Abstract
The lipooligosaccharide from Neisseria gonorrhoeae (GC), consists of lipid A, an oligosaccharide core and three branches, α, β, and γ. We report the cloning of the gene (lgtG, lipooligosaccharide glycosyl transferase G) encoding the glucosyl transferase of GC that initiates the β chain which consists of a lactosyl moiety. This gene contains a homopolymeric tract of cytidine [poly(C)] and we demonstrate that changes in the number of Cs in poly(C) account for the variation of β chain expression in different GC strains. Biochemical analyses and mass spectrometry clearly attribute the reactivity of mAb 2C7 to the presence of the lactosyl β chain. In addition, we demonstrate that in the absence of the lactosyl group, a phosphoethanolamine is added to generate a new antigenic epitope as evidenced by the gain of reactivity to mAb 2-L1–8. These results show that, like the α chain, the β chain of lipooligosaccharide is subject to antigenic variation.
About 62 million cases of gonorrhea, caused by Neisseria gonorrhoeae [gonococcus (GC)], occurred worldwide in 1996 (1) and an estimated 800,000 cases occur every year in the United States (2). In case of inadequate treatment, 10–15% of women infected with gonorrhea develop pelvic inflammatory disease (PID) and at least 15% of infertility is due to tubal damage caused by PID.
The cell wall lipooligosaccharide (LOS) from GC is much smaller than its lipopolysaccharide counterparts in enteric pathogens like Escherichia coli and Salmonella typhimurium. However, like the LOS of another mucosal pathogen, Haemophilus influenzae, GC LOS is able to undergo manifold phase variations which play a critical role in the process of infection (3–5). Several GC LOS biosynthetic genes contain poly(G) tracts of 10 or more bases. These tracts increase or decrease with a frequency of 10−2 to 10−3 and produce frameshifts to turn the genes off or on (6–11). Depending on the on/off status of different LOS glycosyl transferases (products of lgt genes), different biosynthetic pathways are used to generate different LOS structures that likely provide the pathogen with selective advantage in different niches of the human body (4).
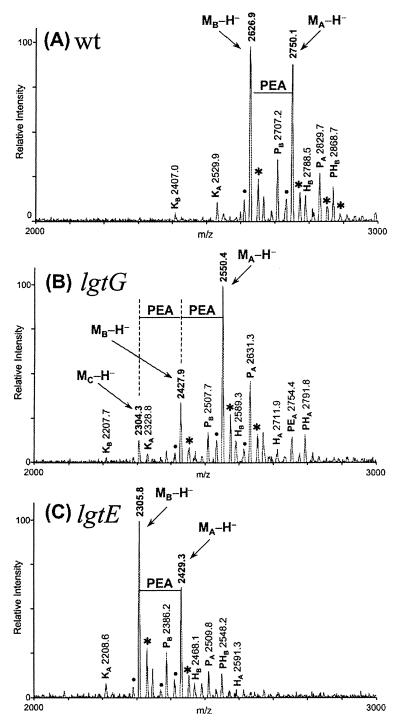
The prototype LOS of GC is depicted in Fig. 1. It consists of a core and three branches (5, 12, 13). The LOS core is relatively invariable between different strains and is synthesized following a pathway similar to that of enteric lipopolysaccharide (14–18). The longest branch, α chain, is attached to the first heptose (Hep1) of the core. β and γ chains are both attached to the second heptose of the core (Hep2). The genes encoding the glycosyl transferases of the α chain had been cloned and characterized to explain the variability of this chain (6–9, 19). The invariable γ chain consists only of a GlcNAc which is attached to C2 of Hep2.
Figure 1.
Structure of LOS from GC. Alternative LOS structures are indicated by dotted lines. The genes that show phase variation are underlined. The positions of bond formation on the sugars are given in small font. “n” denotes a yet undetermined position of bond formation. The epitopes for mAbs used in this study are also shown. The “?” next to lgtE indicates that it is not known whether it plays a direct or indirect role in that particular galactosyl bond formation.
The β chain comprises a lactosyl group with Glc being attached to C3 of Hep2. This chain is absent in some strains (12, 20) and present in others (5), but phase variation of this structure has not been reported previously. It has been suggested that the mAbs 2C7 and 3G9 react with β chain (21, 22). lgtE, the galactosyl transferase that attaches a Gal to the first Glc of the α chain, was also shown to play a role in linking the Gal with the β chain Glc (19). However, the β-glucosyl transferase adding the α1–3 bond between the Glc and Hep2 had not been identified previously.
We report cloning of the gene (lgtG) encoding the glucosyl transferase that forms the α1–3 link between the Glc and Hep2. Furthermore, by molecular genetic, biochemical, and mass spectrometric characterization, we established unequivocally that the β chain of gonococcal LOS is the 2C7 epitope and elucidated additional aspects of the chemistry of LOS synthesis, its variation, and antigenicity.
MATERIALS AND METHODS
Chemicals, Enzymes, Bacterial Strains, and Plasmids.
