Abstract
Herpesviruses exist in two states, latency and a lytic productive cycle. Here we identify an immediate-early gene encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV)/human herpesvirus eight (HHV8) that activates lytic cycle gene expression from the latent viral genome. The gene is a homologue of Rta, a transcriptional activator encoded by Epstein–Barr virus (EBV). KSHV/Rta activated KSHV early lytic genes, including virus-encoded interleukin 6 and polyadenylated nuclear RNA, and a late gene, small viral capsid antigen. In cells dually infected with Epstein–Barr virus and KSHV, each Rta activated only autologous lytic cycle genes. Expression of viral cytokines under control of the KSHV/Rta gene is likely to contribute to the pathogenesis of KSHV-associated diseases.
The recently discovered gamma-herpesvirus, Kaposi’s sarcoma associated herpes virus (KSHV) or human herpesvirus eight (HHV8), can be cultivated in vitro in human B cell lines derived from primary effusion lymphomas (PEL) (1–4). In these cell lines the virus is predominantly in a latent state, but lytic gene expression can be activated by treatment of the B cells with chemical-inducing agents, such as sodium butyrate or phorbol esters (4–6).
The purpose of our experiments was to determine whether a KSHV-encoded gene plays an essential role in this transition between latency and productive infection. Among gamma-herpesviruses, the mechanism of the latency to lytic cycle switch has been studied most intensively in cultured B cells infected with Epstein–Barr virus (EBV) (7, 8). In this system the viral immediate early gene product variably known as ZEBRA, Zta, or EB1, which contains the EBV BZLF1 ORF, is capable of driving the entire lytic cycle in B cells and in epithelial cells (9–12). The product of the BRLF1 gene, called Rta, synergizes with ZEBRA to activate early genes in B cells and activates the lytic cascade in epithelial cells (13–17). Other members of the gamma-herpesvirus family, including herpes- virus saimiri (HVS), bovine herpesvirus 4, and equine herpesvirus 2, contain well-conserved homologues of the Rta protein (18–20). Some of these proteins have been shown to act as transcriptional activators by using reporter gene constructs, but their role in the viral life cycle has not been elucidated. Homologues of EBV BZLF1 have not been found in other members of the gamma-herpesvirus family.
MATERIALS AND METHODS
Cell Culture.
The studies used five B cell lines derived from primary effusion lymphomas. These were BC-1 (3), D5, a single cell subclone of BC-1 (6), BCBL-1 (4), and two cell lines MH-B2 and HH-B2 established in our laboratory by cocultivation of primary effusion lymphoma pleural fluid cells with gamma-irradiated primary human peripheral blood mononuclear cells. BC-1 and D5 are dually infected with KSHV and EBV. All the other cell lines contain only KSHV. Cells were grown in RPMI 1640 medium supplemented with 15% fetal bovine serum at 37°C in the presence of 5% CO2/95% air.
Cloning of KSHV/Rta and KSHV/K8 Genomic DNA.
Total cellular DNA was harvested from BC-1 cells, partially digested with Sau3A, and ligated with the cosmid vector SuperCos-1 (Stratagene). The library was screened by using as probes two PCR fragments of the KSHV genome originally discovered by the representational difference analysis technique (1). Through genomic walking, 89 KSHV-specific clones were identified. The ends of these clones were sequenced. A 7-kb region of the cosmid containing the Rta gene was sequenced by primer walking without subcloning. The sequence was deposited in the GenBank database on Sept. 19, 1996, with accession no. U71368. The genomic DNA containing the KSHV/Rta gene was amplified from total BC-1 cell DNA by PCR by using primer A (5′-CGCGGATCCACAAAAATGGCGCAAGATGACAAGG-3′) and primer B (5′-CGAATTCTGTAGGTTAACTCCACTTTGCACC-3′) (see Fig. 1). These PCR fragments were cloned into the pcDNA3.1 expression vector (Invitrogen) and designated as KSHV/gRta. Primer A and primer B were used to generate PCR products from total BC-1 DNA in a separate reaction. These PCR products were cloned in pRTS15, another eukaryotic expression vector containing the simian virus 40 promoter (kind gift of S. D. Hayward, Johns Hopkins University, Baltimore, MD). One cloned PCR product from this reaction designated KSHV/gRTAmut contained mutations at amino acids 132 and 133 and a stop codon at amino acid 134.
