Abstract
Ace is an adhesin to collagen from Enterococcus faecalis expressed conditionally after growth in serum or in the presence of collagen. Here, we generated an ace deletion mutant and showed that it was significantly attenuated versus wild-type OG1RF in a mixed infection rat endocarditis model (P<0.0001), while no differences were observed in a peritonitis model. Complemented OG1RFΔace (pAT392::ace) enhanced early (4 h) heart valve colonization versus OG1RFΔace (pAT392) (P = 0.0418), suggesting that Ace expression is important for early attachment. By flow cytometry using specific anti-recombinant Ace (rAce) immunoglobulins (Igs), we showed in vivo expression of Ace by OG1RF cells obtained directly from infected vegetations, consistent with our previous finding of anti-Ace antibodies in E. faecalis endocarditis patient sera. Finally, rats actively immunized against rAce were less susceptible to infection by OG1RF than non-immunized (P = 0.0004) or sham-immunized (P = 0.0475) by CFU counts. Similarly, animals given specific anti-rAce Igs were less likely to develop E. faecalis endocarditis (P = 0.0001) and showed fewer CFU in vegetations (P = 0.0146). In conclusion, we have shown for the first time that Ace is involved in pathogenesis of, and is useful for protection against, E. faecalis experimental endocarditis.
Author Summary
Enterococcus faecalis was recognized as a common cause of infective endocarditis (IE) by the early 1900s. It is still third in community-onset IE, but is the second most common cause of hospital-associated IE. Complications due to E. faecalis IE include congestive heart failure, septic emboli and death and current management involves a combination of antimicrobials, often with surgery. Emergence of antimicrobial resistance has created the need for alternative strategies (such as immunoprophylaxis) that target in vivo expressed virulence-associated surface proteins. One such E. faecalis protein is Ace, which is antigenic during human IE and mediates attachment of E. faecalis cells to host extracellular matrix proteins collagen and laminin. Using a rat model, we now show that ace contributes to E. faecalis IE pathogenesis and demonstrate that Ace is expressed at high levels during IE even though produced at low levels under laboratory conditions; both active and passive immunization based on the collagen-binding domain of Ace conferred significant protection against IE. These observations, along with data that human antibodies against Ace inhibit collagen adherence of E. faecalis, indicate that Ace is an important virulence-associated factor and a promising target for prophylactic and possibly therapeutic strategies against E. faecalis IE.
Introduction
Enterococci are gram-positive cocci of intestinal origin first reported as a cause of infective endocarditis (IE) in 1899 [1]. They were recognized as the 3rd most common cause of IE as early as the 1920's, and have remained the 3rd most common cause of community onset IE since then with Enterococcus faecalis accounting for >90% of isolates from enterococcal IE when identified to the species level [1],[2],[3],[4],[5]. Over the past 20 years, enterococci have also become the 2nd–3rd most common organisms isolated from nosocomial (healthcare-associated) infections including UTIs, bacteremia, intraabdominal and wound infections, endocarditis, sepsis in neonates, among others [1],[3]. Indeed, among causes of endocarditis, enterococci (predominantly E. faecalis) have been variably reported as the #1 and #2 cause [6],[7]. Since healthcare-associated infections, particularly those caused by antibiotic resistant bacteria, result in enormous increases in hospital stays and costs, enterococci clearly represent an important drain on healthcare dollars. In one study, the attributable mortality of enterococcal bacteremia was 31% [8], emphasizing the clinical, not just the financial, seriousness of these infections.
The first step in infective endocarditis is vascular tissue colonization, which can be mediated by cell-wall anchored adhesins such as MSCRAMMs (for microbial surface components recognizing adhesive matrix molecules) [9] of gram-positive bacteria. Our previous in silico analyses of the E. faecalis genome identified a family of genes encoding MSCRAMM-like proteins containing one or more regions of ca. 150 aa segments with deviant Ig-like fold(s), characteristic of the Staphylococcus aureus MSCRAMMs [10]. One of these, called Ace (for Adhesin to collagen of E. faecalis), has been studied in detail. Genetic and biochemical analyses showed that Ace mediates adherence of E. faecalis cells to bovine and rat collagen type I (CI), human collagen type IV (CIV), and mouse laminin [11],[12],[13], as well as human dentin [14].
Crystal structure analysis of the ligand-binding segment of Ace showed that the Ace A domain is composed of two sub-domains, N1 and N2, each adopting an Ig-like fold [15]. Subsequent point and truncation mutation analyses suggested that Ace binds to collagen by a mechanism called the “Collagen Hug”[15], a variant of the “Dock, Lock and Latch” ligand-binding mechanism shown for Staphylococcus epidermidis fibrinogen (Fg) adhesin SdrG [16],[17]. The ace gene is ubiquitous [18] in E. faecalis and conserved among diverse isolates albeit with at least four variants due to variation in the number of repeats of the B domain [19]. Conditional in vitro production of Ace (i.e., markedly enhanced production after growth at 46°C, growth in brain-heart infusion plus 40% serum (BHIS) or growth in the presence of collagen versus growth in BHI broth at 37°C) by different strains correlates with conditional adherence of these E. faecalis strains to collagens and laminin [19],[20]. Most sera from patients with E. faecalis IE show reactivity with rAce, indicating that different strains express Ace during human infection and that it is antigenic in vivo [19]. Furthermore, anti-Ace antibodies (affinity purified from human serum or animals immunized with rAce) were shown to inhibit in vitro adherence of E. faecalis strains to collagen and laminin [11],[19]. In a recent study, anti-Ace40 (ligand-binding A-domain of Ace) monoclonal antibodies were shown to completely inhibit binding of Ace40 to human CI and collagen type VI and inhibited binding of Ace-coated fluorescent beads to epithelial cell lines, thus suggesting Ace as a potential therapeutic target antigen against E. faecalis infections [21].
In the present study, we have studied the role of Ace in the pathogenesis of E. faecalis endocarditis by generating an ace deletion mutant in E. faecalis strain OG1RF, by complementing this mutant (OG1RFΔace), by comparing these isogenic strains with OG1RF for adherence to various extracellular matrix (ECM) proteins and for their ability to infect aortic valves in a rat endocarditis model. Finally, we also determined the importance of Ace as a protective antigen against experimental endocarditis in a rat model by using active and passive immunization.
Results
Characterization of the Δace mutant and complementation construct
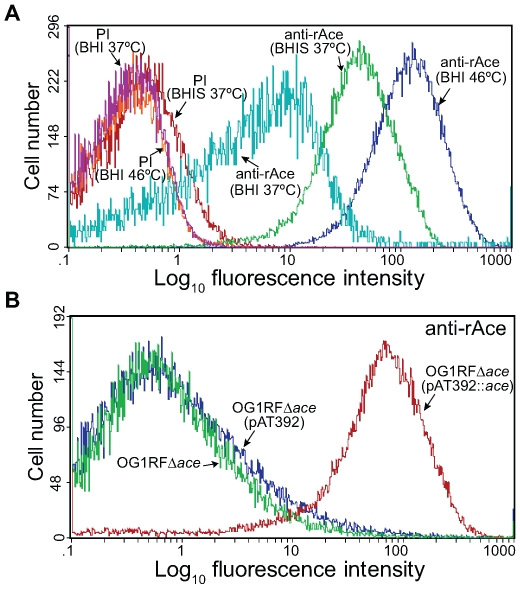
Our previous disruption mutant of ace was found to be unstable in vivo (see below). We therefore constructed an allelic replacement ace deletion mutant of OG1RF (TX5467, OG1RFΔace::cat; resistant to chloramphenicol 10 µg/ml). Deletion of ace from OG1RF was verified by sequencing confirming the correct deletion of ace from −23 to + 2200 (including the RBS, complete ace gene, and 34 bp downstream of ace), and by pulsed field gel electrophoresis (PFGE) and hybridizations (Table 1). Growth (OD600) of the Δace mutant was similar to wild-type (WT) OG1RF in BHI (data not shown). We have previously shown, using western blotting and RT-PCR, that ace is expressed at higher levels when grown in BHIS at 37°C or in BHI at 46°C [11] than in BHI at 37°C. Here, we assessed surface localization of Ace in OG1RF at 10 h using flow cytometry analyses with affinity-purified anti-rAce Igs. The mean fluorescence intensity levels for different culture conditions increased progressively with cells grown in BHI at 37°C, BHIS at 37°C and BHI at 46°C (Figure 1A), consistent with our previous western and immunofluorescence microscopy data [11],[20]. The percentages (%) of Ace-expressing cells in BHIS cultures of OG1RF, OG1RFΔace, OG1RFΔace (pAT392) (empty vector control), and OG1RFΔace (pAT392::ace) (complementation) were >70%, <5%, <5%, and >90%, respectively, demonstrating the inability of OG1RFΔace to produce Ace and the efficient complementation of OG1RFΔace by pAT392::ace. In these experiments, pAT392-containing strains were grown without added gentamicin, the same conditions that we used for preparing inocula for the rat endocarditis experiments. When BHIS was supplemented with gentamicin, expression of Ace increased to >95% of cells in OG1RFΔace (pAT392::ace) (Figure 1B), likely due to improved plasmid stability (see below).