All chemicals were obtained from Sigma and all enzymes were purchased from New England Biolabs, if the source is not mentioned. Bacterial strains and plasmids used are listed in Table 1. In solid and liquid media, GC was grown using conditions described previously (6, 23) and 2 μg/ml erythromycin was added for selection when needed.
Table 1.
A list of strains and plasmids used
| Strains/plasmids | Relevant genotype/phenotype/description | Source |
|---|---|---|
| N. gonorrhoeae strains | ||
| F62 | Full LOS α chain but no β chain. Defective lgtG with 12 C poly(C) | 12, 36 |
| F62lgtG | F62 mutant* with ermC′(6) inserted in lgtG | This work |
| F62Δ5 | lgtA-D deleted F62. Truncated lactosyl α chain | 6 |
| F62Δ8-1 | 239-bp ApoI fragment deleted in lgtA. Truncated lactosyl α chain | This work |
| F62Δ8-1(3G9+) | Spontaneous mutant of F62Δ8-1 that reacts with mAb 3G9 | This work |
| F62Δ8-1(3G9+)lgtG | F62Δ8-1(3G9+) with lgtG deletion. Also see Fig. 3 | This work |
| FA1090 | Full LOS. Complete α and β chains. Functional lgtG with 11 C poly(C) | M. Cohen† and (36) |
| FA1090lgtG | NdeI fragment (538–632 in Fig. 2) deleted in FA1090 lgtG. No β chain | This work |
| 15253 | Full β but truncated lactosyl α chain. lgtB-D deleted and defective lgtA | 5, 19, 37 |
| 15253lgtG | 515253 without β chain due to ermC′ disruption in lgtG | This work |
| 15253lgtE | lgtE mutant of 15253. Both α and β chain contain only one Glc each | 19 |
| 1291 | No β but full α chain. Similar to F62 LOS. 12 C poly(C) in lgtG | 36 |
| MS11 | No β but full α chain like F62 and 1291. 10 C poly(C) in lgtG | J. Swanson‡ and ref. 37 |
| 4318 | Reacts with mAb 3G9. β chain present | P. F. Sparling† |
| Plasmids | ||
| pLgtG7 | 2.3-kb Tsp509I insert with 15253 lgtG in EcoRI site of pBluescriptII SK(+) | This work |
| pLgtG7-erm | Klenow enzyme-treated NotI fragment carrying ermC′ of pHSS6-erm (6) ligated with BstBI digested and T4DNA polymerase-treated pLgtG7 | This work |
| pK18 | GC uptake sequence cloned into BglII site of E. coli vector pK18 (38) | 38 |
| pK18-LgtG18 | pK18 with DNA fragment isolated from strain 4318 | This work |
| pK18-LgtG18ΔNdeI | 94-bp NdeI deletion (538–632 of Fig. 2) in lgtG of pK18-LgtG18 | This work |
All mutants are isogeneic.
University of North Carolina, Chapel Hill, NC.
Rocky Mountain Laboratory, MT.
Recombinant DNA Methods.
A library of GC 15253 was constructed by ligating ≈2- to 4-kb Tsp509I partial digestion fragments of genomic DNA into EcoRI-digested λZapII vector (Stratagene). The phage library was screened by hybridization with a random prime-labeled PCR product generated by oligonucleotide pair LgtG-1 and LgtG-2 (Fig. 2). One positive clone was converted to its corresponding pBluescript SKII(+) phagemid, pLgtG7 (Table 1), by using the manufacturer’s in vivo excision protocol.
Figure 2.
DNA sequence of lgtG locus from GC strain 15253. RBS 1 and RBS 2 indicate two possible ribosome-binding sites (marked by double underlines) corresponding to two alternative translational starts (thick underlines), START 1 and START 2, respectively. poly(C) (black box) indicates the phase variable 11 consecutive Cs. The FA1090 sequence has 11 Cs also. The number of Cs in F62 and 1291 is 12 and the ORF terminates at STOP 2 instead of STOP 1, the putative translational terminator (white box) in 15253/FA1090. In MS11, 10 Cs are present and translation terminates at STOP 3. The locations of the relevant PCR primers are underlined by arrows. The ermC′ cassette was introduced into the indicated BstBI restriction enzyme site (italic font) for insertional inactivation of lgtG ORF. The NdeI sites were used to create pK18-LgtG18ΔNdeI. −10? and −30?, possible −10 consensus and −30 consensus of σ70-binding site (gray boxes).
Another library from strain 4318 (Table 1) was made into the AccI site of pK18 (Table 1) by cloning 2- to 4-kb genomic DNA fragments isolated following partial digestion with HinPI, TaqI, and HpaII.
