Abstract
Chronic obstructive pulmonary disease (COPD) is a major cause of morbidity and mortality worldwide. COPD is thought to arise from the interaction of environmental exposures and genetic susceptibility, and major research efforts are underway to identify genetic determinants of COPD susceptibility. With the exception of SERPINA1, genetic associations with COPD identified by candidate gene studies have been inconsistently replicated, and this literature is difficult to interpret. We conducted a systematic review and meta-analysis of all population-based, case–control candidate gene COPD studies indexed in PubMed before 16 July 2008. We stored our findings in an online database, which serves as an up-to-date compendium of COPD genetic associations and cumulative meta-analysis estimates. On the basis of our systematic review, the vast majority of COPD candidate gene era studies are underpowered to detect genetic effect odds ratios of 1.2–1.5. We identified 27 genetic variants with adequate data for quantitative meta-analysis. Of these variants, four were significantly associated with COPD susceptibility in random effects meta-analysis, the GSTM1 null variant (OR 1.45, CI 1.09–1.92), rs1800470 in TGFB1 (0.73, CI 0.64–0.83), rs1800629 in TNF (OR 1.19, CI 1.01–1.40) and rs1799896 in SOD3 (OR 1.97, CI 1.24–3.13). In summary, most COPD candidate gene era studies are underpowered to detect moderate-sized genetic effects. Quantitative meta-analysis identified four variants in GSTM1, TGFB1, TNF and SOD3 that show statistically significant evidence of association with COPD susceptibility.
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide (1). Although cigarette smoking is the primary risk factor for the development of COPD, family studies support the hypothesis that genetic variation contributes to COPD susceptibility (2,3). The only gene that has been definitively proven to influence COPD susceptibility is SERPINA1, the gene that encodes the alpha-1 antitrypsin protein (4).
The search for other genes implicated in the development of COPD has been inconclusive. There have been promising findings from over 100 published COPD candidate gene studies, but most of these findings have not been consistently replicated. As a result of the volume and heterogeneity of this research, it is difficult to synthesize and interpret these findings.
In order to help address this problem, we have assembled an online compendium of population-based, case–control studies in COPD genetics that is based on a comprehensive, regularly updated literature search for COPD genetic studies (www.tuftscaes.org/copddb). This online database is freely accessible and provides up-to-date, comprehensive information on the genetic loci that have been tested for association with COPD. In this report, we summarize the characteristics of the identified studies and perform quantitative meta-analyses of genetic loci that have been studied in three or more independent study populations.
RESULTS
Literature search
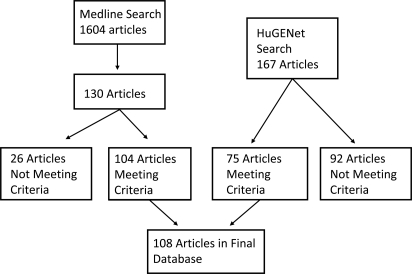
The results of our literature search are presented in Figure 1. The Medline search yielded 1604 publications. Abstract level review narrowed this list to 130 publications. Article level review led to the further exclusion of 26 articles. A parallel search using the Human Genome Epidemiology (HuGE) Navigator tool identified four additional studies that met inclusion criteria, resulting in a final database of 108 population-based, case–control articles pertaining to COPD genetic associations.
Figure 1.
Results of Medline and HuGENet search for case–control COPD genetic association studies. The Medline search yielded 1604 articles. Abstract level review resulted in 130 articles, which were narrowed to 104 articles after full text review. Studies included in the database met the following criteria: peer-reviewed, English language publications of case–control studies of genetic associations with COPD in humans with a total sample size of 10 or more.
Comparing the Medline search to the HuGE Navigator search, 96% of all articles were identified by the Medline search whereas 69% of the articles were identified by the HuGE Navigator search. Of the four articles missed by the Medline search, three were missed due to errors in the abstract screening process and one publication published in May of 2008 did not appear in our initial Medline search. Of the 33 articles missed by the HuGE Navigator search, 22 were published before 2002. The HuGE Navigator database was constructed in 2001.
Analysis of case and control populations
The 108 publications contained information on 82 unique case populations, 96 unique control populations and 100 unique case–control comparisons. The total number of studied individuals was 11 401 cases and 23 775 controls. For the study populations in which the necessary data were available, cases compared with controls were older (median case age 66 years, controls 59) and had more smoking exposure (median pack-years in cases 41, controls 36). Women were underrepresented in these studies, and case and control groups were not balanced by gender (cases 80% male, controls 70.5%).