Table 1. Bacterial strains and plasmids used in this study.
| Strains/Plasmids | Relevant characteristicsa | Reference or source |
| Strains | ||
| E. faecalis | ||
| OG1RF | Laboratory strain; Rif r, Fusr, Chls, Gens, Kans | [61] |
| TX5256 | ace insertion disruption mutant of OG1RF; Rif r, Fusr, Kanr, Chls, Gens | [11] |
| TX5467 | OG1RFΔace::cat, ace deletion mutant of OG1RF; Rif r, Fusr, Kans, Chlr, Gens | This study |
| TX5647 | TX5467 harboring pTEX5646 (for complementation with the ace gene); Rif r, Fusr, Chlr, Genr | This study |
| TX5648 | TX5467 harboring pAT392 (control for complementation); Rif r, Fusr, Chlr, Genr | This study |
| E. coli | ||
| DH5α | E. coli host strain for routine cloning | Stratagene |
| XL1-Blue | E. coli host strain for routine cloning | Stratagene |
| LMG194 | E. coli strain for expression of recombinant proteins | Invitrogen |
| TX5254 | LMG194 (pBAD::ace); 1008 bp OG1RF ace (coding for complete A domain) cloned into pBAD/HisA expression vector; Ampr | [11] |
| TX5428 | DH5α (pTEX5428); Chlr, Genr | This study |
| TX5646 | XL1-Blue (pTEX5646); Genr | This study |
| Plasmids | ||
| pAT392 | Shuttle expression vector (Genr Spcr oriR pUC oriR pAMβ1 oriT RK2 P2) | [51] |
| pTEX5500ts | Shuttle plasmid, ts in Gram+ hosts; Chlr, Genr | [50] |
| pTEX5646 | Construct for complementation; a 2,186-bp fragment containing ace cloned into pAT392 downstream of the P2 promoter | This study |
| pTEX5428 | Plasmid for ace deletion with flanking regions of the ace gene cloned on either side of the cat gene into pTEX5500ts; Chlr, Genr | This study |
aChl, chloramphenicol; Fus, fusidic acid; Gen, gentamicin; Kan, kanamycin; Rif, rifampicin; and ts, temperature-sensitive. Superscript “s” designates sensitivity and “r” designates resistance; “r” is defined for enterococci as MIC >500 for Gen and >2000 for Kan.
Figure 1. Flow cytometry analysis of cell surface expression of Ace by E. faecalis OG1RF, its isogenic ace deletion mutant and its in trans complemented ace deletion mutant.
(A) Comparison of the effect of growth for 10 h under different conditions on expression levels of Ace in OG1RF using both pre-immune Igs (PI) and anti-rAce Igs. (B) Analysis of Ace expression by the OG1RF ace deletion mutant and the effect of its in trans complementation. OG1RFΔace, ace deletion mutant; OG1RFΔace (pAT392::ace), complemented ace deletion mutant; OG1RFΔace (pAT392), ace deletion mutant with the empty vector. Reactivity to affinity-purified specific anti-rAce Igs is shown for each isogenic strain. Bacteria were analyzed using side scatter as the threshold for detection. Binding by specific anti-rAce Igs is indicated as log fluorescence intensity on the X-axis. For each histogram, 50,000 events of bacterium-sized particles were counted.
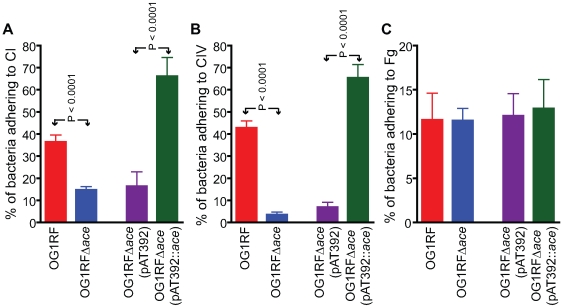
OG1RF and its isogenic Δace mutant as well as the complementation constructs were tested for their ability to adhere to immobilized ECM proteins and BSA. Consistent with our previous demonstration of adherence of OG1RF to CI, CIV and Fg after growth in BHIS at 37°C and to CI and CIV after growth in BHI at 46°C (but not in BHI [11] at 37°C), we observed here that OG1RF adhered to CI and CIV when grown in BHIS at 37°C, unlike OG1RFΔace which showed markedly reduced adherence to CI (from ∼36 to 15%) (Figure 2A), and CIV (43 to 3%) (Figure 2B), but no change in adherence to Fg ((Figure 2C). This corroborates our earlier data with a mutant with an insertional disruption of ace [11],[20]. Introduction of the ace gene in trans into OG1RFΔace resulted in even greater adherence to collagens than WT (>1.5-fold higher), whereas OG1RFΔace electroporated with pAT392 retained its reduced adherence phenotype (Figure 2); these results are consistent with Ace expression data from flow cytometry analysis.
Figure 2. Adherence of E. faecalis OG1RF and its derivatives to immobilized collagens.
(A) Adherence to collagen type I (CI). (B) Adherence to collagen type IV (CIV). (C) Adherence to fibrinogen (Fg). Mean % of cells adhering ± SD from two independent experiments representing 12 wells/sample are shown.
In vivo surface expression of Ace
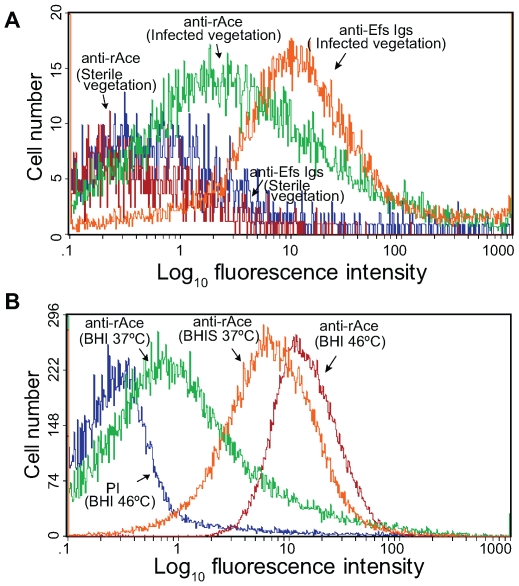
To determine if Ace is produced during infection, we performed flow cytometry analyses on extracts directly processed from IE vegetations infected with OG1RF grown in BHI at 37°C. Forward and side scatter pattern analyses of particles from processed vegetations and comparisons with those from in vitro grown OG1RF cells indicated that most of the gated particles detected by flow cytometry are likely OG1RF bacterial cells, thus confirming the removal of the majority of host tissue particles from the vegetations during the processing steps described in methods. Sterile processed vegetations from non-infected rats probed with anti-rAce-specific Igs (negative control) showed labeling of a minor fraction (<3%) of bacterium-sized particles (Figure 3A), while processed vegetations from OG1RF infected rats probed with Igs from an antiserum raised against formalin-killed E. faecalis strain HH22-whole-cells (positive control) bound 85% of bacterium-sized particles, further indicating that the majority of these particles were E. faecalis cells (Figure 3A). Affinity-purified anti-rAce-specific Igs bound to ∼40 to 45% of bacterium-sized particles from different rat endocarditis vegetations infected with OG1RF (Figure 3), demonstrating that Ace is actively expressed in host vegetations during IE.
Figure 3. Flow cytometry analysis of Ace surface expression by E. faecalis OG1RF cells derived from vegetations of rat experimental infective endocarditis.
(A) Surface expression of Ace by bacterium sized particles derived from vegetations. Vegetations were produced as described in methods, and some rats were injected (i.v.) with BHI-grown OG1RF. Processed sterile and infected vegetations were incubated with specific anti-Ace A-domain Igs or Igs purified from antiserum raised against heat-killed E. faecalis strain HH22-whole-cells (anti-Efs), followed by incubation with R-phycoerythrin-conjugated antibody. Specific binding by anti-Ace or anti-Efs antibodies is indicated as log fluorescence intensity on the X-axis. Each histogram represents 5,000 (non-infected vegetations) to 25,000 (infected vegetations) events of bacterium-sized particles. (B) For comparison, OG1RF cells grown in vitro in BHI at 37°C (inoculum used for infection) and 46°C (the growth condition that exhibited most in vitro Ace expression) as well as in BHIS at 37°C, stained with pre-immune Igs and anti-Ace and the same batch of R-phycoerythrin-conjugated secondary antibody, are shown in panel B.
In vivo testing of WT OG1RF and ace mutants in a mixed infection competition assay
Although our initial mixed-infection competition experiments showed a clear advantage for the WT over an ace disruption mutant TX5256 [11],[20] to develop IE in rat model (data not shown), subsequent experiments identified instability of this single cross-over ace disruption mutant during in vivo growth. Hence, we generated an OG1RFΔace mutant for further in vivo testing.