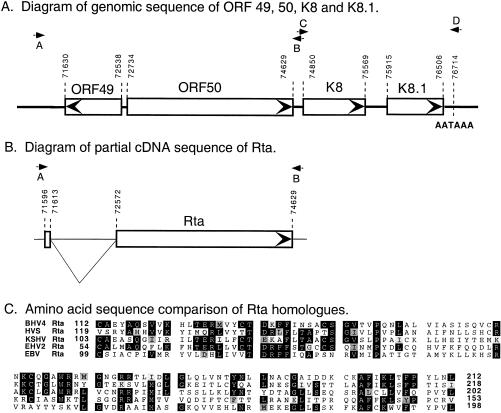
Figure 1.
KSHV gene encoding the R transactivator (Rta). (A) Diagram of the genomic region containing the Rta gene. ORFs are shown in open boxes; the direction of the ORFs is indicated with an arrow inside the box. ORF 50 contains a portion of the Rta gene. Downstream of ORF 50 is an ORF (K8) that has weak homology with EBV BZLF1 (Z). Numbers indicate nucleotide positions in the KSHV sequence (24). The position of primers used for PCR amplification are shown with arrows; their sequences are found in Materials and Methods. A and B indicate positions of primers used to amplify the genomic region containing KSHV/Rta. C and D indicate positions of primers used to amplify genomic DNA containing ORF K8. The single polyadenylation site in this region, AATAAA, is located at position 76,714. (B) Diagram of a partial Rta cDNA clone used for transfection studies. The sequences between 71,613 and 72,572 were removed as an intron; the two open boxes indicate the fused Rta ORF. The start of the Rta mRNA present in infected cells is at position 71,513; the putative initiator methionine is at 71,596. KSHV/Rta is expressed as a 3.6-kb mRNA that contains the indicated mRNA splice involving Rta as well as several downstream splicing events involving K8. (C) Comparison of predicted amino acid sequences of the KSHV/Rta gene with homologues in other members of the gamma-herpesvirus group. Shown is a comparison of KSHV/Rta sequence from aa 103 to 202.
Genomic DNA containing the KSHV/K8 ORF was amplified by PCR by using primer C (5′-CCACCATGGCCAGAATGAAGGACATACCTACTAAG-3′) and primer D (5′-GGTTTTGTGTTACACTATGTAGGG-3′) and cloned into pcDNA3.1.
Cloning of a Predicted KSHV/Rta cDNA.
Based on positional analogies with EBV and HVS, a splicing event was predicted to occur upstream of ORF 50 in the genomic sequence. Canonical splice donor and splice acceptor sequences were identified at nucleotides 71764 and 72570, respectively (see Fig. 1). To clone this predicted cDNA, total cellular RNA was harvested from BCBL-1 cells 18 h after treatment with 20 ng/ml 12-O-tetradecanoylphorbol 13-acetate (TPA). Poly(A)+ RNAs were selected by using the PolyATtract mRNA isolation system (Promega). An Rta-specific cDNA was generated by reverse transcription–PCR by using primers A and B (see Fig. 1). The Rta cDNA was sequenced directly as a PCR fragment. (The GenBank accession no. is U71367.) The cDNA was cloned into pcDNA3.1 vector.
Cloning of a Full-Length KSHV/Rta cDNA.
