Abstract
Epitope mapping of an anti-polyhistidine monoclonal antibody has been performed by in vitro selection using mRNA display with a random, unconstrained 27-residue peptide library. After six rounds of selection, peptides were identified that contain 2 to 5 consecutive, internal histidines and are biased for arginine residues, without any other identifiable consensus. The epitope was further refined by constructing a high complexity, unidirectional fragment-library from the final selection pool. Selection by mRNA display minimized the dominant peptide from the original selection to a 15-residue functional sequence. Other peptides recovered from the fragment-library selection reveal a separate consensus motif (ARRXA) C-terminal to the histidine-track. Kinetics measurements made by surface plasmon resonance, using purified Fab fragments to prevent avidity effects, demonstrate that the selected peptides bind with 10- to 75-fold higher affinities than a hexahistidine peptide. The highest affinity peptides (KD = ~10 nM) encode both a short histidine-track and the ARRXA motif, suggesting that the motif and other flanking residues make important contacts adjacent to the core polyhistidine-binding site and can contribute >2.5 kcal/mol of binding free energy. Besides epitope mapping, the fragment-library construction methodology described here is applicable to the development of high complexity protein or cDNA expression libraries for the identification of protein-protein interaction domains.
Keywords: epitope mapping, in vitro selection, mRNA display, nested deletion, peptide libraries
Introduction
Epitope mapping, the identification of regions of an antigen recognized by an antibody, is an important subset of protein-protein interaction analysis that is relevant in a wide range of disciplines where antibodies are used as molecular reagents. Conventional methods for epitope mapping involve the synthesis or expression of numerous overlapping polypeptides followed by probing for antibody reactivity (Lenstra et al., 1990; Frank, 1992; Frank and Overwin, 1996; Kramer et al., 1999; Reineke et al., 1999). Although these methods can achieve very fine-mapping (single amino acid resolution) of antibodies, they involve tedious, time-consuming, and often cost-intensive steps. These techniques also require a priori knowledge of one of the interacting partners (i.e. the antigen sequence).
Display technologies such as phage (Scott and Smith, 1990) and cell surface display on E. coli or yeast (Boder and Wittrup, 1997; Georgiou et al., 1997) permit the assay of millions of polypeptides simultaneously for the identification of functional properties. In these systems, each display vehicle expresses multiple copies of a single polypeptide sequence on its surface. Active peptides are recovered by affinity selection (e.g. by biopanning or fluorescence-activated cell sorting) and identified by DNA sequencing of the library inserts. Random peptide libraries (Miceli et al., 1994; Parhami-Seren et al., 1997; Murthy et al., 1998), antigen- or gene-fragment libraries (Kuwabara et al., 1999; Christmann et al., 2001; Mullaney et al., 2001), or a combination of both (Fack et al., 1997; Coley et al., 2001) have previously been used for the epitope mapping of a wide variety of monoclonal antibodies (mAbs) (reviewed in Irving et al., 2001). Generally, these libraries suffer from low starting complexities and do not always achieve fine-mapping of antibodies unless the epitope is short (~5 residues) and well-defined. Peptide selection in combination with immunoassay of overlapping synthetic peptides has been used to fully delineate the physicochemical requirements for functional epitopes and accessory factors that influence binding affinity (du Plessis et al., 1994; Fack et al., 1997; Choulier et al., 2001).