In an initial mono-infection experiment (n = 2) with our ace deletion mutant (OG1RFΔace), TX5467, we first determined the expression of cat (encoding chloramphenicol acetyl transferase) in OG1RFΔace, which carries this chloramphenicol resistance marker gene in place of ace, by analyzing individual colonies for chloramphenicol resistance and by high stringency hybridization using intragenic cat and ace DNA probes. We found that ∼10% of the colonies recovered from vegetations were chloramphenicol (10 µg/ml) susceptible even though they were cat probe positive and ace probe negative, indicating that, although the cat gene was present, silencing of its expression had occurred in these colonies. Previously, it has been shown that some antibiotic resistance genes of Escherichia coli are silenced in vivo; specifically, expression of an intact antibiotic resistance gene was switched off during the course of gut colonization in pigs, a phenomenon suggested to be helpful for bacterial fitness [22]. Therefore, for our mixed infection animal experiments, all the results reported here are based on high stringency hybridization with ace and cat probes using ∼200 CFUs/rat vegetation for all the rats used. Of interest, we also tested for chloramphenicol resistance but did not observe further cat silencing.
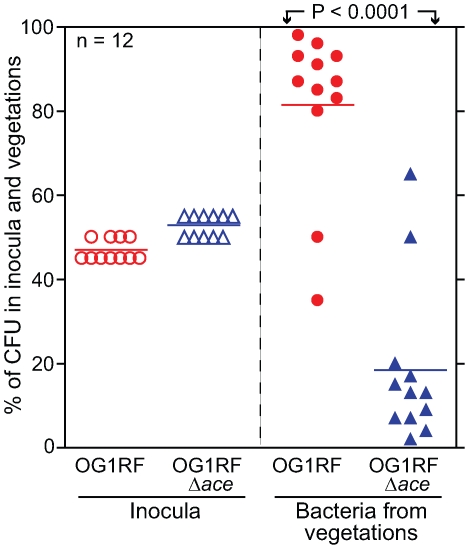
In the mixed infection competition assay, all 12 rats were infected with an approximately 1∶1 mixture (as predicted by OD600) of BHI-grown OG1RF (determined geometric mean (GM) CFU 3.8×107/rat, representing 47% of the inoculum): OG1RFΔace (GM CFU 4.4×107/rat, representing 53% of the inoculum) (Figure 4). Bacterial CFUs from vegetations on aortic valves were recovered at ∼72 h from all 12 rats and are shown in Figure 4. The mean percentage (%) of OG1RF in the total CFU of bacteria recovered was 81.5% versus 18.5% for OG1RFΔace (P<0.0001), thus demonstrating a clear advantage of OG1RF versus OG1RFΔace at 72 h for heart valve colonization in rats. The mean virulence index or competitive index [23],[24], which is a sensitive measure of the relative degree of virulence attenuation of a particular mutant in a mixed infection with the WT strain, was calculated using the equation shown in Materials and Methods. The mean virulence index of the ace mutant relative to WT in vegetations was 0.077; this indicates that ace has an important role in this endovascular infection.
Figure 4. E. faecalis OG1RF and OG1RFΔace (TX5467) in a competition (mixed infection) assay in the rat endocarditis model.
Percentages of OG1RF and OG1RFΔace present in inocula and recovered from vegetations 72 h post infection of 12 rats are shown. Horizontal bars represent the means (P<0.0001 by paired t test) for percentages of total bacteria in the vegetations of OG1RF versus OG1RFΔace. Empty circles and empty triangles represent percentages of OG1RF and OG1RFΔace in inocula, respectively, while solid circles and solid triangles represent percentages of OG1RF and OG1RFΔace in vegetations, respectively.
In vivo testing of the complemented OG1RFΔace (pAT392::ace) construct and OG1RFΔace (pAT392) in the mono-infection model
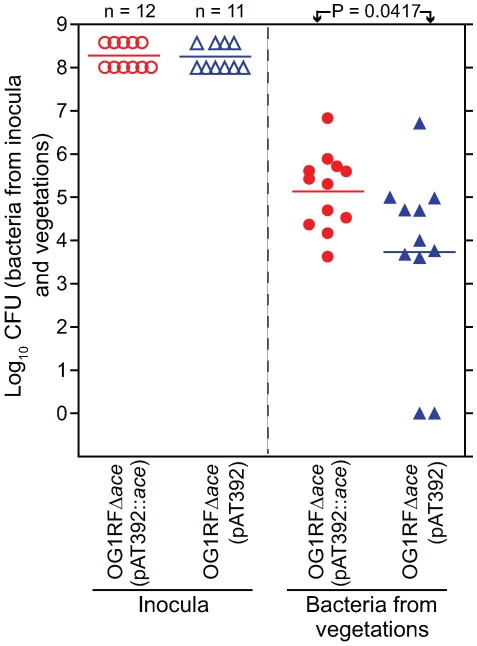
In initial mono-infection experiments with complementation constructs and testing 24 h after inoculation, we observed loss of the plasmid from cells recovered from vegetations, with 94%-98% loss from OG1RFΔace (pAT392::ace) (7 rats) and 14%–100% loss from OG1RFΔace (pAT392) (8 rats) (data not shown). We also tried growing both constructs in BHIS supplemented with gentamicin for the preparation of inocula for infection, but in vivo loss of the plasmid still occurred 24 h after inoculation. To minimize in vivo growth time and to determine the role of Ace in the early stage of valve colonization in rats, we tested both OG1RFΔace (pAT392::ace) and OG1RFΔace (pAT392) in the rat model 4 h after inoculation. Two independent mono-infection experiments were done and the combined results are shown in Figure 5. Rats inoculated with OG1RFΔace (pAT392::ace) (n = 12) showed 1.4±0.6 log10 more CFU/gm than OG1RFΔace (pAT392) (n = 11) (P = 0.0417) (Figure 5), thus demonstrating that Ace has a significant role in early colonization of heart valves in E. faecalis rat IE. Reduced time in vivo also resulted in much less loss of the plasmid from each construct. In the case of OG1RFΔace (pAT392::ace), gentamicin susceptible colonies were recovered in only 2/12 rats (2/2 colonies from one and 3/3 colonies from the other were gentamicin susceptible), while with OG1RFΔace (pAT392), 3–100% of colonies (among 8–48 tested) recovered from 5/11 rats were gentamicin susceptible. These results corroborated the above described complementation of ace surface expression (Figure 1B) and restoration of in vitro adherence of OG1RFΔace (pAT392::ace) to CI and CIV to similar levels as observed for OG1RF.
Figure 5. Complemented OG1RFΔace (pAT392::ace) [TX5647] and OG1RFΔace (pAT392) [TX5648] in early (mono-infection) colonization of aortic valves in the rat model.
In the panel on the left, empty circles and empty triangles represent OG1RFΔace (pAT392::ace) and OG1RFΔace (pAT392) inocula, respectively. In the panel on the right, solid circles and solid triangles represent OG1RFΔace (pAT392::ace) and OG1RFΔace (pAT392) in vegetations, respectively. Data are expressed as log10 CFU/gm recovered from the vegetations 4 h post infection of 12 rats (OG1RFΔace (pAT392::ace) and 11 rats (OG1RFΔace (pAT392), respectively. Horizontal bars represent the geometric mean titers. Significantly enhanced (by a mean ± SD increase of 1.4±0.6 log10 CFU/gm) vegetation titer by OG1RFΔace (pAT392::ace) versus OG1RFΔace (pAT392) (P = 0.0417) by unpaired t test for mean log10CFU/gm is shown.