A full-length cDNA was generated by using a PCR-based cDNA amplification strategy. Polyadenylated RNA was isolated from BC-1 cells that had been treated with N-butyrate for 4 h. Double-stranded cDNA was synthesized with avian meyloblastosis virus reverse transcriptase and cDNA synthesis primer [a modified lock-docking oligo(dT) primer; CLONTECH]. After ligating the cDNAs with an adaptor, the 5′ and 3′ portions were obtained by using the Marathon cDNA amplification kit (CLONTECH). Nested primers designed to clone the KSHV/Rta transcript were primer 1 (5′-CGACACCTGGTACCTCTTTGGG-3′, 74184–74205); primer 2 (5′-CATGTTTCAGGGCCCGCTTCGTCTAACA-3′, 74358–74331); primer 3 (5′-ATGCGCAGAGGCATCCCAAGGCATTATT-3′, 72859–72886) and primer 4 (5′-CAGCCCGGCGGTATCGTACGTGTTGTAG-3′, 73117–73090). To obtain the 5′-rapid amplification of cDNA ends (RACE) fragment of the transcript, the cDNA pool was amplified first with primer 2 and AP1 from the Marathon cDNA kit. The PCR products were then amplified with primer 4 and AP2 from the kit. Similarly, the 3′ portion was obtained through two PCRs with primer 3 and AP1 in the first and primer 1 and AP2 in the second reaction. A 0.6-kb and a 2.0-kb DNA fragment were obtained in the 5′ and 3′ RACE reactions, respectively. The central portion was generated by PCR with primers 2 and 3. These three PCR products were cloned into a T/A-type PCR cloning vector, pCR2.1 (Clontech) and sequenced.
DNA Sequence Analysis.
The DNA sequences of cosmids and PCR products were determined by primer walking with an automatic sequencer. DNA sequence data was compiled and analyzed by using gelassemble, blast, frames, bestfit, and gap of Wisconsin sequence analysis package GCG, version 8 (Genetics Computer Group, Madison, WI).
Transfection.
PEL B cell lines were transfected with 10 μg of plasmids containing KSHV/Rta or other sequences by the electrophoration method (Bio-Rad). The efficiency of transfection was measured by determining the percentage of cells expressing green fluorescent protein (GFP). The gene for enhanced GFP in the plasmid pEGFP-C (CLONTECH) was under the control of the cytomegalovirus immediate early promoter, the same promoter controlling the expression of KSHV/Rta on the pcDNA3 plasmid. Plasmid pEGFP-C was transfected into PEL B cells with or without pcDNA3/KSHV/Rta. After 48 h, the percentage of cells positive for green fluorescence was counted. The presence of pcDNA3/KSHV/Rta did not affect the percentage of cells expressing GFP. Five to eight percent of the electrophorated cells expressed transfected DNA.
RNA Analysis.
Total cellular RNA and Northern blots were prepared as described (21, 22). The blots were hybridized with probes specific for KSHV ORF 50, viral interleukin 6 (IL-6), polyadenylated nuclear RNA (PAN RNA), K8 and small viral capsid antigen (sVCA), EBV BMRF1, and the H1 component of RNase P (23). A complete description of the probes is available on request. To examine sensitivity to cyclohexamide the abundance of RNAs from cells that were treated for 13 h with a combination of butyrate and cyclohexamide was compared by phosphorimagery to RNAs from cells that had been treated with butyrate alone.
Immunofluorescence Assay for sVCA.
An indirect immunofluorescence assay was performed 2 days after BCBL-1 cells were transfected with 10 μg of pcDNA3.1 vector or KSHV/gRta. Cells were reacted with a 1:5 dilution of pre-immune rabbit serum or serum from a rabbit immunized with purified recombinant sVCA (14) followed by a 1:40 dilution of fluorescein-labeled sheep antibody to rabbit Ig. The number of fluorescent cells per 8,000 cells was counted.
RESULTS
Cloning the KSHV/Rta Gene and a Predicted cDNA.
We identified the KSHV homologue of the Rta gene in clones of a cosmid library of genomic DNA from BC-1 cells, a PEL B cell line that is dually infected with KSHV and EBV (3, 21). The major coding portion of the KSHV/Rta gene was present in ORF 50 of the KSHV genome (Fig. 1A) (24). Based on positional analogies with EBV and HVS and conserved mRNA processing events that remove sequences equivalent to KSHV ORF 49 (11), we deduced that the initiator methionine might be present in a small exon upstream of ORF 50 (Fig. 1B). A partial cDNA corresponding to this prediction was isolated from a cDNA pool prepared 18 h after lytic cycle induction. This splicing event introduced a new initiator methionine and an additional 60 aa to the N terminus of ORF 50. We confirmed that this splicing event was a regular feature of KSHV/Rta transcription because 5′-RACE generated a homogeneous product containing the predicted splicing event. The 5′ end of this product was at nucleotide position 71513; thus there are 83 nucleotides of 5′-untranslated mRNA before the start of the KSHV/Rta ORF.