The lgtG knockouts were constructed either by introducing an ermC′ (erythromycin resistance marker) cassette into pLgtG7 or by deleting 94 bp (from 538 to 632 bp in Fig. 2) by digesting pK18-LgtG18 with NdeI. The resulting plasmids were designated as pLgtG7-erm and pK18-LgtG18ΔNdeI (Table 1), respectively. pLgtG7-erm DNA was spot transformed (24) into piliated bacteria of strains F62, FA1090, and 15253 and the transformants were selected on GC agar containing 2 μg/ml erythromycin. The ΔNdeI deletion was introduced nonselectively into the chromosome of F62Δ8–1(3G9+). The nonselective knockout was verified by loss of mAb 3G9 activity and Southern blot hybridization experiments that showed the incorporation of the specific deletion.
Gel Electrophoresis of LOS.
The tricine/SDS-PAGE was performed according to published protocols (6, 19). Two hundred to 400 ng LOS was loaded per lane.
Colony Immunoblots and Immunological Dot Blots.
After overnight growth, colonies were transferred to a nitrocellulose membrane (Schleicher & Schuell) and screened (25) for reactivity to mAb 3G9 (26). mAb 3G9 and mAb 2C7 (used below) have essentially the same epitope specificity.
Dot blot was performed following a published protocol (6) on LOS isolated (19) from GC by using, in addition to mAb 2C7, mAbs 2–1-L8 (27) 1B2 and TH2 (28) which were generously provided by W. Zollinger (Walter Reed Army Institute for Research, Washington, DC), ATCC (T1B-189), and S. I. Hakamori (Pacific Northwest Research Foundation, Seattle, WA), respectively.
Analysis of Sugar Composition.
Sugar analyses were performed by high pH anion exchange chromatography (HPAE) using a CarboPac PA10 column in a DX-500 (Dionex) HPAE system with pulse amperometric detection (PAD) (5, 19). Two hundred micrograms of purified LOS was hydrolyzed following the HPAE-PAD monosaccharide analysis protocol suggested by the manufacturer. The solvent system used was 18 mM NaOH for 35 min at a rate of 1 ml/min. By measuring the peak area of a standard 2.0 nmol mixture of monosaccharides (Dionex), area per nanomole values for GlcN, Gal, and Glc were determined. The value for GlcN was multiplied by 1.613 to account for loss during hydrolysis (19).
O-Deacylation of LOS.
To 0.5–2.5 mg of lyophilized LOS, 100–225 μl of anhydrous hydrazine was added. The suspension was incubated at 37°C for 25 min. Five-tenths to 1.25 ml of chilled (−20°C) acetone was added dropwise. The precipitated O-deacylated LOS was centrifuged (16,000 × g) for 25 min at 4°C, washed with chilled acetone, resuspended in 1 ml of deionized distilled water, and lyophilized overnight.
Mass Spectrometry.
O-deacylated LOS was analyzed by matrix-assisted laser desorption ionization (MALDI) according to the methods of Gibson et al. (29). For MALDI analysis, O-deacylated LOS (0.5 mg in 100 μl of water) was desalted with cation exchange resin, AG 50W-X8 (Bio-Rad), using compact reaction columns (United States Biochemical). The LOSs were mixed with 2,5-dihydroxybenzoic acid (0.1 M in acetonitrile and water, 1:1, vol/vol) and analyzed by a Voyager-DE STR Biospectrometry Workstation, MALDI time-of-flight (ToF) mass spectrometer (PerSeptive Biosystems, Framingham, MA). Negative ion mass spectra were collected and calibrated using the manufacturer’s default calibration.
RESULTS
The lgtG gene (Fig. 2) was identified independently by two of the collaborating laboratories using different strategies and when this was recognized it was decided to present the results together. One method relied on the identification of the gene responsible for the expression of the carbohydrate epitope reactive with mAb 3G9. The other strategy was to query the yet incomplete genomic sequence of GC for a homolog of the E. coli rfaG known to catalyze the addition of Glc to position 3 of Hep2.
Analyses of LOS Obtained from F62Δ8–1(3G9+), a Variant of Strain F62 That Can React with mAb 3G9, and Its Derivatives.
When colonies of N. gonorrhoeae F62 were tested for their ability to react with mAb 3G9, a background haze of mAb reactivity was always seen, suggesting that the expression of the mAb 3G9-reactive LOS might also be subject to variable expression (data not shown). To test this hypothesis, lawns of F62 and various isogeneic LOS mutants were prepared and tested for their ability to react with mAb 3G9. Individual mAb 3G9-reactive colonies were identified at a frequency of about 1/3000 colonies examined (data not shown). A derivative of F62 that was defective in lgtA expression, F62Δ8–1 (Table 1 and Fig. 3A), produced colonies with the most robust mAb 3G9 reactivity. One of these F62Δ8–1-derived 3G9 reactive colonies, F62Δ8–1(3G9+), was saved for further analysis.
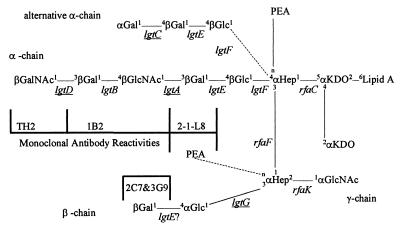
Figure 3.