Active immunization with rAce and in vivo protection against E. faecalis experimental IE in rats
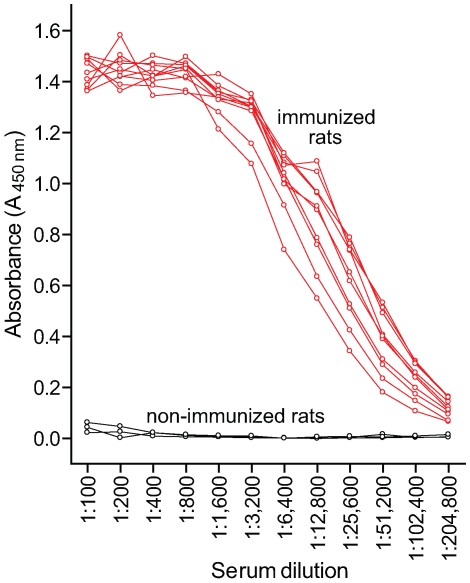
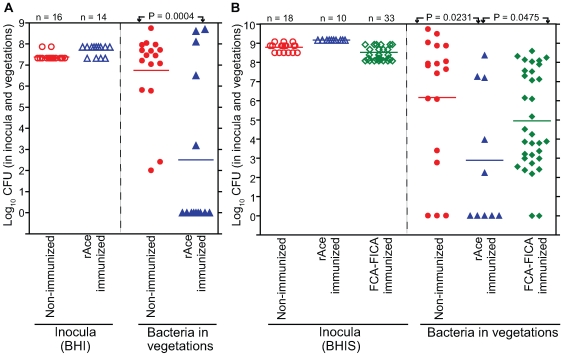
Since Ace was found to be an important virulence factor in rat experimental IE, an Ace-specific immune response might hinder the development of IE. To study this, rats were vaccinated s.c. thrice with 99% pure 100 µg rAce or PBS or Freund's complete adjuvant – Freund's incomplete adjuvant (FCA – FICA) and were challenged with 107 to 109 CFU of E. faecalis OG1RF per rat (see methods). Comparison of anti-Ace antibody levels of 10 immunized and three non-immunized animals by ELISA showed that all 10 immunized rats tested had high levels of anti-Ace titers (1: >50,000), whereas no anti-Ace antibodies were detected in any of the three control rats (Figure 6). Sixteen of 16 no-treatment control rats (100%) developed E. faecalis endocarditis after challenge with 10 times the ID50 of BHI-grown OG1RF compared with 5 of 14 rats (35%) in the rAce active-immunization group (P = 0.0001) (Figure 7A). The no-treatment control rats showed a mean of 4.2±1.0 log10 more CFU/gm than the rAce active-immunized (P = 0.0004) in vegetations recovered from heart valves. In an independent experiment, in order to mimic in vivo growth conditions more closely and because we had found that Ace is expressed at higher levels by OG1RF when grown in BHIS [11], we used BHIS-grown OG1RF for the preparation of inocula. rAce (n = 10) rats were inoculated with a higher inoculum of 1.4×109 CFU/rat (∼100 times the ID50), while non-immunized controls (n = 18) were inoculated with 3.1×108 –1.1×109 CFU/rat. Fifteen of 18 non-immunized control animal (83%) developed E. faecalis endocarditis compared with 5 of 10 rats (50%) in the rAce-immunized group with a mean ± standard deviation (SD) increase of 3.2±1.3 log10 CFU (P = 0.0231) in the non-immunized group (Figure 7B), showing significantly reduced bacterial titers in rAce-immunized group even when given a higher inoculum. Thirty-three sham (FCA - FICA)-immunized control group rats inoculated with 1.2–3.8×108 CFU/rat also showed significantly higher bacterial counts with a mean ± SD increase of 2.0±1.0 log10 CFU/gm (P = 0.0475) versus the rAce active-immunized treatment group (n = 10) (Figure 7B). These results demonstrate reproducibility of in vivo protection by active immunization with rAce against E. faecalis experimental endocarditis in rats using two different growth conditions to prepare the inocula.
Figure 6. Comparison of serum anti-Ace titers in immunized and non-immunized rats.
Rats were immunized and boosted twice with 100 μg rAce. Antibody levels were measured by ELISA. Mean serum anti-Ace titers were plotted for each antibody dilution tested.
Figure 7. rAce active-immunization in rat endocarditis model.
(A) BHI-grown OG1RF. In the panel on the left, empty circles and empty triangles represent OG1RF used for non-immunized and rAce active-immunized rats, respectively. In the panel on the right, solid circles and solid triangles represent OG1RF recovered from the vegetations, 48 h post infection. Horizontal bars represent the geometric means. Significantly fewer rats were infected by OG1RF in rAce active-immunized (5/16) versus non-immunized (16/16) (P = 0.0001 by Fisher's exact test). Vegetations showed 4.2±1.0 log10 more OG1RF CFU/gm (mean ± SD) from non-immunized versus rAce active-immunized rats (P = 0.0004, by unpaired t test). (B) BHIS-grown OG1RF. In the panel on the left, empty circles, empty triangles and empty diamonds represent OG1RF used for non-immunized, rAce active-immunized and sham-immunized rats, respectively. In the panel on the right, solid circles, solid triangles and solid diamonds represent OG1RF recovered from the vegetations, 48 h post infection. Horizontal bars represent the geometric means. Significantly reduced vegetation bacterial counts of OG1RF in rAce active-immunized (n = 18), versus non-immunized (n = 10) (P = 0.0231) and versus sham-immunized (n = 33) (P = 0.0475) by unpaired t test are shown. Rats (non-immunized) showed a mean ± SD increase of 3.2±1.3 log10 OG1RF CFU/gm from vegetations versus rAce active-immunized rats while rats (FCA-FICA immunized) showed a mean ± SD increase of 2.0±1.0 log10 OG1RF CFU/gm from vegetations versus rAce active-immunized rats.
In vivo protection against E. faecalis experimental endocarditis in rats using anti-rAce Igs for passive immunization
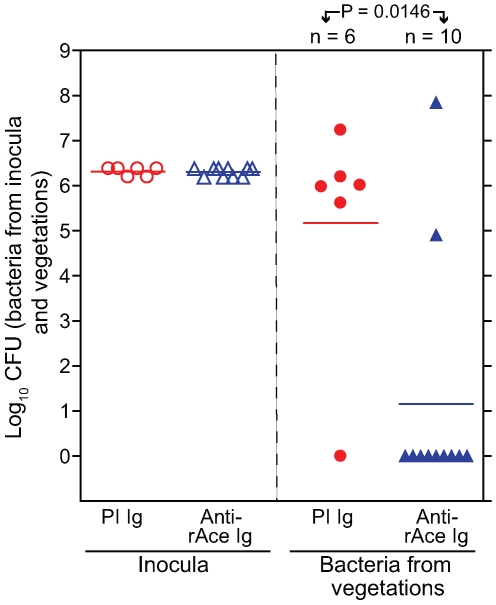
Five of 6 (83%) of rats administered purified control Igs (2 mg/kg) from pre-immune serum 1 h prior to inoculation of OG1RF developed E. faecalis IE compared with 2/10 rats (20%) of the group given anti-rAce Igs (2 mg/kg) affinity-purified from immune serum (P = 0.0001) (Figure 8). Mean bacterial titers recovered from control rat aortic vegetations showed 3.8±1.4 log10 more CFU/gm than the group given anti-rAce Igs (P = 0.0146) (Figure 8). Thus, these results corroborated the protection results seen above in active immunization using rAce against E. faecalis IE in rats.
Figure 8. Passive immunization (anti-rAce Ig versus PI Ig) in rat endocarditis model.
In the panel on the left, empty circles and empty triangles represent OG1RF inocula for pre-immune (PI) Ig treated and affinity purified specific anti-rAce Ig treated rats, respectively. In the panel on the right, solid circles and solid triangles represent OG1RF recovered from rat vegetations 24 h post infection. Horizontal bars represent the geometric means. Significantly fewer rats were infected by OG1RF in rAce Ig (2 mg/kg) (2/10) versus PI Ig (2 mg/kg) (5/6) treated rats (P = 0.0001 by Fisher's exact test). Rats (PI Ig treated) showed a mean ± SD increase of 3.8±1.4 log10 OG1RF CFU/gm from vegetations versus anti-rAce Ig treated rats (P = 0.0146 by unpaired t test).
Mouse peritonitis model
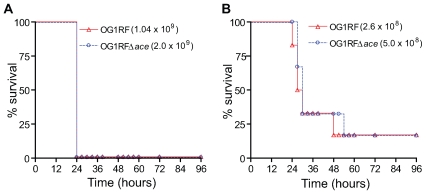
In this in vivo model, both OG1RF and OG1RFΔace caused animal mortality at similar rates with all the inocula tested (data with two inocula were shown in Figure 9) showing that OG1RFΔace was not attenuated versus OG1RF.
Figure 9. Kaplan-Meier survival plots of wild-type OG1RF and the ace mutant in the mouse peritonitis model.
(A) Survival plots of OG1RF and OG1RFΔace using 109 inocula (B) Survival plots of OG1RF and OG1RFΔace using 108 inocula. Six mice were tested with each inoculum of each of the strains shown.
Discussion
Infective endocarditis, which affects the endothelial lining of the heart, is among the most severe of the wide range of enterococcal infections encountered in humans, presenting a major therapeutic challenge and resulting in considerable mortality even when treated with antibiotics [5],[7],[8],[25]. Development of endocarditis can be initiated by injury to the valvular endothelium, which disrupts the normal valve structure and exposes underlying tissues, including ECM material. Deposition of host proteins, such as fibrin, as well as platelets at the site of injury then leads to the formation of a sterile thrombotic vegetation. This endovascular lesion may become colonized by circulating bacteria, leading to the growth of an infected vegetation. Valvular and aortic tissues are rich in collagen [26], and collagen is also found in sterile vegetations [26]. Previous studies have demonstrated that Ace plays a major role in the in vitro adherence of E. faecalis isolates to immobilized collagen [11],[12],[13]. Therefore, we reasoned that collagen could serve as a potential adhesion target for enterococci during bacteremia and that Ace could mediate bacterial attachment to these collagen-containing sites. To date, no studies have demonstrated a role for Ace in endocarditis and only very recently has a report appeared showing that Ace is important in a murine urinary tract infection model [27]. However, our previous demonstration of the role of host-derived cues (i.e., using moieties typically encountered in the host, such as serum or collagen [19],[20]), for induction of both Ace expression and adherence of E. faecalis cells to collagen, suggested that both of these phenotypes are elicited by close association of this organism with a mammalian host/tissue. Moreover, we found that 90% of patients with prior E. faecalis endocarditis have Ace-specific antibodies in their sera, implying that Ace is expressed in vivo during the infection and that it is immunogenic [19]. For these reasons, we chose an experimental IE model in this study to explore the role of Ace in E. faecalis pathogenesis.