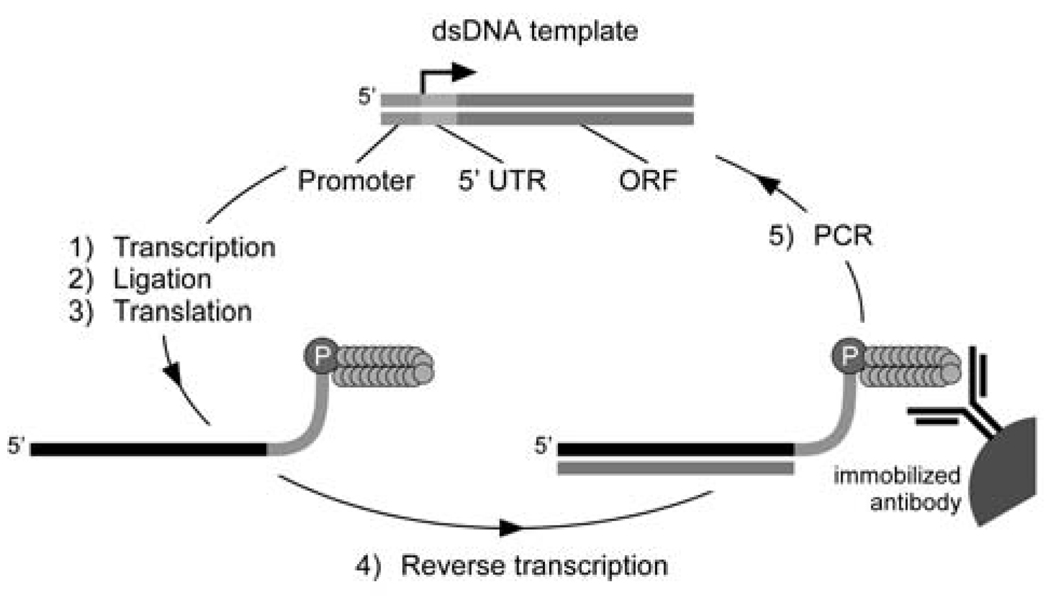
More recently, entirely in vitro techniques for protein selection such as ribosome (Mattheakis et al., 1994; Hanes and Plückthun, 1997; He and Taussig, 1997) and mRNA display (Roberts and Szostak, 1997) have emerged. In mRNA display, peptides are covalently attached to the 3’-end of their encoding mRNA via a tethered puromycin moiety. Pools of RNA-peptide fusions are selected for binding via their attached peptides and recovered fusions are RT-PCR-amplified for the next round of selection and/or cloned for DNA sequencing (Figure 1). The mRNA display system generates libraries that are robust (functional in a wide variety of conditions), encode high complexities (>1013 unique sequences, compared with ~108–109 for techniques requiring an in vivo transformation step), and lack avidity effects as only one peptide is displayed per mRNA sequence. By accessing larger libraries, extremely rare sequences (such as long, discontinuous epitopes or peptides with better functional properties) can be selected and amplified (Takahashi et al., 2003). Epitope-like consensus motifs that define the core determinants of binding for the trypsin active site and for the anti-c-Myc antibody, 9E10, have previously been identified using mRNA display with a random peptide library (Baggio et al., 2002).
Fig. 1.
In vitro selection scheme using mRNA display. The starting dsDNA pool (top, center) which encodes the peptide library is transcribed in vitro. Purified mRNA is enzymatically ligated to a puromycin-DNA oligo prior to RNA-peptide fusion formation via in vitro translation. Purified RNA-peptide fusions are reverse transcribed and affinity selected onto the immobilized antibody target. Eluted cDNA is used as the template for PCR for the next cycle of selection.
A further advancement of mRNA display technology is described here, where a unidirectional nested deletion library is constructed. A number of methods have been described for generating gene- or fragment-libraries from DNA, typically involving degenerate oligonucleotide priming (Whitcomb et al., 1993; Hampson et al., 1996; Santi et al., 2000), random fragmentation of DNA (Gupta et al., 1999), or iterative removal of bases from either end of the gene (Henikoff, 1984; Milavetz, 1992; Pues et al., 1997), followed by ligation to a vector or PCR for subsequent amplification of the library. These techniques have been employed for a variety of purposes, including epitope mapping and the determination of protein interaction domains (Fack et al., 1997; Kuwabara et al., 1999; Christmann et al., 2001; Coley et al., 2001; Mullaney et al., 2001; McPherson et al., 2002). Because of the random nature of library construction, the majority of sequences in these libraries are non-viable due to frame shifts and ligations in the anti-sense orientation. Techniques have been described to maintain gene orientation using a pair of degenerate primers with constant 5’ sequences used sequentially in the amplification of cDNA (Hampson et al., 1996) or mRNA (Hammond et al., 2001; McPherson et al., 2002). However, these methods are technically challenging and may be prone to poor library coverage due to biased hybridization to target sequences (Telenius et al., 1992; Zhang and Byrne, 1999).
As mRNA display facilitates selection from peptide libraries larger than previously possible, improvements are needed for generating libraries with broad coverage while maintaining high sequence complexity. The method described here uses a partial DNase I digestion to fragment the DNA pool randomly. These fragments are then directionally amplified, maintaining the sense orientation, and used to generate an mRNA display library. We first developed a pool of active members by performing in vitro selection with a random peptide library against a His6-tagged protein immobilized by an anti-polyhistidine mAb. Due to the weak affinity of the mAb for the cited His6 epitope, we inadvertently selected for peptide sequences with high affinity for the antigen-binding region of the mAb. This pool of mAb-binding peptides was subsequently used as the template for a nested deletion library. A 35-residue “winning” peptide was minimized to a 15-mer sequence using the mRNA display fragment-library. Selected peptides were analyzed by surface plasmon resonance (SPR) and demonstrated 10- to 75-fold higher affinities than the cited epitope. The fragment-library selection also revealed a new motif important for high affinity binding, demonstrating how sequence length may be an important factor in delineating an epitope. The nested deletion construction methods should be highly applicable toward the isolation of minimal protein interaction domains from cDNA or protein expression libraries using mRNA display.