According to the amino acid sequence predicted from the cDNA, KSHV/Rta is 691 aa in length. The strongest homology of KSHV/Rta with corresponding genes in other gamma-herpesviruses is found in aa 103–202 of KSHV/Rta, a region that would lie within the known DNA binding and dimerization region of the EBV Rta protein (Fig. 1C). In this region there is 24% identity between KSHV/Rta and EBV/Rta, with no gaps. Over the whole protein the identity is 22% with gaps. Downstream of ORF50 in the KSHV genome is an ORF K8 with weak homology (22% identity with gaps) to the EBV BZLF1 ORF.
Transcription of KSHV/Rta.
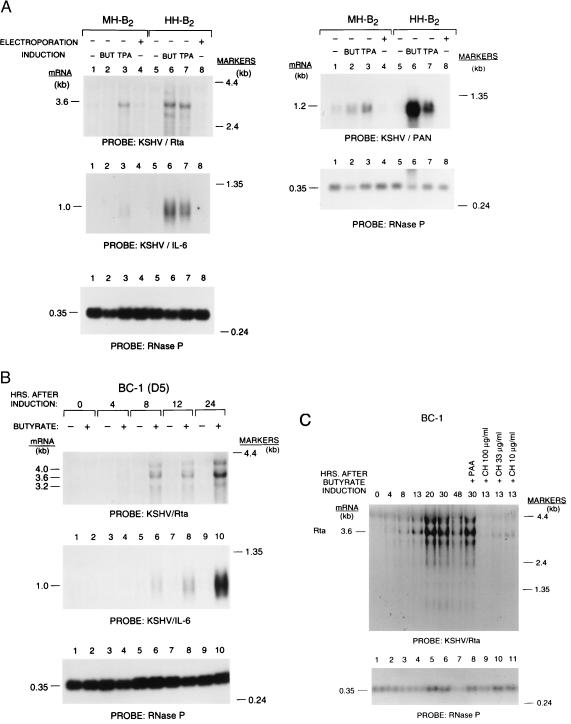
Fig. 2 shows that KSHV/Rta was transcribed after treatment of three PEL B cell lines with inducing chemicals. The extent of expression of three KSHV/Rta transcripts of 4.0, 3.6, and 3.2 kb in response to chemical induction was characteristic of each cell line. In the EBV-negative cell line HH-B2 these transcripts were strongly induced by either TPA or butyrate; in the EBV-negative MH-B2 cell line they were only weakly induced by TPA, and the 4.0- and 3.2-kb transcripts were not evident (Fig. 2A). In EBV-positive cell line BC-1 the Rta transcripts were induced by N-butyrate (Fig. 2B) but not by TPA (data not shown).
Figure 2.
Expression of KSHV/Rta, PAN RNA, and KSHV/IL-6. (A) Chemical induction of KSHV/Rta and KSHV/IL-6 and PAN RNA in two EBV-negative cell lines derived from primary pleural effusion lymphoma. Cells were untreated or treated with TPA or N-butyrate (BUT) or electrophorated without DNA. Twenty-four hours after treatment RNA was prepared and analyzed with probes specific for KSHV Rta, IL-6, and PAN RNA. Each lane contains the RNA equivalent from 2 × 106 cells. (B) Kinetics of expression following lytic cycle induction. BC-1 clone D5 cells were treated with N-butyrate; at the indicated intervals after treatment RNA was prepared and analyzed by Northern blotting. The probes were specific for the second exon of KSHV/Rta, for KSHV/IL6, and for the H1 RNA of RNase P (20). (C) Resistance of the 3.6-kb KSHV/Rta mRNA to the action of cyclohexamide. RNA harvested at intervals after addition of butyrate was analyzed by Northern blotting by using a probe from ORF 50. Aliquots of cells were treated with different amounts of cyclohexamide (CH) and harvested 13 h after induction (lanes 9–11). Another aliquot of cells was treated with phosphonoacetic acid (PAA) and harvested 30 h after induction.