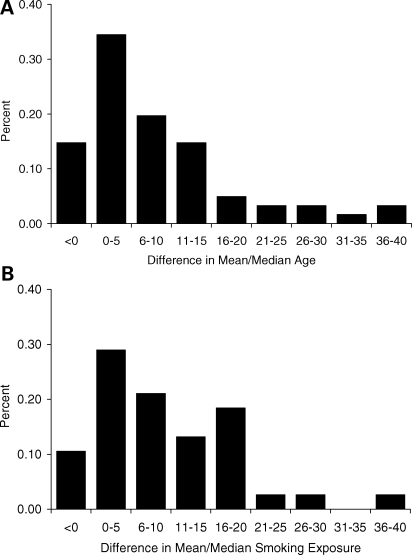
We further described the within-study differences between cases and controls for age and smoking exposure. Data for mean/median age and pack-years of smoking exposure were available in 61 and 41% of all case–control comparisons, respectively. In this subset, the difference in mean/median age was 5 years (cases minus controls) and exceeded 10 years in 31% of the comparisons. The mean/median difference in smoking exposure was 7 pack-years and exceeded 10 pack-years in 39% of the comparisons. The distribution of these differences is displayed in Figure 2.
Figure 2.
Distribution of the difference in mean/median age (A) and pack-year tobacco exposure (B) between case and control groups. Data taken from subset of studies reporting age and pack-year data for both cases and controls (61 and 41% of all studies, respectively). Difference is calculated as mean/median in cases minus controls.
Phenotypes studied
The description of studied COPD phenotypes is presented in Table 1. The majority of studies (72%) used spirometric criteria alone in their COPD definition. 12% of studies used emphysema or chronic bronchitis in their COPD definition, though most of the emphysema studies also reported spirometric measures. (All chronic bronchitis studies included spirometry based on our study inclusion criteria.) Most studies used an forced expiratory volume in one second (FEV1) level of less than 80% of predicted to define COPD, and the bulk of the remaining studies used a cutoff of 70% of predicted. Nearly all studies using FEV1/forced vital capacity (FVC) in the COPD definition specified a cutoff of 70%.
Table 1.
Description of phenotypic definitions for COPD
| COPD definitiona | n (%) |
|---|---|
| Spirometry | 72 (72) |
| FEV1 and FEV1/FVC | 49 (68) |
| FEV1 only | 12 (17) |
| FEV1/FVC only | 8 (11) |
| Not stated | 3 (4) |
| Emphysema | 6 (6) |
| Chronic bronchitis | 6 (6) |
| Not stated | 16 (16) |
aEmphysema definitions may also include spirometry. By our study definition, chronic bronchitis also includes evidence of obstruction.
Genes studied
A total of 72 genes were studied in our publication database. For each study, the median number of variants studied per gene was one, with a range of 1–27. The most heavily studied genes were TNF and EPHX1, with 49 and 30 tests for association between a genetic variant and disease, respectively. Of the ten most-studied variants, five are known inflammatory genes, two are detoxification genes, two are involved in protease/anti-protease activity and one is involved in beta-adrenergic signaling. A list of studied genes in included in Supplementary Material, Table S1.
Power assessment
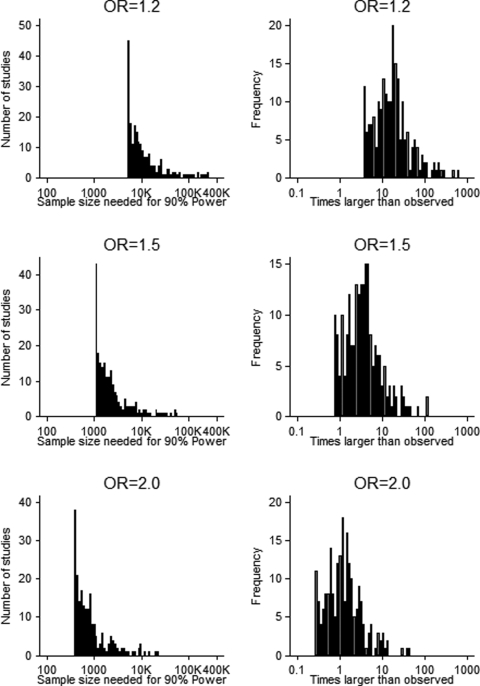
The results of our power assessment are presented in Figure 3. The unit of analysis was each tested genetic association within each study. For each tested association, the percentage of tests that were adequately powered to detect ORs of 1.2, 1.5 and 2.0 was 0%, 9.2% (19 out of 207) and 41.1% (85 out of 207), respectively. We calculated how many times larger the sample size would need to be in order to have 90% power to detect ORs of 1.2, 1.5 and 2.0, and the median required increase in sample size was 17-fold, 3-fold and 1.2-fold larger, respectively.
