo.131.6.1446616. [DOI] [PubMed] [Google Scholar]
- 55.O’Sullivan DC, Szestak TA, Pell JM. Regulation of IGF-I mRNA by GH: putative functions for class 1 and 2 message. Am J Physiol Endocrinol Metab. 2002;283(2):E251–E258. doi: 10.1152/ajpendo.00016.2002. [DOI] [PubMed] [Google Scholar]
- 56.Ohtsuki T, Otsuki M, Murakami Y, Hirata K, Takeuchi S, Takahashi S. Alternative leader-exon usage in mouse IGF-I mRNA variants: class 1 and class 2 IGF-I mRNAs. Zoolog Sci. 2007;24(3):241–247. doi: 10.2108/zsj.24.241. [DOI] [PubMed] [Google Scholar]
- 57.Oishi Y, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1(1):27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 58.Manabe I, Owens GK.CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J Clin Invest. 20011077823–834. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.