The most abundant transcript of 3.6 kb was resistant to inhibition by cyclohexamide (Fig. 2C). Because this mRNA does not require de novo protein synthesis, KSHV/Rta is an immediate early gene. Preliminary data (S-F.L. and Y.Y., unpublished data) indicates that this 3.6-kb mRNA represents a multiply spliced bicistronic mRNA containing ORF 50 and K8. The two exons of KSHV/Rta are illustrated in Fig. 1B; in addition, there are several splicing events involving K8 and downstream sequences.
Other KSHV encoded lytic cycle transcripts, for example the virally encoded IL-6 and the abundant PAN RNA, were also induced by chemicals (21, 25–27) (Fig. 2 A and B). PAN RNA is an abundant transcript colocalized in the nucleus with Sm antigen, a component of the mRNA splicing apparatus (28–30). PAN RNA does not appear to encode protein. On the basis of their sensitivity to cyclohexamide, kinetics of expression, and resistance to inhibitors of viral DNA synthesis, viral IL-6 and PAN RNA were classified as viral early genes (data not shown). The extent of induction of PAN RNA and viral IL-6 mRNA by chemical stimuli was parallel to the extent of induction of KSHV/Rta (Fig. 2A). There was a strong response of both KSHV/Rta and KSHV/IL-6 in HH-B2 cells and a weak response of the two genes in MH-B2 cells.
We examined the kinetics of induction of lytic cycle mRNAs (Fig. 2B). In three replicate experiments, KSHV/Rta mRNA was stimulated 2.2-to 6.0-fold by 4 h after N-butyrate treatment at a time when KSHV/IL-6 mRNA and KSHV/K8 mRNA remained at uninduced baseline levels. These results suggested that viral IL-6 and K8 were kinetically downstream of KSHV/Rta.
Activation of KSHV Lytic Gene Expression by KSHV/Rta.
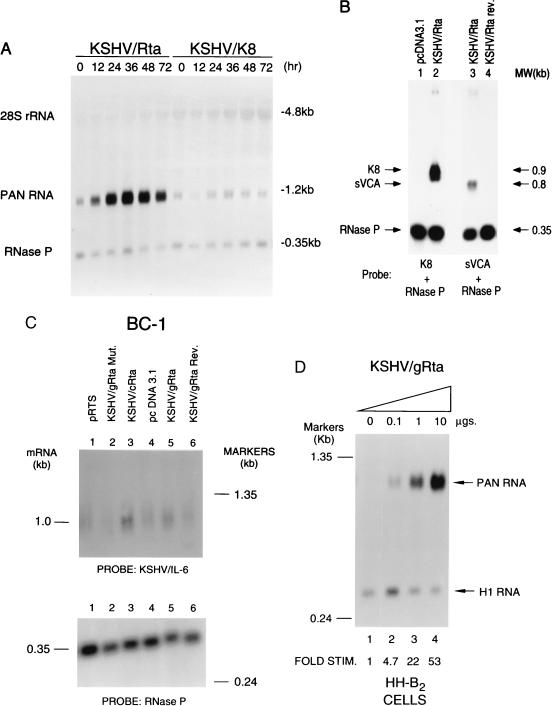
To explore the hypothesis that KSHV/Rta might be a lytic cycle activator expression plasmids containing the KSHV/Rta gene were transfected into cultured PEL B cell lines. The effect on expression of KSHV lytic cycle genes was monitored by Northern blot analysis and quantitated by phosphorimaging. Expression of PAN RNA was progressively stimulated following transfection of BC-1 cells with KSHV/Rta (Fig. 3A). The peak stimulation of PAN RNA was 9-fold 36 h after transfection of KSHV/Rta (Fig. 3A). Activation of expression of PAN RNA as a marker for early lytic gene expression was proportional to the input dose of KSHV/Rta expression plasmid. In EBV-negative HH-B2 cells transfected with 0.1, 1.0, and 10 μg of KSHV/Rta the stimulation of PAN RNA was 4.7-, 22-, and 53-fold when measured 24 h after transfection (Fig. 3D). The extent of induction reflected KSHV/Rta activity in the 5–8% of the cells which were determined to express transfected DNA by using plasmids encoding a green fluorescent protein marker. KSHV/Rta also induced expression of several other early genes including KSHV/K8 gene (Fig. 3B), KSHV/IL-6 (Fig. 3C), and KSHV/MIP-II (ORF K4) (data not shown).