Involvement of lgtG and its poly(C) tract on LOS biosynthesis of GC. (A) SDS-PAGE analysis of LOS isolated from F62Δ8–1 and its derivatives. The lanes represent LOS isolated from: 1, F62Δ8–1; 2, F62Δ8–1(3G9+); 3, F62Δ8–1(3G9+)lgtG; and 4, F62Δ8–1(3G9+)lgtG transformed with pK18-LgtG18 (Table 1). (B) DNA sequencing gel containing the C lanes of the three derivatives of pK18-LgtG18. (C) Photoreproduction of the region of a GC-agar plate containing spots of various F62Δ8–1-derived cells. The small dots inside the spots represent actual colonies after 24 hr of growth. Rows 1 through 3 represent cells that were transformed with constructs that contained 10 Cs, 12 Cs, and 11 Cs, respectively, in poly(C). Row 4 represents cells that were transformed with N. gonorrhoeae strain 4318 (Table 1) chromosomal DNA. (D) Colony immunoblot of the cells shown in C using mAb 3G9.
Furthermore, using F62Δ8–1 as the host, we screened a genomic library from mAb 3G9-reactive GC 4318 (Table 1) to obtain a plasmid clone, pK18-LgtG18, that possessed the ability to transform F62Δ8–1 to 3G9 reactivity. This plasmid contained about 2.6 kb of gonococcal DNA which included the entire lgtG sequence.
To verify that pK18-LgtG18 possessed the ability to transform F62Δ8–1 to mAb 3G9 reactivity, a site-specific deletion of pK18-LgtG18 was made by re-ligating the larger of the two DNA fragments that were produced by digesting this plasmid with NdeI. The resulting plasmid, designated as pK18-LgtG18ΔNdeI (Table 1), lacked segments 538–632 of the sequence shown in Fig. 2. This deletion was introduced nonselectively into the chromosome of F62Δ8–1(3G9+) by a spot transformation procedure (24). All transformants that had acquired the deletion, namely, F62Δ8–1(3G9+)lgtG, failed to bind mAb 3G9. To verify that the change in LOS expression was due to this deletion, several F62Δ8–1(3G9+)lgtG bacteria were retransformed with pK18-LgtG18 (Table 1). Each of these F62Δ8–1(3G9+)lgtG organisms transformed with pK18-LgtG18 regained the original mAb 3G9 reactivity along with the complete lgtG locus. Based on these observations, we concluded that lgtG was responsible for the variation seen in LOS expression. An example of the LOS profiles produced by each of these strains is shown in Fig. 3A.
Changes in the Poly(C) Tract Are Responsible for β Chain Variation.
PCR was used to isolate variants of pK18-LgtG18 that contained differing numbers of cytosines in the poly(C). Jennings et al. (10) showed that Taq-polymerase frequently makes errors when replicating polymeric strings of cytosines/guanines. Using oligonucleotide primers 196-1040R and 196-1530F (Fig. 2), we isolated variants of pK18-LgtG18 that contained differing numbers of Cs. We then tested to see whether each of these plasmids could transform F62Δ8–1 into reactivity with mAb 3G9. The data presented in Fig. 3 B–D indicate that the only construct capable of transforming this strain to mAb reactivity was the one that contained 11 Cs.
A GC ORF with Homology to E. coli rfaG.
In E. coli, addition of αGlc to position 3 of Hep2 is catalyzed by the enzyme encoded by rfaG. A BLAST (30) search of the GC strain FA1090 Data Bank at the University of Kansas (Norwalk, KS), yielded a potential homolog of E. coli rfaG with a score of ≈34% identity and ≈52.5% homology. This region was named lgtG (LOS glycosyl transferase G) following the nomenclature of other neisserial LOS glycosyl transferases (6). lgtG mapped to contig 249 (as of June 15, 1998, caution: the contig numbers keep changing because the genome sequence is not yet complete). PCR oligonucleotides LgtG-1 and LgtG-2 (Fig. 2) were designed based on the data bank sequence. Using the LgtG-1/LgtG-2 amplicon as a probe, the genomic library from GC strain 15253 (known to have β chain) was screened to obtain pLgtG7 (Table 1) that contained a ≈2.3 kb insert. The insert was sequenced and the part containing the lgtG ORF is shown in Fig. 2. The lgtG ORF in strain 15253 contains a poly(C) of 11 Cs roughly in its middle. There are two possible start sites (Fig. 2), of which, the first ATG is less likely to be used due to its larger (10 bp) distance from the corresponding ribosome-binding site (RBS) and also because the RBS overlaps with a potential −10 σ70 consensus.
We compared the lgtG sequence from strain 15253 with the lgtG sequence from the genome project of GC FA1090 (University of Kansas, Norwalk). These two sequences are almost identical. The 15253 lgtG ORF (Start 2 to Stop 1 of Fig. 2) matches bp 17194–18249 of contig 249 from strain FA1090 of University of Kansas, Norwalk. Both of the strains produce the β chain and both contain 11 Cs in poly(C). We also sequenced the lgtG region of strains F62, MS11, and 1291 which lack the β chain. F62 and 1291 contain 12 Cs and MS11 has 10 Cs. The premature terminations induced by these frameshifts are shown in Fig. 2.