We first looked for direct evidence showing that Ace is expressed during E. faecalis infection. E. faecalis OG1RF cells recovered directly from infected vegetations showed surface expression of Ace with a much higher (∼3×) mean fluorescence intensity compared to cells grown in vitro in BHI at 37 °C, indicating that the host environment in vegetations, similar to collagen and serum [19],[20], can induce production of Ace and its localization on the cell surface.
As anticipated, in vitro ECM protein adherence results with OG1RFΔace corroborated our previous results with an insertionally inactivated ace [11],[20]. Collagen adherence of the ace deletion mutant was restored by complementation in trans and the adherence of the complemented strain was 1.5-fold above the level of the WT parent strain, likely due to the higher number of Ace molecules displayed on the surface of the complemented strain, as determined by flow cytometry.
Deletion of ace resulted in significant attenuation in the ability of the mutated E. faecalis OG1RF strain to compete successfully with its isogenic WT parent in infection of vegetations in a mixed-inoculum rat IE model. To the best of our knowledge, this is the first demonstration that Ace contributes to E. faecalis virulence in endocarditis. When we complemented the ace mutant in trans, significantly more colonization of heart valves was observed at 4 h after infection by this strain than by an isogenic strain containing an empty vector. Thus, these results confirmed our 72 h results with the ace deletion mutant and, furthermore, provide evidence that Ace plays an important role during the initial attachment and colonization stage of IE development, possibly by mediating adherence of E. faecalis cells to exposed collagen at the site of endovascular injury. This is consistent with the high surface expression levels of Ace in the complemented strain shown by our flow cytometry analysis. While stably maintained by the majority of E. faecalis cells during early colonization (4 h), the high instability of the complementation vector after extended growth in vegetations (94–98% of cells had lost the plasmid by 24 h) reduced its effect at later stages of endocarditis. The residual ability of OG1RFΔace to cause endocarditis in some rats indicates that Ace is not absolutely required for E. faecalis to cause endocarditis; this is in agreement with published studies that showed a role for additional factors in causing E. faecalis IE [28],[29],[30]. While the precise mechanism of action of Ace for initiating, maintaining and/or propagating IE has yet to be elucidated, we infer that the difference in virulence of OG1RFΔace may be due to its reduced ability to adhere to collagen. However, we cannot exclude the possibility of another ligand or another function of this protein.
Interestingly, deletion of ace did not result in observable effects in the mouse peritonitis model in terms of either time to death or total mortality, suggesting that ace is not important for this infection or that the direct administration of a large inoculum of bacteria into the peritoneal cavity may bypass an early infection stage where Ace might be involved. These results also indicate that deletion of ace did not affect growth or survival of OG1RF in vivo in general, consistent with the similar growth rate and viability of the ace deletion mutant and WT when grown in vitro.
We have recently shown that Acm, a collagen adhesin from E. faecium, is an important factor for endocarditis caused by that species [23]; this is similar to a previous observation with Cna of S. aureus [31], which is also involved in other infections, such as septic arthritis [32]. These MSCRAMMs share a large degree of sequence conservation in their collagen-binding domains; similar proteins are present in several other species of gram-positive pathogens, such as Streptococcus equi [33], Arcanobacterium pyogenes [34], Bacillus anthracis [35] and Streptococcus gallolyticus [36], and they possibly share a similar collagen-binding mechanism, called the “Collagen Hug” that has been characterized for Cna and Ace [15],[37]. Therefore, it seems plausible that this family of proteins has been preserved or acquired across different gram-positive species/genera as a generalized mechanism to provide a binding function, although the ligand in the ecological niches where enterococci are found in nature and the purpose for these adhesins is not known. Recently, we described Ebp pili as another important factor for E. faecalis endocarditis [29], as well as urinary tract infections and biofilm formation [29],[38], a further indication of the significance of surface proteins of the MSCRAMM family for E. faecalis pathogenesis.
Our results with active immunization of rats using the collagen-binding domain of Ace showed that only 25% of immunized rats developed endocarditis, while the infection rate in the untreated group was 100%. Protection was also evident when bacterial counts were evaluated. Consistent with these results, prophylactic treatment of rats with affinity-purified anti-Ace antibodies raised against the collagen-binding domain of Ace significantly reduced bacterial numbers in vegetations, demonstrating that passive transfer of Ace-specific antibodies confers significant protection against E. faecalis IE in rat. The differences in pre-infection procedures between the active- and passive-immunized groups preclude direct comparison of results from these two methods. Based on the results presented in this study, it seems likely that these preventive strategies specifically target the initial attachment and colonization stage of endocarditis by blocking collagen adherence of E. faecalis cells. Consistent with this hypothesis, we have previously shown that Ace-specific polyclonal antibodies purified from immunized rabbits or from humans with a prior E. faecalis endocarditis infection were effective in inhibiting adherence of Ace-expressing E. faecalis isolates to collagen [11],[19]. Furthermore, a recent study that generated monoclonal antibodies against rAce showed that some of the mAbs completely inhibited binding of rAce to collagen and Ace-coated fluorescent beads to epithelial cell lines [21].The ace gene is ubiquitously present among isolates of E. faecalis and its encoded amino acid sequence, especially within the collagen-binding domain, is highly conserved [19]. Therefore, targeting ace could potentially offer protective immunization against a large spectrum of genetically diverse E. faecalis isolates, an advantage over other virulence-associated factors, such as aggregation substance, hemolysin and gelatinase, which were found to be produced by <45% of endocarditis isolates [39] and for which protective efficacy has not been shown [40]. So far, only one E. faecalis antigen, the capsular polysaccharide, has shown promise as a potential vaccine candidate, as passive and active immunization against it lowered bacterial counts in kidneys, spleens and livers in a mouse i.v. infection model [41]. To our knowledge, our study is the first report of an immunization strategy that reduces E. faecalis colonization of aortic valves and shows protection against the development of E. faecalis endocarditis, thus, suggesting Ace as a promising alternative target for prophylaxis of E. faecalis endocarditis in high risk patients. However, the ability of OG1RF to cause IE in some of the rAce-immunized rats and also in some anti-Ace antibody-treated rats indicates that targeting multiple MSCRAMMs may be necessary for a robust protection of E. faecalis IE. Consistent with this, a recent study [42] showed full vaccine protection against abscess formation or lethal challenge with S. aureus strains when a combination of four MSCRAMM antigens were used versus a moderate reduction in bacterial load when used as individual vaccine antigens.
In summary, we have demonstrated here that i) deletion of the ace gene resulted in significant attenuation of the ability of E. faecalis to colonize aortic valves and cause endocarditis in an experimental rat IE model, coinciding (ii) with reduced in vitro adherence by the ace deletion mutant to collagen types I and IV; we have also shown that (iii) Ace is actively expressed within host vegetations during endocarditis and that (iv) both active and passive immunization against the collagen-binding domain of Ace conferred significant protection against endocarditis and reduced the numbers of bacteria found in vegetations. Taken together, these results demonstrate that Ace is an important virulence-associated factor and a likely target for prophylactic and therapeutic strategies against E. faecalis endocarditis. Since Ace-like proteins are widespread among streptococci and staphylococci, future cross-protection studies may reveal novel opportunities for the development of vaccines or immunotherapeutics that may be useful for the prevention and treatment of gram-positive infective endocarditis.
Materials and Methods
Ethics statement
The rat endocarditis model and surgical procedures were performed in accordance with the institutional policies and the guidelines stipulated by the animal welfare committee, University of Texas Health Science Center at Houston (AWC, UTHSC). This study was reviewed and approved by the University Institutional Review Board (AWC approval # HSC-AWC-08-067).
Bacterial strains, plasmids, materials, standard molecular techniques, and growth conditions
E. coli and E. faecalis strains and all plasmids used in this study are listed in Table 1. All constructs were given TX numbers and plasmids from these constructs were assigned respective pTEX numbers (Table 1). E. coli strains were grown in Luria-Bertani media (Difco Laboratories, Detroit, Mich.). Enterococci were grown either in BHI, BHIS, Todd-Hewitt (TH) broth/agar (Difco Laboratories) or Enterococcosel™ Agar (EA) (Becton Dickinson) at 37°C, unless a different growth temperature is specified. The following antibiotic concentrations were used with E. faecalis: chloramphenicol 10 µg/ml, kanamycin 2000 µg/ml, rifampicin 100 µg/ml and gentamicin 125 µg/ml. With E. coli, the concentrations used were chloramphenicol 10 µg/ml, kanamycin 50 µg/ml, and gentamicin 25 µg/ml. Resistance of enterococci to gentamicin and kanamycin was defined as MICs >500 and >2000 µg/ml, respectively [18].