Figure 3.
Activation of KSHV lytic cycle genes by KSHV/Rta. (A) KSHV/Rta stimulates expression of PAN RNA. Replicate aliquots of 2.5 × 107 BC-1 cells were transfected with 10 μg of KSHV/genomic Rta or KSHV/K8 (8, 10, 12). RNA prepared at the times indicated after transfection was analyzed by Northern blotting by using probes specific for PAN RNA and the H1 component of RNase P. (B) KSHV/Rta stimulates expression of KSHV/K8 and KSHV/sVCA (ORF 65). The analysis of KSHV/K8 was carried out in BC-1 cells; RNA was harvested 48 h after transfection. The analysis of KSHV/sVCA was carried out in BCBL-1 cells; the RNA was prepared 96 h after transfection. (C) KSHV/Rta cDNA activates expression of viral IL-6. BC-1 cells were transfected with plasmids containing KSHV/Rta in the genomic configuration (g), in the cDNA configuration (c), in the genomic configuration in the reverse orientation (rev), or in the genomic configuration with a stop codon mutation (mut). The expression plasmids pRTS and pcDNA3.1 were used as negative controls. RNA prepared 24 h after transfection was analyzed by Northern blotting with probes specific for viral IL-6, PAN RNA and RNase P. (D) Response of PAN RNA expression following transfection of different amounts of KSHV/gRta expression plasmid into HH-B2 cells.
KSHV encoded sVCA, a late gene encoded in ORF 65, was also induced following transfection of KSHV/Rta (Fig. 3B). In an immunofluorescence assay, KSHV/Rta caused a 3.2-fold increase in the number of cells expressing sVCA. Correcting for transfection efficiency we estimated that about 20% of cells that received KSHV/Rta expressed this KSHV late viral protein. Because expression of sVCA protein is dependent on lytic viral DNA replication (22), KSHV/Rta may be sufficient to drive the lytic cycle through DNA replication into late gene expression.
Both genomic and cDNA constructs containing KSHV/Rta were comparably active in stimulating expression of viral IL-6 and PAN RNA. Plasmids containing KSHV/Rta in the reverse orientation relative to the cytomegalovirus immediate early promoter were inactive at stimulation of viral IL-6 or sVCA (Fig. 3C, Fig. 4 and data not shown). A construct containing a stop codon at KSHV/Rta aa 134 was likewise inactive at stimulation of expression of PAN RNA and viral IL-6 mRNA (Fig. 3C).
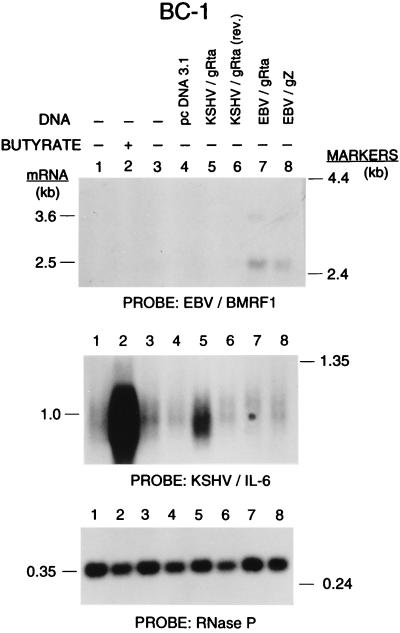
Figure 4.