Figure 3.
Power analysis of COPD candidate gene studies. For each tested genetic association in our database, we calculated the sample size necessary for 90% power to detect genetic effect sizes of 1.2, 1.5 and 2.0 based on the observed minor allele frequency and case:control ratio (left column). The right column shows how many times larger the necessary sample size is compared with the observed sample size for a given genetic effect.
Meta-analyses
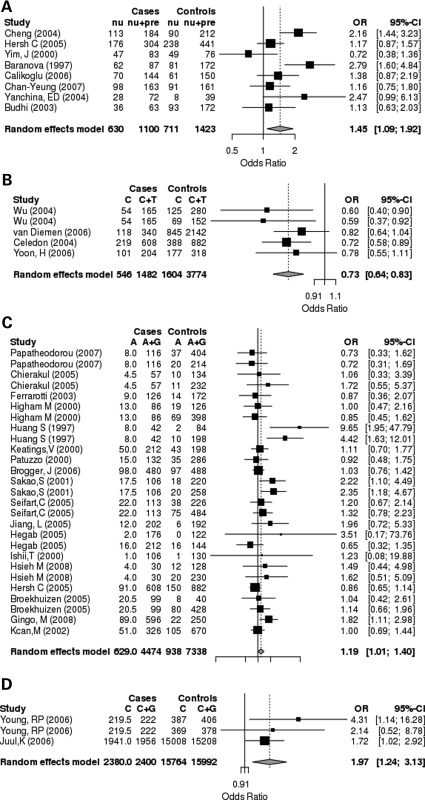
The 100 unique case–control comparisons yielded 207 unique associations between specific genetic variants and COPD susceptibility. Of these, 27 variants had been studied in three or more independent study populations. The quantitative meta-analysis results for these variants are displayed in Table 2. The association with COPD susceptibility reached nominal statistical significance for four variants, GSTM1 null, TNF −308 GA (rs1800629), TGFB1 +29 TC (rs1800470) and SOD3 (rs1799895). Forest plots for each significant association are shown in Figure 4.
Table 2.
Primary meta-analysis results
| Gene | Polymorphism | rs number | Contrast | OR | Studiesa | Allelesb | I2 |
|---|---|---|---|---|---|---|---|
| ACE | Indel | del→ ins | 1.12 (0.92–1.38) | 6 | 1746 | 0.00 | |
| ADRB2 | +46 AG | rs1042713 | A to G | 1.11 (0.75–1.65) | 5 | 2340 | 0.77 |
| ADRB2 | +79 CG | rs1042714 | C to G | 0.89 (0.51–1.41) | 5 | 2362 | 0.82 |
| EPHX1 | His139Arg | rs2234922 | A to G | 1.06 (0.91–1.23) | 15 | 9380 | 0.39 |
| EPHX1 | Tyr113His | rs1051740 | C to T | 1.11 (0.95–1.30) | 15 | 9374 | 0.65 |
| GSTP1 | Ile105Val | rs1695 | A to G | 1.09 (0.80–1.48) | 10 | 6228 | 0.83 |
| IL13 | +2044 GA | A to G | 1.00 (0.76–1.31) | 4 | 1276 | 0.00 | |
| IL13 | −1055 CT | C to T | 0.73 (0.35–1.56) | 3 | 1072 | 0.83 | |
| IL1B | −31 TC | C to T | 0.95 (0.62–1.46) | 4 | 1066 | 0.54 | |
| IL1B | −511 CT | C to T | 1.40 (0.94–2.07) | 5 | 884 | 0.71 | |
| IL4 | −33 CT | rs2070874 | C to T | 1.05 (0.79–1.38) | 4 | 1192 | 0.00 |
| IL6 | −174 GC | C to G | 0.98 (0.77–1.25) | 5 | 1354 | 0.00 | |
| LTA | +252 AG | A to G | 1.00 (0.78–1.28) | 4 | 1596 | 0.00 | |
| MMP9 | −1562 CT | C to T | 0.69 (0.44–1.09) | 4 | 1700 | 0.42 | |
| SERPINA3 | Ala9Thr | A to G | 0.80 (0.62–1.03) | 5 | 2676 | 0.45 | |
| SOD3 | Arg213Gly | rs1799895 | C to G | 1.97 (1.24–3.13) | 3 | 18392 | 0.00 |
| TGFB1 | −509 CT | rs1800469 | C to T | 1.05 (0.70–1.58) | 4 | 5030 | 0.85 |
| TGFB1 | +29 TC | rs1800470 | C to T | 0.73 (0.64–0.83) | 5 | 5256 | 0.00 |
| TIMP2 | +853 GA | rs2277698 | A to G | 0.59 (0.23–1.48) | 3 | 2102 | 0.89 |
| TNF | +489 GA | rs1800610 | A to G | 1.26 (0.96–1.66) | 6 | 2848 | 0.00 |
| TNF | −1031 TC | rs1799964 | C to T | 0.89 (0.67–1.17) | 3 | 1670 | 0.00 |
| TNF | −238 GA | rs361525 | A to G | 0.82 (0.57–1.19) | 5 | 3672 | 0.04 |
| TNF | −308 GA | rs1800629 | A to G | 1.19 (1.01–1.40) | 27 | 11812 | 0.34 |
| TNF | −857 CT | rs1799724 | C to T | 1.04 (0.77–1.40) | 3 | 1606 | 0.00 |
| TNF | −863 CA | rs1800630 | A to C | 0.91 (0.67–1.24) | 3 | 1646 | 0.00 |
| GSTM1 | Null | null → wt | 1.45 (1.09–1.92) | 8 | 2523 | 0.62 | |