Materials
All antibiotics were obtained from Sigma (St. Louis, Mo.). Tran 35S-label and bovine serum albumin (BSA) were purchased from MP Biomedicals Inc. (Irvine, Calif.). C I and CIV were from Sigma and Fg was from Enzyme Research Laboratories (South Bend, Ind.). Oligonucleotide primers were purchased from Invitrogen (Carlsbad, Calif.) or IDT (Coralville, Iowa) or Sigma and their sequences are provided in Table 2. Restriction enzymes and DNA modification enzymes were mostly from Invitrogen and New England BioLabs, Inc. (Beverly, Mass.). All other chemicals used in the investigation were of molecular biology grade.
Table 2. Primers used in this study.
| Primer Name | Sequence (5′→3′)a | Function | Amplicon |
| AceDelF1 | CGCGGATCCTGCCGAGTGACAGGCATTCTGTATTGC | Deletion mutant generation | Upstream fragment of ace |
| AceDelR1 | CCCAAGCTTCTCTTATTTTTTCCACTTAGTGGTCTT | Deletion mutant generation | Upstream fragment of ace |
| AceDelF2 | AAAACTGCAGTTTCTATTATCTGGAGATAAATTGCTG | Deletion mutant generation | Downstream fragment of ace |
| AceDelR2 | CCGGAATTCTTCCAAGCGCTGATAGGCTACTTTATC | Deletion mutant generation | Downstream fragment of ace |
| AceUpF1 | CCAAACATATTGCCACTTAAATCTCTA | Mutant confirmation | |
| AceDnR1 | CACACATCTTTTAATGAAATTGTTTGA | Mutant confirmation | |
| AceComF1 | GCGGAGCTCAGAAGGGTGAATAATTTTTTATGAC | Complementation | Complete ace |
| AceComR1 | GCGGGATCCTTAATTCTTTCTGATTTGTAGATAAC | Complementation | Complete ace |
aIntroduced restriction sites are underlined.
Standard molecular techniques
Chromosomal DNA from E. faecalis isolates was prepared following the hexadecyltrimethyl ammonium bromide method described earlier [43]. Plasmid DNA was isolated from E. coli using the Wizard Plus SV minipreps DNA purification system (Promega Corporation, Madison, Wis.) and, from E. faecalis, by a previously described method [44]. General recombinant DNA techniques such as ligation and agarose gel electrophoresis were performed using standard methods [45]. When necessary, DNA fragments were purified with low melting temperature agarose gel followed by purification using QIAquick-gel extraction kit (Qiagen Inc., Valencia, Calif.). PCR reactions were performed with a Perkin-Elmer GeneAmp PCR system 9700 using the optimized buffer B (1 × buffer: 60 mM Tris-HCl [pH 8.5], 15 mM ammonium sulfate and 2 mM MgCl2) obtained from Invitrogen. PCR-generated fragments were purified using the Wizard PCR DNA Cleanup System (Promega Corporation). Recombinant plasmids were generated in E. coli DH5α or XL1-blue. Electroporation of E. coli and E. faecalis was carried out using a Gene Pulser (Bio-RAD Laboratories, Richmond, Calif.) as described previously [46],[47]. Agarose plugs containing genomic DNA were digested with SmaI and PFGE was performed using a previously described method [1]. Southern blotting was performed using Hybond-N+ nylon membrane and 0.4 N sodium hydroxide solution. Preparation of colony lysate blots was described elsewhere [48]. The RadPrime DNA Labeling System (Invitrogen) was used for labeling DNA probes with [α-32P] dCTP (GE Healthcare, Piscataway, N.J.) and hybridizations were carried out using high stringency conditions [48],[49]. DNA sequencing reactions were performed by the Taq dye-deoxy terminator method and an automated ABI Prism sequencer (Applied Biosystems, Foster city, Calif.).
Construction of an ace deletion mutant in OG1RF and its complementation
An E. faecalis ace mutant (OG1RFΔace::cat) was constructed by allelic replacement using pTEX5500ts as described earlier for E. faecium [50]. We used a replacement strategy in this study to facilitate distinguishing between WT and OG1RFΔace during in vivo animal experiments with mixed cultures; bioinformatics and mRNA analyses of the ace locus predicts the absence of a polar effect of ace deletion by the cat replacement (unpublished data). E. faecalis OG1RFΔace was constructed by allelic replacement using pTEX5500ts as described earlier for E. faecium [50]. A 1027-bp DNA fragment (AceDelUp) encompassing the region upstream of ace was amplified from OG1RF genomic DNA template using primers AceDelF1 and AceDelR1 (Table 2), digested with BamHI and HindIII, and ligated with similarly digested pTEX5500ts. Similarly, a 989-bp DNA fragment (AcedelDn) encompassing the region downstream of ace was amplified from the same genomic DNA template using primers AceDelF2 and AceDelR2 (Table 2). The AceDelDn PCR product digested with PstI and EcoRI was ligated to similarly digested pTEX5500ts::AceDelUp and was then transformed into E. coli DH5α to obtain TX5428. The plasmid from this construct, pTEX5428 (pTEX5500ts::AceDelUp+AceDelDn), was introduced into electrocompetent cells of OG1RF and cells were then plated on gentamicin plates at the permissive temperature (28°C). A single gentamicin and chloramphenicol resistant colony from these plates was grown overnight at 42°C, then plated on chloramphenicol plates and incubated at 37°C. After confirming the specific single crossover integration (OG1RFaceUp::pTEX5428) by PCR (with primer sets AceUpF1 and CmR as well as AceDnR1 and CmF), one of the integrants was picked, grown for eight overnight serial passages at 42°C, and then plated on BHI to select for plasmid excision by double crossover recombination. The colonies from these BHI plates were then replica plated to chloramphenicol plates and gentamicin plates to identify colonies that retained the cat gene but not the vector.
To complement OG1RFΔace in trans, an ∼2 kb fragment containing the ace open reading frame plus its ribosome-binding site (amplified using primers aceComF1 and aceComR1; Table 2) was cloned under the control of the P2 promoter of the shuttle vector, pAT392 [51]. This in vitro-ligated construct for complementation (designated as pTEX5646) was transformed into E. coli XL1-Blue to obtain TX5646 and was then introduced into electrocompetent cells of TX5467 to obtain TX5647 (OG1RFΔace (pAT392::ace). Surface expression of Ace in OG1RFΔace (pAT392::ace) was determined by flow cytometry (see below).
Growth curves
Overnight cultures were inoculated into BHI broth at a dilution of 1:100. The cultures were then grown at 37°C with shaking in an orbital shaker and aliquots were removed hourly from 0 to 12 h and at 24 h, for determining the absorbance at 600 nm (OD600) with a spectrophotometer.
In vitro adherence assay
Adherence of E. faecalis to CI, CIV, Fg and BSA was determined in four independent experiments using Tran 35S-labeled bacteria by a previously described assay [11].
Expression and purification of (His)6 tagged Ace A domain
Construction of the recombinant plasmid pTEX5254 (complete ace A domain of OG1RF cloned into pBAD/HisA expression vector) was described previously [11]. Expression cultures of TX5254 were induced with arabinose and the N-terminally His6 tagged proteins were purified using nickel affinity chromatography and anion exchange chromatography, as described previously [11],[52]. Protein concentrations were determined by absorption spectroscopy at 280 nm using calculated molar absorption coefficient values [53].
Ace specific polyclonal antibodies
Expression and purification of (His)6-tagged recombinant Ace A domain was done using a previously described construct and methods [11]. Goat polyclonal serum against recombinant rAce A domain (rAce) was generated by Bethyl Laboratories (Montgomery, TX). Ace A-domain specific antibodies were eluted from rAceA coupled to cyanogen bromide-activated Sepharose 4B, according to the manufacturer's protocol (Amersham Biosciences, Piscataway, N.J.). The antibodies were concentrated by ultrafiltration with a 10,000-Da molecular-weight-cutoff filter (Millipore, Bedford, Mass.), dialyzed against PBS and concentrations were determined by absorption spectroscopy.
Protein extraction and Western blotting
Surface protein extracts from E. faecalis isolates were prepared using mutanolysin (Sigma) as described earlier [11]. Protein extracts were electrophoresed in 4–12% NuPAGE Bis-Tris gels (Invitrogen) under reducing conditions in MOPS buffer, and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were then incubated with either affinity-purified anti-Ace A-domain specific immunoglobulins (Igs) [11] or pre-immune rabbit serum Igs followed by horseradish peroxidase-conjugated anti-goat antibodies. The blots were then developed with Supersignal West Pico Chemiluminescent substrate (PIERCE, Rockford, Ill.). Purified recombinant Ace A-domain was used as a positive control.