Specificity of reactivation of two gamma-herpesviruses, KSHV and EBV, in dually infected primary effusion lymphoma cells. BC-1 cells were transfected with KSHV/Rta or EBV/Rta or EBV/Z. RNAs prepared 24 h after transfection were analyzed for transcripts of KSHV/IL-6 and EBV/BMRF1.
Comparable amounts of a plasmid containing KSHV/K8 did not induce expression of PAN RNA (Fig. 3A) or viral IL-6 (data not shown), nor did KSHV/K8 synergize with KSHV/Rta in induction of PAN RNA or viral IL-6 mRNA (data not shown).
Specificity of Activation of KSHV and EBV.
The availability of BC-l, a PEL line dually infected with both KSHV and EBV, offered a unique system in which to explore a question that, to our knowledge, has not before been examined experimentally, namely the specificity of the reactivation pathway controlled by the lytic cycle switch genes of the two gamma-herpesviruses (6). Following transfection of BC-1 cells, KSHV/Rta stimulated expression of KSHV/IL-6 but did not alter expression of the EBV BMRF1 gene, a marker for early EBV lytic cycle gene expression (Fig. 4). By comparison, both EBV/Z and EBV/Rta activated expression of EBV BMRF1 without altering expression of KSHV/IL-6. In the BCBL-l PEL line infected only with KSHV, neither EBV/Rta nor EBV/Z activated KSHV genes (data not shown). Thus each early lytic cycle activator specifically stimulated expression of its autologous virus.
DISCUSSION
A viral gene, Rta, that has homologues among other gamma-herpesviruses, activates lytic cycle gene expression in KSHV. Based on what is known about the homologues in EBV and HVS, viral proteins of this group are likely to act as transcriptional activators (11, 13, 19, 31, 32). These proteins are known to bind DNA specifically, but they may also interact with DNA via other proteins (33). The gamma-herpesvirus Rta proteins appear to be unique: the region involved in DNA binding do not contain any well-characterized DNA binding motifs and the Rta proteins are not homologues of any known cellular transcriptional activators. Therefore, they may be attractive targets for antiviral therapy.
KSHV/Rta is a potent activator. When transfected into three different PEL B cell lines it caused a 3.7- to 53-fold increase in expression of PAN RNA, a 2.0- to 3.4-fold increase in abundance of the mRNA for viral IL-6, and a similar increase in the number of cells expressing capsid antigens. Because only 5–8% of the cells were transfected, the effects of KSHV/Rta were comparable to treatment with inducing chemicals that affect all of the cells. Downstream genes of both early and late kinetic class were activated by KSHV/Rta. However, it is not yet clear whether transfection of KSHV/Rta can lead to increased viral DNA synthesis or viral particle formation. Recent work has shown that EBV/Rta can activate the EBV lytic cascade in epithelial cells (17), and we find that it also disrupts latency in several B lymphoid cell lines (42). Thus, Rta homologues can activate lytic cycle gene expression of both subfamilies of gamma-herpesviruses that are latent in lymphoid cells. This functional homology is consistent with conservation of Rta sequences and localization in the genome (Fig. 1).
Among the downstream targets of KSHV/Rta action are virally encoded cytokines and chemokines such as IL-6 and macrophage inflammatory proteins (MIPs). In a companion study of Kaposi’s sarcoma biopsies that used in situ hybridization KSHV/Rta, KSHV/IL-6, MIPs, PAN RNA, and sVCA have all been found to be expressed in ≈1–5% of Kaposi’s sarcoma tumor cells (refs. 27 and 34, and our unpublished results). Thus these genes are expressed to high level in vivo only in cells that have undergone a latent to lytic switch.
Proinflammatory cytokines have long been considered to be essential components of the pathogenesis of KSHV-associated diseases such as Kaposi’s sarcoma, multicentric Castleman’s disease, primary effusion lymphoma, and multiple myeloma (35–41). Our findings provoke a scenario for pathogenesis by KSHV in which lytic cycle activation and accompanying expression of viral proinflammatory cytokines is a pivotal event. Thus an appreciation of the mechanism of control of the viral lytic cycle by KSHV/Rta would appear to be necessary for unraveling the complex pathogenesis of KSHV-associated diseases.