Materials and methods
General
Enzymes were purchased from New England Biolabs unless otherwise noted. Other reagents and solvents were obtained from Sigma-Aldrich or VWR International. All buffer components for RNA and RNA-peptide fusions were made with diethyl pyrocarbonate-treated ddH2O. DNA oligos were synthesized at the Caltech Biopolymer Synthesis and Analysis Facility and were desalted by OPC purification with the exception of DNA template 130.2 which was synthesized at the W. M. Keck Foundation Biotechnology Resource Laboratory (http://keck.med.yale.edu) and purified by urea-PAGE. Oligo and peptide concentrations were determined by UV spectrophotometry using a calculated extinction coefficient (http://paris.chem.yale.edu/extinct.html). Protein concentrations were determined by UV absorbance at 205 nm (Scopes, 1974). The values obtained with this method were within 5% of those obtained using a calculated extinction coefficient at 280 nm.
mRNA display library construction
The anti-sense DNA template 130.2 (5’-AGC GCA AGA GTT ACG CAG CTG (SNN)27 CAT TGT AAT TGT AAA TAG TAA TTG TCC C, S = C or G, N = A, C, G, or T) was PCR-amplified with primers 47T7FP (5’-GGA TTC TAA TAC GAC TCA CTA TAG GGA CAA TTA CTA TTT ACA ATT AC) and mycRP (5’-AGC GCA AGA GTT ACG CAG CTG) to produce the initial template containing a T7 promoter, a 5’-untranslated region (UTR), an ATG methionine start codon, 27 random amino acids each encoded by NNS, and a constant 3’-end that encoded the peptide, QLRNSCA. In vitro transcription, purification of the mRNA templates, and ligation of the puromycin linker oligo were performed essentially as described (Liu et al., 2000). Transcription reactions were pretreated with RNAsecure (Ambion) to inhibit RNase activity and library DNA was removed by DNase I (Epicentre) digestion prior to purification of the mRNA pool. The ligation was performed with the puromycin-DNA linker, pF30P (5’-A21[S9]3ACC-P, S9 = spacer phosphoramidite 9, P = puromycin, 5’-phosphorylated with phosphorylation reagent II, Glen Research) and a splint oligo (5’-TTT TTT TTT TTN AGC GCA AGA GT ) (Liu et al., 2000). Puromycin-conjugated templates (mRNA-F30P) were purified by urea-PAGE.
RNA-peptide fusion preparation and selection
Purified mRNA-F30P templates were translated in rabbit reticulocyte lysate (Red Nova lysate, Novagen) as per the manufacturer’s instructions with optimized conditions (100 mM KOAc, 0.5 mM MgOAc, and 0.5 µM mRNA-F30P) and additional L-Met (0.5 mM final, 1 mL total reaction volume) or 35S-Met labeling (150 µL reaction, New England Nuclear, now PerkinElmer Life Sciences). Following the incubation step at 30 °C, KOAc and MgCl2 were added to 585 mM and 50 mM (final), respectively, and the reactions were incubated on ice for 15 min to facilitate RNA-peptide fusion formation. Radioactively labeled and non-labeled RNA-peptide fusions were pooled and subsequently purified with oligo dT-cellulose (New England Biolabs) as described (Liu et al., 2000). Purified fusions were concentrated (Microcon YM-30, Millipore) and reverse transcribed as per the manufacturer’s instructions (Superscript II, Invitrogen) with the mycRP primer.
The matrix preparation and all selection steps were performed at 4 °C. The reverse-transcribed fusions, in 1 mL of selection buffer (50 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 10 mM NaF, 30 µM AlCl3, 0.05% Tween 20, 1 mM β-mercaptoethanol (β-ME), and 5 µM GDP), were precleared by rotating with 20 µL of protein G-sepharose (4B Fast Flow, Sigma) for >1 h. The supernatant was transferred to the target matrix (80 µg of His6-Giα1 (Lee et al., 1994) immobilized by 40 µg of anti-polyhistidine mAb (H1029, Sigma) on 20 µL of protein G-sepharose) and rotated for 1 h. The matrix was washed with 3 × 1 mL selection buffer and the bound RNA-peptide fusions were eluted with 2 × 200 µL 4% acetic acid through a 0.45 µm spin filter (SpinX, Costar). Washes and an aliquot of the elution were scintillation counted (LS 6500, Beckman Coulter) to determine the amount of bound fusions.