Flow cytometry
Bacteria
Surface expression of Ace on OG1RF or OG1RFΔace (pAT392::ace) cells was detected by flow cytometry using affinity purified Ace A-domain (ligand-binding domain)-specific antibodies, as described earlier [54]. Bacteria grown for 10 h in appropriate conditions were probed with pre-immune or affinity purified anti-rAceA specific-antibodies followed by donkey anti-goat IgG F(ab')2 fragment conjugated with R-phycoerythrin (Jackson Immunoresearch Laboratory, West Grove, Pa.). The cells were fixed in paraformaldehyde and analyzed with a Coulter EPICS XL AB6064 flow cytometer (Beckman Coulter, Fullerton, Calif.) and System II software.
Endocarditis vegetations
Sterile vegetations were produced in 11 rats, as described below, and nine rats were injected (i.v.) with OG1RF. Vegetations harvested after 48 h from the nine infected and two non-infected rat heart valves were processed to remove host tissue debris, using a previously described method [23]. Processed samples in groups of three infected vegetations were mixed and then divided into aliquots for labeling with a) Igs purified from antiserum raised against formalin-killed E. faecalis HH22-whole-cells (positive control) and b) affinity-purified anti-rAce-specific Igs. To assess possible cross-reactivity of anti-rAce Igs with host tissue, the non-infected vegetation sample was probed with affinity-purified anti-rAce-specific Igs. Forward scatter (for analysis of particle sizes in the sample) and side scatter (for analysis of cell granularity or internal complexity) of vegetation processed cells were analyzed and compared with the in vitro grown E. faecalis OG1RF cells.
Antibody titers
Anti-Ace antibody titers in rat sera were determined by ELISA as described by [19] with some modifications. Briefly, 96-well plates (Immulon 4HBX, Thermo Fisher Scientific, Waltham, Mass.) were coated with 1 μg of rAce in 0.05 M carbonate-bicarbonate buffer, pH 9.6. Rat sera were tested in duplicate with serial dilutions from 1∶100 to 1∶240,800, followed by detection with peroxidase-conjugated anti-rat secondary antibody (Jackson ImmunoResearch Laboratory, West Grove, Pa.) and TMB peroxidase substrate (Bethyl Laboratories, Montgomery, Tex.). The reaction was stopped with 2 M H2SO4. Antibody titers were expressed as the highest serum dilution with an A450nm ≥0.10 at 3 min after addition of the substrate [55].
Testing the effect of E. faecalis OG1RF, ace mutants, complemented mutants and immunizations in experimental endocarditis
Aortic valve endocarditis was produced in rats by following previously published methods [23],[30],[55],[56],[57],[58]. In brief, for induction of endocarditis, white Sprague-Dawley rats (∼200 gm) were used. The animals were anesthetized with isoflurane for placement of intravascular catheters. The right carotid artery was exposed and a sterile polyethylene catheter was inserted through a small incision and advanced to ∼4 cm into the left ventricle. Proper positioning was assured by sensing resistance and vigorous pulsation of the line.
Mixed infection competition assay using OG1RF and ace mutants
For testing OG1RF and OG1RFΔace in a mixed infection competition assay, bacteria (∼1∶1 by OD) were inoculated via the catheter 20 min after catheter placement; the catheter was heat-sealed and left in place during the course of the experiment and the skin was closed with sutures. Bacterial geometric mean (GM) CFUs determined for OG1RF and OG1RFΔace from the inocula were 3.8×107/rat and 4.4×107/rat, respectively. Animals were sacrificed 72 h after infection. Hearts were aseptically removed and aortic valves were examined. The platelet-fibrin vegetations formed on aortic valves were excised, weighed, homogenized in 1 ml of saline and dilutions were plated onto BHI and EA media. Ninety-six colonies/rat were picked into microtiter plate wells containing BHI, grown for 4–6 h and then replica plated onto EA and BHI supplemented with chloramphenicol 10 µg/ml, or BHI supplemented with kanamycin 2000 µg/ml in the case of the ace disruption mutant, to verify the phenotypic markers of OG1RF and OG1RFΔace. DNA lysates from colonies obtained from the OG1RFΔace plus OG1RF mixed infection were hybridized under high stringency conditions [48], using intragenic DNA probes of ace and cat in order to generate the percentage (%) of OG1RF and OG1RFΔace colonies of the recovered bacteria from vegetations. Rats with sterile cultures of undiluted vegetation homogenates (∼1 ml) were considered to have had no induction of endocarditis. Data were expressed as percentages (%) of WT and mutant per vegetation. For vegetations showing only OG1RF colonies, for example, when 96/96 colonies were cat negative and ace positive and 0/96 colonies were cat positive and ace negative, the proportion of OG1RFΔace in the vegetation was assigned the value of 1/97 (∼1%) (i.e., assuming the next colony picked would have been a mutant).
Mono-infection testing using complemented OG1RFΔace (pAT392::ace) and OG1RFΔace (pAT392)
For testing of OG1RFΔace (pAT392::ace) versus OG1RFΔace (pAT392) in the mono-infection model, rats were inoculated (i.v.) 24 h post-catheterization and were sacrificed at 4 h or 24 h post infection. Due to the instability of pAT392 seen in in vivo experiments, inocula were then grown in the presence of gentamicin (125 μg/ml) and animals were sacrificed at 24 h post infection. To minimize the loss of pAT392 over time and to determine the role of Ace in early colonization of aortic valves by OG1RFΔace (pAT392::ace) versus OG1RFΔace (pAT392), we also sacrificed a group of 11 to 12 animals administered with bacteria grown in BHIS at 4 h post inoculation. Rats with sterile cultures of undiluted vegetation homogenates were considered to have had no induction of endocarditis. In order to determine the in vivo stability of plasmid pAT392 in complemented OG1RFΔace (pAT392::ace) and OG1RFΔace (pAT392), ∼95 random colonies recovered from vegetations were picked into microtiter wells and were replica plated onto BHI supplemented with gentamicin 125 μg/ml versus BHI to differentiate between gentamicin resistant (the marker of pAT392) [51] and gentamicin susceptible colonies.
Estimation of WT OG1RF ID50 in rat IE model
ID50 of OG1RF was determined for BHI and BHIS grown cultures. Twenty one rats were inoculated (i.v.) with either BHI or BHIS grown OG1RF in a range of 1.6×104–2.6×108 and 7.2×105–1.1×109 CFU/ rat, respectively. The ID50 was determined by the method of Reed and Muench [59].
Immunizations
For active immunization, animals were divided into three groups: (a) rAce active-immunized, (b) non-immunized and (c) FCA – FICA sham-immunized. Immunizations were done following previously published methods [55],[56]. In brief, animals in group (a) received an initial dose of 100 μg of rAce in FCA (week 1) followed by a second and third dose of 100 μg of rAce in FICA at a two weeks interval. Animals in group (c) received FCA (week 1) followed by a second and third dose of FICA at a two weeks interval. Surgeries and catheter placement were done as described above for testing the ace mutant. Twenty-four hours post catheterization, animals were inoculated (i.v.) using BHI- or BHIS-grown OG1RF and were sacrificed at 48 h post infection. Three independent experiments were done and results were combined.
For the passive immunization group, animals were injected (i.v.) with 2 mg/kg of affinity purified Ace Igs purified from immunized goat serum 24 h post-catheterization and 1 h prior to bacterial inoculation; controls received 2 mg/kg of purified Igs from pre-immune goat serum. Animals were sacrificed at 24 h post infection. Two independent experiments were done and results were combined.
Post euthanasia procedures
Hearts were aseptically removed from all euthanized animals. The vegetations on aortic valves were excised, weighed, homogenized in 1 ml of saline and dilutions were plated onto BHI, EA and BHI supplemented with rifampicin 100 µg/ml for bacterial CFU recovery. Rats with sterile cultures of undiluted vegetation homogenates were considered to have had no induction of endocarditis.
PFGE
Randomly picked colonies recovered from vegetations of infected rats were also tested by PFGE to reconfirm the infecting organism [49]. In brief, agarose plugs containing genomic DNA were digested with SmaI and PFGE was performed using a previously described method with ramped pulse times of 5 s and 45 s.
Mouse peritonitis model
OG1RF and OG1RFΔace were tested following our previously published method [60]. In brief, mice were injected intraperitoneally with appropriate dilutions of bacteria (BHI or BHIS), premixed with sterile rat fecal extract (SRFE) and were observed for 5 days for survival. Two-fold inocula (range of ∼1×108–1×109 CFU/ml) of both test bacteria were used to compare animal survival/mortality. LD50 was determined using six mice per group and by the method of Reed and Muench [59].
Statistics
To compare the mean ± SD values of the adherence results, an unpaired t test was used. Percentages (%) of OG1RF versus OG1RFΔace present in mixed infection vegetations were analyzed by the paired t test. Similar to the method previously described for E. faecalis and E. faecium endocarditis using a mixed infection [23],[29], the mean virulence index of the mutant relative to WT was calculated using the following equation:
 |
Mean virulence index for the mutant should be 1.0, if the WT and the mutant have the same level of virulence, and lower values would indicate increasing levels of attenuation. Differences in bacterial log10 CFU (geometric mean) from vegetations of rAce-immunized versus non-immunized and FCA-FICA-immunized controls were analyzed by the unpaired t test. Fisher's exact test was used for comparing the total number of infected/non-infected rats in the rAce-immunized group versus control groups. Graph Pad Prism version 4.00 for Windows (GraphPad Software, San Diego, Calif.) was used for statistical analysis.