Acknowledgments
We are grateful to D. Hayward for a plasmid containing EBV/Rta; Y. Chang, P. Moore, and D. Ganem for gifts of BC-1 and BCBL-1 cells; C. Metroka for patient samples; E. Grogan and D. Shedd for technical assistance; and T. Ragoczy for helpful discussions. This work was supported by National Institutes of Health Grants CA70036, CA12055 and T32CA09159.
ABBREVIATIONS
- KSHV
Kaposi’s sarcoma associated herpes virus
- PEL
primary effusion lymphomas
- EBV
Epstein–Barr virus
- HVS
herpes virus saimiri
- TPA
12-O-tetradecanoylphorbol 13-acetate
- RACE
rapid amplification of cDNA ends
- IL-6
interleukin 6
- sVCA
small viral capsid antigen
- PAN RNA
polyadenylated nuclear RNA
Footnotes
References
- 1.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 4.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 5.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller G. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 8.Kieff E. Epstein–Barr Virus and Its Replication. New York: Raven; 1996. [Google Scholar]
- 9.Countryman J K, Miller G. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manet E, Gruffat H, Trescol B M C, Moreno N, Chambard P, Giot J F, Sergeant A. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marschall M, Leser U, Seibl R, Wolf H. J Virol. 1989;63:938–942. doi: 10.1128/jvi.63.2.938-942.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu N, Sakuma S, Ono Y, Takada K. Virology. 1989;172:655–658. doi: 10.1016/0042-6822(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 15.Cox M A, Leahy J, Hardwick J M. J Virol. 1990;64:313–321. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick J M, Lieberman P M, Hayward S D. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalani S, Holley-Guthrie E, Kenney S. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, et al. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Santen V L. J Virol. 1993;67:773–784. doi: 10.1128/jvi.67.2.773-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomonte P, Bublot M, van Santen V, Keil G, Pastoret P P, Thiry E. Vet Microbiol. 1996;53:79–89. doi: 10.1016/s0378-1135(96)01236-9. [DOI] [PubMed] [Google Scholar]
- 21.Sun R, Lin S F, Gradoville L, Miller G. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartkiewicz M, Gold H, Altman S. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 24.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore P S, Boshoff C, Weiss R A, Chang Y. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 26.Nicholas J, Ruvolo V, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 27.Zhong W, Wang H, Herndier B, Ganem D. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baserga S J, Steitz J A. The RNA World. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 359–381. [Google Scholar]
- 29.Guthrie C, Patterson B. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 30.Sharp P A. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutsch D E, Marcu K B, Kenney S C. Cell Mol Biol. 1994;40:747–60.1. [PubMed] [Google Scholar]
- 34.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ensoli B, Barillari G, Gallo R C. Immunol Rev. 1992;127:147–155. doi: 10.1111/j.1600-065x.1992.tb01412.x. [DOI] [PubMed] [Google Scholar]
- 36.Miles S A. Cancer Treat Res. 1992;63:129–140. doi: 10.1007/978-1-4615-3088-6_6. [DOI] [PubMed] [Google Scholar]
- 37.Akira S, Kishimoto T. Semin Cancer Biol. 1992;3:17–26. [PubMed] [Google Scholar]
- 38.Lichtenstein A, Berenson J, Norman D, Chang M P, Carlile A. Blood. 1989;74:1266–1273. [PubMed] [Google Scholar]
- 39.Brandt S J, Bodine D M, Dunbar C E, Nienhuis A W. J Clin Invest. 1990;86:592–599. doi: 10.1172/JCI114749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Hon F D, Ehlers M, Rose-John S, Ebeling S B, Bos H K, Aarden L A, Brakenhoff J P. J Exp Med. 1994;180:2395–2400. doi: 10.1084/jem.180.6.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 42.Ragoczy, T., Heston, L. & Miller, G. (1998) J. Virol., in press. [DOI] [PMC free article] [PubMed]