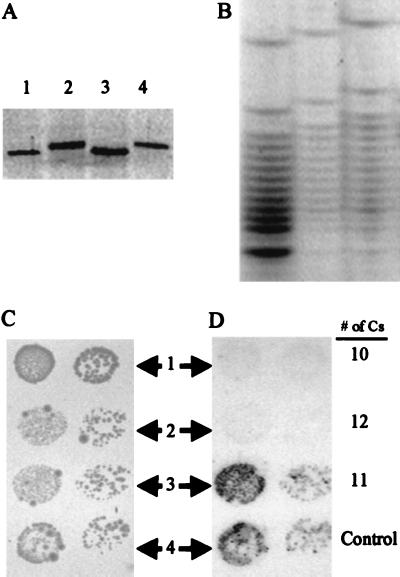
Knockout of the lgtG in GC with β Chain Results in a Smaller LOS That Fails to React with mAb 2C7 while Gaining Reactivity to mAb 2–1-L8.
pNGLgtG7-erm (Table 1) containing a lgtG knockout was transformed into GC strains F62, FA1090, and 15253 to obtain erythromycin-resistant transformants. The resulting lgtG mutants were confirmed by PCR reaction using 7F-2/7R-1 primer pair (Fig. 2). The LOS from the lgtG mutants of F62, FA1090, and 15253 were examined by SDS-PAGE (Fig. 4A). LOS from 15253lgtG and FA1090lgtG showed increased mobility compared with their wild-type counterparts. Also, the 15253lgtG LOS had a mobility similar to F62Δ5 (Table 1) which has a lactosyl α chain (6). The lgtG knockout in F62 did not produce any observable difference in its LOS.
Figure 4.
Comparison of size, immunoreactivity, and monosaccharide composition of LOSs from lgtG mutants with that from their parent strains and other mutants. (A) Silver-stained SDS-PAGE of different LOS preparations as indicated. (B) Immunoblot of LOSs from wild-type GC strains and their lgtG mutants. mAbs are in columns and GC strains are in rows. The epitopes for the mAbs are indicated in Fig. 1. (C) Monosaccharide analysis of GC LOS by HPAE-PAD. The top chromatogram is of a standard mixture of monosaccharides containing 2 nmol of each component. The other chromatograms are hydrolysates of indicated GC LOSs.
LOS from the lgtG mutants and their wild-type parents were tested with several anti-LOS antibodies (Fig. 4B). The reactions are very similar for α chain-specific antibodies TH2 and 1B2. However, there was a clear loss of 2C7 reactivity due to lgtG knockout in strains FA1090 and 15253. This loss of reactivity is marked by a simultaneous gain in 2–1-L8 reaction by FA1090lgtG and 15253lgtG. Also, a Western blot (data not shown) showed that 2–1-L8 reacted only with the smaller (the lower band in Fig. 4A) of the two LOS isoforms of the FA1090lgtG. This LOS isoform appears to be of the same molecular weight as that of F62Δ5 and 15253lgtG LOS, both of which react with 2–1-L8.
Monosaccharide Composition Analysis Indicates a Loss of a Glc and a Gal in 15253lgtG.
The chromatograms obtained from the HPAE-PAD monosaccharide analysis is given in Fig. 4C and the corrected monosaccharide molar ratios obtained from these chromatograms are shown in Table 2. For 15253lgtG, a clear decrease in Glc and Gal with respect to GlcN was evident. Based on these chromatograms, roughly, 1:2:2 and 1:0:2 Glc:Gal:GlcN ratios (Table 2) were obtained from the 15253wt and 15253lgtE LOS, respectively. These values match well with previously published results (19). However, for 15253lgtG, Glc:Gal:GlcN ratio is 1:1:1. This observation is consistent with the loss of one Glc and one Gal that would result from the loss of the lactosyl β chain in 15253lgtG.
Table 2.
Molar ratios of GlcN, Gal, and Glc in gonococcal LOS
| GlcN | Gal | Glc | |
|---|---|---|---|
| 15253wt | 1.14 | 1.79 | 2 |
| 15253lgtG | 1.06 | 0.89 | 1 |
| 15253lgtE | 1.23 | – | 2 |
Mass Spectrometry of 15253lgtG LOS Indicates Loss of Two Hexoses and Gain of a Phosphoethanolamine (PEA) as a Result of lgtG Knockout.