| GSTT1 | Null | null → wt | 0.92 (0.73–1.15) | 5 | 1408 | 0.00 |
Meta-analyses performed for variants studied in three or more independent study populations. Random-effects meta-analysis performed using an allele-based model.
aNumber of case–control comparisons in which a variant has been tested for association with COPD.
bNumber of alleles included in meta-analysis (i.e. # of individuals x 2).
Figure 4.
Forest plots of the four meta-analyses meeting nominal statistical significance. Random effects meta-analysis was performed using an allele-based genetic model. Of 27 studied variants, meta-analyses for GSTM1 null (A), rs1800470 in TGFB1 (B), rs1800629 in TNF (C) and rs1799895 in SOD3 (D) yielded summary effect size estimates with P-values <0.05. In Figure 4A, nu = null, pre = wild-type allele. Fractional allele counts result from situations in which a single case group has been contrasted with multiple control groups. In this instance the case allele counts have been divided by the overall number of contrasts to avoid double counting.
In several sensitivity analyses, the GSTM1 finding remained robust. Exclusion of the first published study (5) resulted in a reduced but still statistically significant association (OR 1.32, CI 1.02–1.72). Leave-one-out meta-analysis did not identify any single study driving the cumulative association. Meta-analysis stratified by smoking exposure yielded a similar strength of association, and the Egger test showed no systematic bias between smaller and larger studies. Meta-analysis stratified by race suggested that the association was stronger in Caucasians than Asians, but the strength of this finding is limited by the small number of studies in each ethnic group. None of the studies in the GSTM1 meta-analyses demonstrated deviation from Hardy–Weinberg equilibrium (HWE) in controls.
The association with TGFB1 +29TC was robust in leave-one-out meta-analysis. Although the strength of effect was similar in separate analyses stratified for race and smoking, the meaning of these analyses is limited by the small number of studies in each strata.
The TNF −308GA association was less robust to sensitivity analyses. The association with COPD susceptibility was no longer statistically significant after removal of the 1997 publication by Huang et al. (6). Similarly, there was no association noted in the meta-analysis that excluded studies reporting on multiple populations, suggesting that perhaps publication bias plays a significant role in these studies. Meta-analysis stratified by race suggested that the association was strong in Asians and weak or absent in Caucasians, and this effect persisted after exclusion of the first study (OR in Asians 2.20, CI 1.60–3.04; in Caucasians 1.03, 0.91–1.17).
Since the SOD3 variant meta-analysis included data from three populations presented in two studies, we did not perform sensitivity analyses for this variant. Despite the small number of studies, this variant has been studied in the largest number of individuals, because the study by Juul et al. (7) includes over 9000 individuals (978 COPD cases, 7604 controls).