Acknowledgments
We thank Karen Jacques-Palaz and L. Charlene Thomson for their technical assistance.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by National Institutes of Health grant R37 AI47923 from the Division of Microbiology and Infectious Diseases, NIAID to BEM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowler VG, Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, et al. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA. 2005;293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 3.Murray BE. Enterococci. In: Gorbach SL, Bartlett JG, Blacklow NR, editors. Infectious Diseases. Second ed. Philadelphia, PA: W. B. Saunders Company; 1998. pp. 1723–1730. [Google Scholar]
- 4.Tleyjeh IM, Steckelberg JM, Murad HS, Anavekar NS, Ghomrawi HM, et al. Temporal trends in infective endocarditis: a population-based study in Olmsted County, Minnesota. JAMA. 2005;293:3022–3028. doi: 10.1001/jama.293.24.3022. [DOI] [PubMed] [Google Scholar]
- 5.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century–a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Guerrero ML, Verdejo C, Azofra J, de Gorgolas M. Hospital-acquired infectious endocarditis not associated with cardiac surgery: an emerging problem. Clin Infect Dis. 1995;20:16–23. doi: 10.1093/clinids/20.1.16. [DOI] [PubMed] [Google Scholar]
- 7.Giannitsioti E, Skiadas I, Antoniadou A, Tsiodras S, Kanavos K, et al. Nosocomial vs. community-acquired infective endocarditis in Greece: changing epidemiological profile and mortality risk. Clin Microbiol Infect. 2007;13:763–769. doi: 10.1111/j.1469-0691.2007.01746.x. [DOI] [PubMed] [Google Scholar]
- 8.Landry SL, Kaiser DL, Wenzel RP. Hospital stay and mortality attributed to nosocomial enterococcal bacteremia: a controlled study. Am J Infect Control. 1989;17:323–329. doi: 10.1016/0196-6553(89)90001-1. [DOI] [PubMed] [Google Scholar]
- 9.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 10.Sillanpaa J, Xu Y, Nallapareddy SR, Murray BE, Hook M. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology. 2004;150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- 11.Nallapareddy SR, Qin X, Weinstock GM, Hook M, Murray BE. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, et al. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 13.Tomita H, Ike Y. Tissue-specific adherent Enterococcus faecalis strains that show highly efficient adhesion to human bladder carcinoma T24 cells also adhere to extracellular matrix proteins. Infect Immun. 2004;72:5877–5885. doi: 10.1128/IAI.72.10.5877-5885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalski WJ, Kasper EL, Hatton JF, Murray BE, Nallapareddy SR, et al. Enterococcus faecalis adhesin, Ace, mediates attachment to particulate dentin. J Endod. 2006;32:634–637. doi: 10.1016/j.joen.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Ponnuraj K, Xu Y, Ganesh VK, Sillanpaa J, et al. The Enterococcus faecalis MSCRAMM Ace binds its ligand by the Collagen Hug model. J Biol Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- 16.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, et al. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J Biol Chem. 2008;283:638–647. doi: 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- 17.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 18.Duh RW, Singh KV, Malathum K, Murray BE. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb Drug Resist. 2001;7:39–46. doi: 10.1089/107662901750152765. [DOI] [PubMed] [Google Scholar]
- 19.Nallapareddy SR, Singh KV, Duh RW, Weinstock GM, Murray BE. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. Infect Immun. 2000;68:5210–5217. doi: 10.1128/iai.68.9.5210-5217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallapareddy SR, Murray BE. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect Immun. 2006;74:4982–4989. doi: 10.1128/IAI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall AE, Gorovits EL, Syribeys PJ, Domanski PJ, Ames BR, et al. Monoclonal antibodies recognizing the Enterococcus faecalis collagen-binding MSCRAMM Ace: conditional expression and binding analysis. Microb Pathog. 2007;43:55–66. doi: 10.1016/j.micpath.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Enne VI, Delsol AA, Roe JM, Bennett PM. Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrob Agents Chemother. 2006;50:3003–3010. doi: 10.1128/AAC.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76:4120–4128. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majcherczyk PA, Barblan JL, Moreillon P, Entenza JM. Development of glycopeptide-intermediate resistance by Staphylococcus aureus leads to attenuated infectivity in a rat model of endocarditis. Microb Pathog. 2008;45:408–414. doi: 10.1016/j.micpath.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Arias CA, Murray BE. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther. 2008;6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 26.Angrist AA, Oka M. Pathogenesis of bacterial endocarditis. JAMA. 1963;183:249–252. doi: 10.1001/jama.1963.63700040009010b. [DOI] [PubMed] [Google Scholar]
- 27.Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, et al. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immun. 2009;77:2832–2839. doi: 10.1128/IAI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nannini EC, Teng F, Singh KV, Murray BE. Decreased virulence of a gls24 mutant of Enterococcus faecalis OG1RF in an experimental endocarditis model. Infect Immun. 2005;73:7772–7774. doi: 10.1128/IAI.73.11.7772-7774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hienz SA, Schennings T, Heimdahl A, Flock JI. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis. 1996;174:83–88. doi: 10.1093/infdis/174.1.83. [DOI] [PubMed] [Google Scholar]
- 32.Patti JM, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, et al. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lannergard J, Frykberg L, Guss B. Cne, a collagen-binding protein of Streptococcus equi. FEMS Microbiol Lett. 2003;222:69–74. doi: 10.1016/S0378-1097(03)00222-2. [DOI] [PubMed] [Google Scholar]
- 34.Esmay PA, Billington SJ, Link MA, Songer JG, Jost BH. The Arcanobacterium pyogenes collagen-binding protein, CbpA, promotes adhesion to host cells. Infect Immun. 2003;71:4368–4374. doi: 10.1128/IAI.71.8.4368-4374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Liang X, Chen Y, Koehler TM, Hook M. Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J Biol Chem. 2004;279:51760–51768. doi: 10.1074/jbc.M406417200. [DOI] [PubMed] [Google Scholar]
- 36.Sillanpaa J, Nallapareddy SR, Qin X, Singh KV, Muzny DM, et al. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis Group, biotype I). J Bacteriol. 2009;191:6643–6653. doi: 10.1128/JB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong Y, Xu Y, Liang X, Keene DR, Hook A, et al. A ‘Collagen Hug’ model for Staphylococcus aureus Cna binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195:1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coque TM, Patterson JE, Steckelberg JM, Murray BE. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis. 1995;171:1223–1229. doi: 10.1093/infdis/171.5.1223. [DOI] [PubMed] [Google Scholar]
- 40.McCormick JK, Hirt H, Waters CM, Tripp TJ, Dunny GM, et al. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect Immun. 2001;69:3305–3314. doi: 10.1128/IAI.69.5.3305-3314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huebner J, Quaas A, Krueger WA, Goldmann DA, Pier GB. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect Immun. 2000;68:4631–4636. doi: 10.1128/iai.68.8.4631-4636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, David DM, Scidman JG, editors. Current Protocols in Molecular Biology. Brooklyn, NY: Green Publishing Associates; 1994. pp. 2.4.1–2.4.2. [Google Scholar]
- 44.Woodford N, Morrison D, Cookson B, George RC. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob Agents Chemother. 1993;37:681–684. doi: 10.1128/aac.37.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- 46.Li X, Weinstock GM, Murray BE. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh KV, Malathum K, Murray BE. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob Agents Chemother. 2001;45:263–266. doi: 10.1128/AAC.45.1.263-266.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh KV, Coque TM, Weinstock GM, Murray BE. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol. 1998;21:323–331. doi: 10.1111/j.1574-695X.1998.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 49.Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, et al. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72:334–345. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D,D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47:1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- 53.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun. 2007;75:5399–5404. doi: 10.1128/IAI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viscount HB, Munro CL, Burnette-Curley D, Peterson DL, Macrina FL. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect Immun. 1997;65:994–1002. doi: 10.1128/iai.65.3.994-1002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peerschke EI, Bayer AS, Ghebrehiwet B, Xiong YQ. gC1qR/p33 blockade reduces Staphylococcus aureus colonization of target tissues in an animal model of infective endocarditis. Infect Immun. 2006;74:4418–4423. doi: 10.1128/IAI.01794-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh KV, Nallapareddy SR, Nannini EC, Murray BE. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immun. 2005;73:4888–4894. doi: 10.1128/IAI.73.8.4888-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Que YA, Haefliger JA, Piroth L, Francois P, Widmer E, et al. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J Exp Med. 2005;201:1627–1635. doi: 10.1084/jem.20050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed LJ, Muench H. A simple method of estimating fifty percent end points. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- 60.Singh KV, Qin X, Weinstock GM, Murray BE. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 61.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]