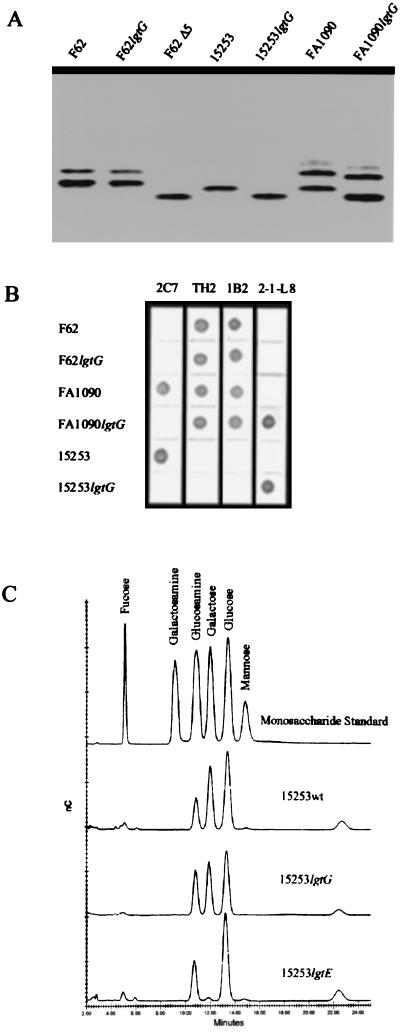
Molecular masses of O-deacy1ated LOS prepared from 15253wt and 15253lgtG were measured by mass spectrometry to analyze the LOS structure changes introduced by lgtG mutation. The 15253wt LOS reveals two major (M-H)− peaks at m/z of 2750.1 and 2626.9 (Fig. 5A). Molecular mass of 275l.1 Da, derived from the peak of 2750.1, was consistent with the calculated mass (Mr) of 2752.5 Da for the hydrazinolyzed 15253wt LOS containing Glc2Ga12G1cNAc1Hep2KDO2PEA1 oligosaccharide (5) and an O-deacylated lipid A. Another peak at 2626.9 indicated the formation of a second LOS fragment with one less PEA. This observation conforms to the characteristic easy fragmentation of PEAs during mass spectroscopy. Similar ± PEA peaks were also observed in MALDI-TOF analysis of Haemophilus ducreyi LOS by Gibson et al (29).
Figure 5.
Negative ion MALDI-TOF-MS spectra of O-deacylated LOS from wild-type (A), lgtG knockout (B), and 1gtE (C) mutants of GC 15253. Measured masses (m/z) for all of the major peaks are given in the spectra. Peaks corresponding to major intact O-deacylated LOS glycoforms are denoted as their deprotonated ions (M-H−). Several less abundant glycoforms were also observed in the spectra and they have either one less ketodeoxyoctulosonate (Kn), one more phosphate group (Pn), one more hexose (Hn), one more phospho-PEA (PEn), or addition of both phosphate and hexose moieties (PHn) to the major glycoforms. Peaks labeled with (•) and (∗) are water loss peaks and sodium-cationized peaks, respectively.
The 15253lgtG mass spectrum demonstrated a predominant (MA-H)− peak at m/z 2550.4 (Fig. 5B). The mass difference (199.7 Da) between this peak and the (MA-H)− peak of 15253wt (Fig. 5A) implied that the O-deacylated LOS of 15253lgtG mutant contains two less hexose residues and one more PEA (162.1 × 2–123.1 = 201.1). The calculated molecular mass for 15253lgtG LOS is 2551.3 Da, matching the measured molecular mass of 2551.4 Da. The peak corresponding to lipid A fragment was observed at m/z 952 in both spectra, which indicated no structural change in lipid A due to the lgtG knockout. These results suggested that the oligosaccharide present in 15253lgtG is Glc1Gal1GlcNAc1Hep2KDO2PEA2. The existence of the two PEA moieties was confirmed by the two additional peaks, (MB-H)− and (MC-H)−, at m/z of 2427.9 and 2304.3 (Fig. 5B). These two peaks corresponded to the fragments having one and two PEAs less respectively than the main glycoform peak (MA-H)−. Three PEA peak spectral patterns were also observed previously in MALDI-TOF analysis of H. ducreyi LOS that contained two PEAs (29). The mass spectrometric data, along with the monosaccharide analyses, indicated that knockout of lgtG in 15253 produced a loss of the lactosyl β chain as well as a concurrent gain of one PEA molecule.
To further confirm this finding, we analyzed the O-deacylated LOS prepared from the lgtE mutant of 15253 (Fig. 5C), which was known to lack two Gals from the 15253wt LOS (19). 15253lgtE yielded a ±PEA mass spectral pattern (Fig. 5C) similar to that of 15253wt (Fig. 5A), indicating only one PEA. The two major peaks at m/z 2429.3 (MA-H)− and 2305.8 (MB-H)− suggest that each of these fragments contains two less Gals compared with their corresponding 15253wt peaks. In l5253lgtE LOS, the truncation of β chain by one Gal did not introduce any additional PEA. In contrast, the lack of synthesis of the whole β chain in 15253lgtG LOS resulted in addition of a PEA. These data corroborate the fact that the lgtG mutation leads not only to a loss of the β chain but also to a simultaneous gain of a PEA.