We examined all meta-analyzed variants for deviation from HWE. Using a threshold of P < 0.05, we identified 12 instances of deviation from HWE in controls. Exclusion of these studies had little effect on most meta-analyses. However, 6 of the 12 instances pertained to the EPHX1 Tyr113His variant, in which there is a known genotyping issue resulting in preferential amplification of the His113 allele and potential misclassification of heterozygotes as His113 homozygotes (8). After excluding these studies, the trend toward significance in the Tyr113His variant was completely attenuated (OR decreased from 1.11 to 0.97), suggesting that this trend was likely due to genotyping error rather than a biological mechanism. Since the genotyping error results in a predictable misclassification of heterozygotes, we repeated our analysis using a dominant genetic model, and the association remained non-significant (OR 1.14, CI 0.92–1.43).
DISCUSSION
Our study is a response to the call from the HuGE Network for field-specific systematic reviews and genetic meta-analyses (9,10), and it is the most comprehensive genetic data synthesis project to date in COPD. Detailed qualitative review of the published literature demonstrated a significant male bias in study sample recruitment, a tendency for case groups to be older and have more smoking exposure than controls, and deficiencies in study reporting, particularly in regard to study sample characteristics and smoking exposure. In addition, the vast majority of studies are dramatically underpowered to detect genetic effect sizes in the range of effects recently identified in GWA studies. In this context, the appropriate application of meta-analysis to achieve increased power can provide a substantial benefit. We identified 27 genetic variants that were suitable for quantitative meta-analysis. Four of these variants, GSTM1 null, rs1800470 in TGFB1, rs1800629 in TNF and rs1799895 in SOD3 are significantly associated with COPD susceptibility. We have made this work publicly available in an online, regularly updated database of COPD genetic associations and cumulative meta-analysis results at www.tuftscaes.org/copddb.
Regarding our meta-analysis findings, the four genetic loci demonstrating statistically significant association with COPD should be prioritized for further study, including additional epidemiologic analysis to confirm or refute these associations, dense genotyping or sequencing to narrow the implicated genomic intervals, and functional studies. When interpreting our negative meta-analyses it is important to note that only nine of our ‘negative’ meta-analyses were adequately powered for ORs of 1.5, and none of our meta-analyses were adequately powered to exclude ORs of ≤1.2.
The deficiencies noted in study reporting are surprising, particularly regarding smoking exposure and basic demographic characteristics, such as age. One of the most common reasons for this was the use of blood donor controls for which little or no smoking and demographic data were available. Given the importance of smoking in the development of COPD and the known association between age and FEV1 decline, it will be essential to address these readily correctable deficiencies in data collection and reporting in future studies.
Two recently published genome wide association study (GWAS) have examined COPD-related phenotypes, but the top hits are in genomic locations that are not represented in our case–control database. One locus (near HHIP) was significantly associated with COPD-related phenotypes in both studies; another locus (near CHRNA3/5) was significantly associated with COPD in one of these studies (11,12). It would be of interest to test our significant meta-analysis associations in these GWA cohorts when the data become available. In the future, we intend to incorporate available GWAS data into our online database, which will serve the dual functions of allowing public access to comprehensive summary GWAS results and integrating GWAS and candidate gene era findings.
There have been four previously published meta-analyses of genetic associations with COPD. These studies pertain to variants in the following genes—TNF (13,14), EPHX1 (13,15) and GSTM1 (16). In addition, the recently published meta-analysis by Smolonska considers 12 genes from well-studied biologic pathways in COPD (17). The significantly associated variants identified in these meta-analyses are as follows: Brogger et al.—EPHX1 Tyr113His, EPHX1 His139Arg and TNF −308GA; Hu et al.—GSTM1 null, EPHX1 Tyr113His, EPHX1 His139Arg, and the fast and slow variants of EPHX1 compared with the normal activity variant; Gingo et al.—TNF 308GA; and Smolonska et al.—the IL1RN variable number tandem repeat (VNTR) polymorphism, three SNPs in TGFB1 (including rs1800470), TNF −308GA (rs1800629) and GSTP1 Ile105Val (rs1695). There were a number of differences from study-to-study in terms of genetic models used, choice of fixed versus random effects meta-analysis, and in stratification variables. We re-analyzed our data using the genetic models of previous meta-analyses and our results were generally consistent with these previous findings, though there were some differences in included/excluded studies. Furthermore, we limited our analysis to single, biallelic polymorphisms, thus we did not analyze the IL1RN VNTR polymorphism or the fast and slow variants of EPHX1.
The differences in our approach compared with the approaches taken by others relate principally to the choice of genetic model and the specification of inclusion/exclusion criteria. We performed allele-based contrasts, because this allows the inclusion of studies that report only allele frequency data. We also applied more restrictive inclusion/exclusion criteria than some previous authors, resulting in the exclusion of four studies for GSTM1, two studies for EPHX1 and 2 studies for TNF that had been included in previous meta-analysis efforts. Of these eight studies, four were published in a non-English language, two included pediatric populations, one drew its case and control populations from a pool of lung cancer patients; and one was excluded because the study population was a subset of a larger study published 1 year later.