The eluted fusions were either desalted by ultrafiltration (Microcon YM-30, Millipore) or frozen and dried by vacuum centrifugation. After resuspension in ddH2O or 10 mM Tris-HCl, pH 8, samples were PCR-amplified for the next cycle of selection and/or for DNA sequencing (TOPO TA cloning, Invitrogen). Subsequent selection rounds were performed similarly, except that smaller translation reactions were used (300 µL non-labeled, 100 µL 35S-Met labeled). Unblocked mAb (without the His6-tagged protein) was used as the target in the 6th round of selection, when it was realized that the peptides were specific for the mAb.
RNA-peptide fusion binding assay
Aliquots of purified 35S-Met labeled RNA-peptide fusions were treated with RNase (DNase-free, Roche) and added to ~15 µL of protein G-sepharose matrix (with or without ~10 µg of anti-polyhistidine mAb) in 1 mL of selection buffer. Mixtures were rotated at 4 °C for 1 h and washed with 3 × 1 mL selection buffer. The percent binding was determined by scintillation counting of the washes and the matrix.
Fragment-library preparation
To generate the fragment-library, first-strand cDNA from a selected library was synthesized with dUTP instead of dTTP nucleotides (Superscript II). After RNase H treatment (Roche) to remove mRNA, the cDNA was purified by spin-column (QIAquick, Qiagen) and randomly digested with DNase I (0.25 U DNase I (Invitrogen) added to 30 pmol cDNA (~1.2 µM final) in ice-cold 1 × DNase I buffer (10 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, and 0.1 mM CaCl2)) at 15 °C for 10 min. DNase I was removed using DNase Removal Reagent (Ambion). A fill-in reaction (Sequenase v2.0, Amersham Biosciences) was performed according to the manufacturer’s instructions with 125 pmol of myc6-N6-FP (5’-ATC TCT GAA GAG GAC CTG NNN NNN) and 200 µM of each dNTP (~0.6 µM cDNA final). First-strand cDNA was digested with uracil-DNA glycosylase (UDG) and ssDNA >50 bases was extracted with QiaEX II (Qiagen) from a 4% agarose gel (Frohlich and Parker, 2001). A second fill-in reaction was performed with 3myc-N6-RP (5’-AAA TGC ACA AGA GTT GCC CTC GNN NNN N) as before. The dsDNA was subsequently agarose gel-purified by spin-column (QIAquick).
PCR using primers T7mycFP (5’-GGA TTC TAA TAC GAC TCA CTA TAG GGA CAA TTA CTA TTT ACA ATT ACA ATG GAA CAG AAA CTG ATC TCT GAA GAG GAC CTG) and psn3mycRP (5’-AAA TGC ACA AGA GTT GCC CTC G) resulted in a smear of products ranging from 100 to 200 bp on an agarose gel. DNA corresponding to 150 to 200 bp was extracted by spin column (QIAquick). Amplification of the dsDNA by PCR using the primers 47T7FP and psn3mycRP produced the initial library for selection. The selection was performed against the anti-polyhistidine mAb as before, except that the puromycin moiety was coupled to the mRNA by UV photo-crosslinking with oligo psn-mycF15P (5’-[Ps]-TGC ACA AGA GTT GA15-[S9]2-CC-P, Ps = Psoralen C6, Glen Research) as described previously (Kurz et al., 2000). The selection buffer used for the fragment selection was 1× PBS, 1 mM β-ME, 1 mM EDTA, 0.05% Tween 20, 0.2% (w/v) BSA, and 1 µg/mL yeast tRNA (Roche). In rounds 2 and 3 of the selection, the matrix was more stringently washed by incubation in buffer containing poly-L-His (0.15 mg/mL) and His6 peptide (60 µM, Covance Research Products) for ~40 min at 4 °C (Boder and Wittrup, 1998).
Direct binding assay of in vitro translated peptides in lysate
Individual clones (in pCR4-TOPO vector, Invitrogen) were PCR amplified with primers 47T7FP and mycRP, in vitro transcribed, urea-PAGE-purified, and in vitro translated (Red Nova Lysate) with 35S-Met labeling as per the manufacturer’s instructions. 4 µL of the translation reaction was added directly into an assay tube (600 µL fragment selection buffer, 10 µL protein G-sepharose, 5 µg anti-polyhistidine mAb). After rotating at 4 °C for 1 h, the sepharose was washed with 6 × 600 µL fragment selection buffer in a 0.45 µm spin filter (SpinX) and bound peptides were eluted with 2 × 20 µL 0.05% SDS. Half of the sample was analyzed via tricine SDS-PAGE along with 2 µL of the original translation reaction for comparison. After electrophoresis, gels were destained (40% methanol and 10% acetic acid) for 20 min, dried under vacuum, and imaged via autoradiography (Storm Phosphorimager, Amersham Biosciences). Peptide band intensities were analyzed with ImageQuant software (Amersham Biosciences).