DISCUSSION
In this report, we describe the cloning of a GC LOS biosynthetic gene encoding the glucosyl transferase that forms the α glycosidic linkage between C3 of Hep2 and C1 of the Glc of β chain. In E. coli and Salmonella typhimurium, this αGlc(1→3)αHep2 bond is formed by the glucosyl transferase encoded by rfaG. However, in spite of displaying sequence similarity with E. coli rfaG, the GC gene fails to complement the rfaG mutant of S. typhimurium (data not shown). Since the GC gene does not add glucose in the appropriate S. typhimurium rfaG LOS core, we designated this gene not as rfaG but as lgtG following the generic nomenclature of other GC LOS glycosyl transferases (6). The structure of the enteric LOS core is considerably different from that of GC. In enterics, Hep2 does not contain a GlcN but a αHep at position 7 and a phosphate in position 4 (31). Additionally, in GC, Hep1 bears a phosphoethanolamine, whereas the enteric Hep1 has either a phosphate or a diphosphorylethanolamine. These differences in substrate LOS structures might explain why GC lgtG does not complement the enteric rfaG mutants.
It is noteworthy that one of the approaches that allowed identification of the gene depended on the availability of raw sequence data from the GC genome project supporting the value of early release of such data. A BLAST (30) search of GenBank/European Molecular Biology Laboratory database using the GC LgtG peptide (352 aa) yields the best score (44% identity) with the yet uncharacterized H. influenzae ORF3 peptide (350 aa) (32). ORF3’s strong homology with GC lgtG suggests its involvement in αGlc(1→3)αHep2 bond formation in H. influenzae. In further support of this hypothesis, ORF3 sequence is not present in the genome sequencing project’s H. influenzae Rd strain (33), which is known to have only a core LOS and lacks the αGlc(1→3)αHep2 bond.
The analysis of the serological reactivity performed in this study as well as elaborate biochemical characterization of several defined mutants and different natural isolates indicate that GC lgtG can be turned off and on. This variation is due to the presence of a hypermutable poly(C) tract in this ORF that encodes three consecutive proline residues. This mechanism is analogous to the previously described instances of mutability of α chain glycosyl transferases which involve poly(G) tracts. However, the protein structural consequences of having three prolines in a row can be distinctly different from having glycines in a row as is the case with poly(G). Whether mutant LgtG structures could tolerate fewer or more prolines remains to be answered.
Like previous examples of LOS phase variation, this on-off switch gives rise to alternate LOS structures. However, this particular variation involves an exchange of the lactosyl moiety by a nonsugar moiety, PEA. It seems likely that the PEA molecule occupies C3 of Hep2, but our data does not exclude the possibility that the PEA may be linked to another acceptor site of the LOS core region. Nevertheless, it is clear that the PEA for β chain exchange leads to substitution of the 2C7 epitope by the 2–1-L8 epitope.
The 2C7 epitope present on the majority of GC evokes a strong immune response following vaccination and natural infection. In addition, unlike α chain epitopes, it fails to react with human glycosphingolipid antigens. By virtue of these attributes, the 2C7 epitope was proposed to be a candidate for potential vaccine (21). Future development of this vaccine approach will need to take into consideration the demonstration that the 2C7 epitope is subject to high frequency variation.
Finally, in spite of being a good target for the host antibodies, the LOS β chain is present in 90% of the strains (21). This implies an important role of this LOS moiety in the pathogenesis of GC. However, β chain is not an absolute requirement for producing the disease because both MS11 (34, 35) and 1291 (E.C.G., unpublished data) can cause infections and the re-isolates still do not express the β chain. Further work is needed to unravel the complex biology of LOS synthesis and the role of the various glycoforms in the host–parasite relationship.
Acknowledgments
We thank James Parker and Pe Jun He for excellent technical support and Dr. Vijay Pancholi for help with HPLC. This work was supported by Public Health Service Grants AI-10615 and AI-26558 (to E.C.G.), AI-32725 (to P.A.R.), and AI-24452 (to D.C.S.). We are indebted to the Gonococcal Genome Sequencing Project and B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, and D. W. Dyer.
ABBREVIATIONS
- lgtG
lipooligosaccharide glycosyl transferase G
- GC
Neisseria gonorrhoeae
- LOS
lipooligosaccharide
- PEA
phosphoethanolamine
- HPAE
high pH anion exchange chromatography
- PAD
pulse amperometric detection
- MALDI
matrix-assisted laser desorption ionization
- ToF
time-of-flight
- RBS
ribosome-binding site
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AF076919).