Our study has the following limitations. First, our approach is limited to population-based case–control studies. The quantitative synthesis of population-based and family-based studies is an area of ongoing research, and in the future it would strengthen our project to incorporate results from family-based studies. However, the vast majority of published genetic association studies in COPD are population-based case–control studies. Second, publication bias may have affected some of our results. This potential bias is difficult to overcome in retrospective meta-analysis. One of the great strengths of GWAS results is that, if the full set of results is available, publication bias can be avoided entirely. In the future, we anticipate including GWAS results in our database. Third, since COPD is a heterogenous disease, it may be more powerful to analyze distinct COPD subtypes than to analyze COPD as a unified entity. Consensus definitions for emphysema and other COPD-subtypes could significantly improve the power of genetic association analyses. Fourth, our study only considers genetic main effects. It is likely that gene-by-smoking interactions are important in determining COPD susceptibility. In the candidate gene era, the number of gene-by-smoking studies is relatively small. Ongoing, large GWAS studies may provide quality data regarding gene-by-smoking interactions and shed significant light on the genetic architecture of COPD. Finally, despite combining all the available published data, our meta-analyses are not adequately powered to detect weak-to-moderately strong associations. Thus, with the availability of more data, it is likely that some of our ‘negative’ meta-analyses will attain traditional thresholds of statistical significance.
In summary, our database is an online resource that will be regularly maintained so that up-to-date meta-analysis results can be freely accessed, and it provides a systematic, comprehensive, and quantitative approach to gauge the cumulative strength of association between individual genetic variants and COPD susceptibility. Similar web-based databases in Alzheimer's disease (18), Parkinson's disease (19) and schizophrenia (20) have been heavily utilized. As our understanding of the complex genetic architecture of COPD evolves, systematic, ongoing evidence synthesis efforts can contribute to the larger research effort by identifying methodological weaknesses (i.e. study reporting and case–control selection), drawing attention to understudied areas (COPD in women), and prioritizing promising variants for future studies (GSTM1 null, rs1800470 in TGFB1, rs1800629 in TNF and rs1799895 in SOD3).
MATERIALS AND METHODS
Literature search and data extraction
We conducted a literature search of the Ovid Medline database on 16 July 2008 to identify all published, population-based, case–control studies of genetic associations with COPD. The search strategy is included in the Supplementary Material.
Studies eligible for inclusion in our database met the following criteria: population-based, case–control studies pertaining to genetic associations with COPD in adult populations, 10 or more study subjects, and published as full manuscripts in an English language journal. We defined a COPD study as any publication in which the case population was described as having COPD, emphysema or chronic bronchitis. Chronic bronchitis studies were included only if cases also had spirometric evidence of airflow obstruction. Family-based studies were excluded, since there are methodological difficulties combining these results with the odds ratio (OR) metrics provided by case–control studies. SERPINA1 variants were included in the online database but excluded from this analysis in order to focus on novel genetic determinants. To verify the sensitivity of our search strategy, we performed a parallel search using the Phenopedia function of the HuGE Navigator (21).
The following data were extracted from eligible publications: publication information, demographic information, number of cases, number of controls, race of study samples, variables pertaining to COPD definition used (spirometric definitions, emphysema and chronic bronchitis), genotype data, adjusted and unadjusted ORs and covariates included in analytic models. For genotype data, double data extraction was performed on 100% of the articles and conflicts were resolved by additional review and discussion until consensus was reached. When data from multiple populations were presented in a single publication, data for each population were extracted and analyzed separately (i.e. multiple control groups, multiple racial groups). Electronic data extraction was performed using Epidata version 3.1 (22).
More detailed information is supplied in the Supplementary Material.
Database and website design
Extracted data are stored in a structured database using SQLite (http://www.sqlite.org/), a light-weight, self-contained Structured Query Language (SQL) database engine. We have implemented an HTML interface to the data via Pylons (http://pylonshq.com/), a Python (http://python.org) web-application framework. This interface allows the user to query the database for studies that meet specific criteria, and subsequently run a meta-analysis on the returned studies. The computer code for this application is open-source (http://github.com/bwallace/copd_db). The meta-analysis is performed using the rmeta (http://cran.r-project.org/web/packages/rmeta/index.html) package. Python dispatches the calls to R via the rpy (http://rpy.sourceforge.net/rpy2.html) interface and then renders the results.