References
- 1.World Health Organization. World Health Report 1997. Geneva: W.H.O.; 1997. [Google Scholar]
- 2.Institute of Medicine. Committee on Prevention and Control of Sexually Transmitted Diseases. The Hidden Epidemic: Confronting Sexually Transmitted Diseases. Washington, DC: National Academy Press; 1997. [Google Scholar]
- 3.Schwan E T, Robertson B D, Brade H, van Putten J P M. Mol Microbiol. 1995;15:267–275. doi: 10.1111/j.1365-2958.1995.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 4.van Putten J P M. Mol Microbiol. 1995;16:847–853. doi: 10.1111/j.1365-2958.1995.tb02312.x. [DOI] [PubMed] [Google Scholar]
- 5.Yamasaki R, Kerwood D E, Schneider H, Quinn K P, Griffiss J M, Mandrell R E. J Biol Chem. 1994;269:30345–30351. [PubMed] [Google Scholar]
- 6.Gotschlich E C. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch C L, Danaher R J, Stein D C. J Bacteriol. 1997;179:982–986. doi: 10.1128/jb.179.3.982-986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danaher R J, Levin J C, Arking D, Burch C L, Stein D C. J Bacteriol. 1995;177:7275–7279. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q-L, Gotschlich E C. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon E R. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 11.Maskell, D. J., Szabo, M. J., Butler, P. D., Williams, A. E. & Moxon, E. R. (1992) J. Infect. Dis. 165, Suppl. 1, S90–S92. [DOI] [PubMed]
- 12.Yamasaki R, Bacon B E, Nasholds W, Schneider H, Griffiss J M. Biochemistry. 1991;30:10566–10575. doi: 10.1021/bi00107a028. [DOI] [PubMed] [Google Scholar]
- 13.Kerwood D E, Schneider H, Yamasaki R. Biochemistry. 1992;31:12760–12768. doi: 10.1021/bi00166a008. [DOI] [PubMed] [Google Scholar]
- 14.Levin J C, Stein D C. J Bacteriol. 1996;178:4571–4575. doi: 10.1128/jb.178.15.4571-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drazek E S, Stein D C, Deal C D. J Bacteriol. 1995;177:2321–2327. doi: 10.1128/jb.177.9.2321-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petricoin E F, Danaher R J, Stein D C. J Bacteriol. 1991;173:7896–7902. doi: 10.1128/jb.173.24.7896-7902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petricoin E F, Stein D C. Infect Immun. 1989;57:2847–2852. doi: 10.1128/iai.57.9.2847-2852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danaher R J, Stein D C. J Bacteriol. 1994;176:6869–6876. doi: 10.1128/jb.176.22.6869-6876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erwin A L, Haynes P A, Rice P A, Gotschlich E C. J Exp Med. 1996;184:1233–1241. doi: 10.1084/jem.184.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John C M, Griffiss J M, Apicella M A, Mandrell R E, Gibson B W. J Biol Chem. 1991;266:19303–19311. [PubMed] [Google Scholar]
- 21.Gulati S, McQuillen D P, Mandrell R E, Jani D B, Rice P A. J Infect Dis. 1996;174:1223–1237. doi: 10.1093/infdis/174.6.1223. [DOI] [PubMed] [Google Scholar]
- 22.Gulati S, McQuillen D P, Sharon J, Rice P A. J Infect Dis. 1996;174:1238–1248. doi: 10.1093/infdis/174.6.1238. [DOI] [PubMed] [Google Scholar]
- 23.Swanson J L. Infect Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn J S, Stein D C. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- 25.Sandlin R C, Apicella M A, Stein D C. Infect Immun. 1993;61:3360–3368. doi: 10.1128/iai.61.8.3360-3368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandrell R, Schneider H, Apicella M, Zollinger W, Rice P A, Griffiss J M. Infect Immun. 1986;54:63–69. doi: 10.1128/iai.54.1.63-69.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudas K C, Apicella M A. Infect Immun. 1988;56:499–504. doi: 10.1128/iai.56.2.499-504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorn J J, Levery S B, Salyan M E, Stroud M R, Cedergren B, Nilsson B, Hakomori S, Clausen H. Biochemistry. 1992;31:6509–6517. doi: 10.1021/bi00143a022. [DOI] [PubMed] [Google Scholar]
- 29.Gibson B W, Engstrom J J, John C M, Hines W, Falick A M. J Am Soc Mass Spectrom. 1997;8:645–658. [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Raetz C R H. In: Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1035–1063. [Google Scholar]
- 32.High N J, Jennings M P, Moxon E R. Mol Microbiol. 1996;20:165–174. doi: 10.1111/j.1365-2958.1996.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 33.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 34.Schneider H, Griffiss J M, Boslego J W, Hitchcock P J, Zahos K M, Apicella M A. J Exp Med. 1991;174:1601–1605. doi: 10.1084/jem.174.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson J L, Robbins K, Barrera O, Corwin D, Boslego J, Ciak J, Blake M S, Koomey J M. J Exp Med. 1987;165:1344–1357. doi: 10.1084/jem.165.5.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West S E H, Clark V L. Clin Microbiol Rev. 1989;2:S92–S103. doi: 10.1128/cmr.2.suppl.s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien J P, Goldenberg D L, Rice P A. Medicine. 1983;62:395–406. [PubMed] [Google Scholar]
- 38.Pridmore R D. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]