Statistical analyses
Comparison of demographic and exposure variables between case and control groups
We compared age, gender and pack-year smoking distributions in cases and controls using t-tests, the Mann–Whitney test or χ2 tests.
Power assessment
For each study for which a 2×2 allele-based table could be constructed, we calculated the number of subjects that would be required for 90% power to detect genetic effect ORs of 1.2, 1.5 and 2.0 for each studied genetic variant using the sampsi function in Stata (Intercooled Stata version 8.2). Our power calculations were based on data drawn from each study, i.e. the minor allele frequency for each variant in the controls and the case:control ratio. The direction of effect (i.e. predisposing or protective for COPD) for each variant was obtained from the observed genotype distribution in cases and controls. We then calculated how many times larger this sample size was compared with the observed sample size.
Meta-Analyses
We calculated crude, study-specific ORs and 95% confidence intervals (CIs) using allele-based contrasts. Our analysis was limited to bi-allelic variants only. The unit of analysis was at the level of case–control contrast, thus a single study reporting results from multiple case and/or control groups could contribute multiple case–control contrasts for the same variant. In instances in which data were available for variant-disease associations in three or more case–control contrasts, we calculated summary ORs and CIs using the random effects method of DerSimonian and Laird (23). In instances where a case population was contrasted with multiple control populations, we included all comparisons but divided the number of cases by the number of contrasts in order to avoid double counting.
We quantified between-study heterogeneity using the Q-statistic (24) and the I2 metric. The I2 metric provides the percentage of observed between-study variance that cannot be attributed to chance. Values greater than 50% are considered to represent large amounts of heterogeneity. When the number of studies is small, both Q and I2-based estimates of heterogeneity should be interpreted with caution due to their wide confidence intervals (25).
Sensitivity analyses
For variants studied in four or more study populations, we conducted pre-specified sensitivity analyses to identify causes of heterogeneity and evaluate the robustness of our findings. We performed the following meta-analyses—leave-one-out, excluding studies when the control group was out of HWE (as defined by P < 0.05), excluding the first published study, excluding studies reporting results from multiple populations, stratified by race, and stratified by smoking balance between cases and controls. We also compared the results of smaller versus larger studies with the Egger test. For the analysis stratified by race, study participants were classified according to the following groups—Caucasian, Asian, African and other. If the racial composition of a study sample was not specifically mentioned, race was assigned according to the country of origin of the publication. For the analysis stratified by smoking balance, studies in which the difference in mean or median pack-years between cases and controls was <10 pack-years were compared with those in which this difference was ≥10 pack-years.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institute of Health Grants T32 HS00060 (P.J.C.), F32 HL094035 (P.J.C.), T32HL007427 (M.H.C.), K12HL089990 (M.H.C.), UL1 RR025752, R01 HL075478 (E.K.S.) and R01 HL084323 (E.K.S.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the Alzheimer Research Foundation for their assistance in developing our online compendium. Alzgene is funded by the Cure Alzheimer's Fund.
Conflict of Interest statement. E.K.S. reports grant support and consulting fees from GlaxoSmithKline for studies of COPD genetics and honoraria from GlaxoSmithKline, Wyeth, Bayer and AstraZeneca for lectures on COPD genetics. M.H.C. reports having received reimbursement from GlaxoSmithKline to attend a conference. None of the other authors declare any competing interests.
REFERENCES
- 1.Lopez A.D., Shibuya K., Rao C., Mathers C.D., Hansell A.L., Held L.S., Schmid V., Buist S. Chronic obstructive pulmonary disease: current burden and future projections. Eur. Respir. J. 2006;27:397–412. doi: 10.1183/09031936.06.00025805. [DOI] [PubMed] [Google Scholar]
- 2.Lewitter F.I., Tager I.B., McGue M., Tishler P.V., Speizer F.E. Genetic and environmental determinants of level of pulmonary function. Am. J. Epidemiol. 1984;120:518–530. doi: 10.1093/oxfordjournals.aje.a113912. [DOI] [PubMed] [Google Scholar]
- 3.Redline S., Tishler P.V., Lewitter F.I., Tager I.B., Munoz A., Speizer F.E. Assessment of genetic and nongenetic influences on pulmonary function. A twin study. Am. Rev. Respir. Dis. 1987;135:217–222. doi: 10.1164/arrd.1987.135.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Stoller J.K., Aboussouan L.S. Alpha1-antitrypsin deficiency. Lancet. 2005;365:2225–2236. doi: 10.1016/S0140-6736(05)66781-5. [DOI] [PubMed] [Google Scholar]
- 5.Baranova H., Perriot J., Albuisson E., Ivaschenko T., Baranov V.S., Hemery B., Mouraire P., Riol N., Malet P. Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum. Genet. 1997;99:822–826. doi: 10.1007/s004390050455. [DOI] [PubMed] [Google Scholar]
- 6.Huang S.L., Su C.H., Chang S.C. Tumor necrosis factor-alpha gene polymorphism in chronic bronchitis. Am. J. Respir. Crit. Care Med. 1997;156:1436–1439. doi: 10.1164/ajrccm.156.5.9609138. [DOI] [PubMed] [Google Scholar]
- 7.Juul K., Tybjaerg-Hansen A., Marklund S., Lange P., Nordestgaard B.G. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006;173:858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez F., Jardi R., Costa X., Juan D., Galimany R., Vidal R., Miravitlles M. Detection of polymorphisms at exons 3 (Tyr113→His) and 4 (His139→Arg) of the microsomal epoxide hydrolase gene using fluorescence PCR method combined with melting curves analysis. Anal. Biochem. 2002;308:120–126. doi: 10.1016/s0003-2697(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis J.P., Gwinn M., Little J., Higgins J.P., Bernstein J.L., Boffetta P., Bondy M., Bray M.S., Brenchley P.E., Buffler P.A., et al. A road map for efficient and reliable human genome epidemiology. Nat. Genet. 2006;38:3–5. doi: 10.1038/ng0106-3. [DOI] [PubMed] [Google Scholar]
- 10.Khoury M.J., Bertram L., Boffetta P., Butterworth A.S., Chanock S.J., Dolan S.M., Fortier I., Garcia-Closas M., Gwinn M., Higgins J.P.T., et al. Genome-Wide Association Studies, Field Synopses, and the Development of the Knowledge Base on Genetic Variation and Human Diseases. Am. J. Epidemiol. 2009;170:269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A., et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilk J.B., Chen T.h., Gottlieb D.J., Walter R.E., Nagle M.W., Brandler B.J., Myers R.H., Borecki I.B., Silverman E.K., Weiss S.T., O'Connor G.T. A genome-wide association study of pulmonary function measures in the framingham heart study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brogger J., Steen V.M., Eiken H.G., Gulsvik A., Bakke P. Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur. Respir. J. 2006;27:682–688. doi: 10.1183/09031936.06.00057005. [DOI] [PubMed] [Google Scholar]
- 14.Gingo M.R., Silveira L.J., Miller Y.E., Friedlander A.L., Cosgrove G.P., Chan E.D., Maier L.A., Bowler R.P. Tumour necrosis factor gene polymorphisms are associated with COPD. Eur. Respir. J. 2008;31:1005–1012. doi: 10.1183/09031936.00100307. [DOI] [PubMed] [Google Scholar]
- 15.Hu G., Shi Z., Hu J., Zou G., Peng G., Ran P. Association between polymorphisms of microsomal epoxide hydrolase and COPD: results from meta-analyses. Respirology. 2008;13:837–850. doi: 10.1111/j.1440-1843.2008.01356.x. [DOI] [PubMed] [Google Scholar]
- 16.Hu G., Yao W., Zhou Y., Hu J., Shi Z., Li B., Ran P. Meta- and pooled analyses of the effect of glutathione S-transferase M1 and T1 deficiency on chronic obstructive pulmonary disease. Int. J. Tuberc. Lung Dis. 2008;12:1474–1481. [PubMed] [Google Scholar]
- 17.Smolonska J., Wijmenga C., Postma D.S., Boezen H.M. Meta-analyses on suspected COPD genes—a summary of 20 years' research. Am J. Respir. Crit Care Med. 2009;180:618–631. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- 18.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 19.Bagade S., Allen N.C., Tanzi R., Bertram L. The PDGene Database. Alzheimer Research Forum. 2001. Available at: http://www.pdgene.org .
- 20.Allen N.C., Bagade S., McQueen M.B., Ioannidis J.P., Kavvoura F.K., Khoury M.J., Tanzi R.E., Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 21.Yu W., Gwinn M., Clyne M., Yesupriya A., Khoury M.J. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 22.Lauritsen J.M., Bruus M. Epidata (version 3). A Comprehensive Tool for Validated Entry and Documentation of Data. Odense, Denmark: The Epidata Association; 2008. [Google Scholar]
- 23.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Cochran W.G. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 25.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.