Peptide synthesis/protein purification
Peptides were synthesized on an ABI 432A Synergy peptide synthesizer (Applied Biosystems) using Fmoc chemistry. Peptides included the sequence GGYK-NH2 at their C-terminus, where K is biotinyl-lysine (biocytin, BAchem) and -NH2 represents C-terminal amidation. The tyrosine residue, used for quantitation by UV absorbance, was omitted from the synthesis for peptides that already contained a tryptophan and/or tyrosine. Crude peptides were deprotected in TFA:thioanisole:1,2-ethanediol (450:25:25 µL, 2 h at room temperature), precipitated with methyl tert-butyl ether, purified to >95% purity by reverse-phase HPLC on a semi-preparative C18 column (250 × 10mm, Vydac), and confirmed by MALDI-TOF mass spectroscopy.
Several peptide sequences were expressed in E. coli as in vivo biotinylated maltose-binding protein (MBP) fusions using a vector derived from pDW363 (Tsao et al., 1996). The MBP gene from pDW363 was amplified by successive PCR (primers 35.3 5’-GGA CTA GTA AAA TCG AAG AAG GTA AAC TGG TAA TC and 35.4 5’-CCA TTG GAT CCT TAA TTA GTC TGC GCG TCT TTC AG, then primers 84.1 5’-GAG CAC TCG AGC GGT GCG AAT TCA AAC AAC ATC GAG GGG CGC GCC GGT GGC ACT AGT AAA ATC GAA GAA GGT AAA CTG GTA ATC and 29.3 5’-CCA TTG GAT CCT TAA TTA GTC TGC GCG TC). The PCR-amplified fragment and pDW363 were digested with XhoI/BamHI, purified, and ligated to produce the pDW363B vector.
DNA templates encoding peptides B and C were amplified by PCR using the universal forward primer 29.4 (5’-TGA AGT CTG GAG TAT TTA CAA TTA CAA TG) and a template-specific reverse primer that added a SpeI site. BpmI/SpeI digested dsDNA was co-ligated into XhoI/SpeI digested pDW363B with DNA linkers (XhoI linker 5’-TCG AGC TCT GGA GGC ATC GAG GGT CGC AT and BpmI linker 5’-GCG ACC CTC GAT GCC TCC AGA GC) to produce the expression vector. Inserts contained an N-terminal bio-tag, peptide B or C, and a C-terminal MBP fusion. The vectors produce a dicistronic mRNA which encode the bio-tag-peptide-MBP fusion and biotin holoenzyme synthetase (birA), an enzyme that attaches biotin to the bio-tag in vivo.
Protein expression with 30 mL cultures of E. coli BL21 cells was performed as described (Tsao et al., 1996). Cells were lysed with B-PER (Pierce) and MBP fusions were purified on monomeric avidin-agarose (Pierce) as per the manufacturer’s instructions. Purified proteins were concentrated and desalted into 1× PBS by ultrafiltration (Centriprep YM-10, Millipore).
Anti-polyhistidine mAb in ascites fluid was affinity purified on protein G-sepharose in 1× PBS/0.1% triton X-100, eluted with 0.1 M citric acid buffer, pH 3, and immediately neutralized with buffer. After concentration and buffer exchange (Centriprep YM-50) into papain buffer (20 mM phosphate, pH 7, 10 mM EDTA), antigen-binding fragments (Fab) were generated and purified using the ImmunoPure Fab Preparation Kit (Pierce) as per the manufacturer’s instructions.
Surface plasmon resonance
SPR measurements were made at 25 °C on a Biacore 2000 (Biacore) equipped with either SA (streptavidin) sensor chips or research-grade CM5 sensor chips (Biacore) with amine-coupled streptavidin (ImmunoPure, Pierce). The CM5-streptavidin chips were prepared in-house by standard NHS/EDC amine coupling (Biacore) and achieved >1100 RU of immobilized streptavidin per flow cell. HBS-EP (20 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P20 (Tween 20)) was used as the running buffer for all experiments. Biotinylated ligands were diluted in HBS-EP to 1 nM and immobilized to individual flow cells (~10 RU for peptides and ~100 RU for proteins). Flow cell 1 was left as a streptavidin negative control in all sensor chips. To collect kinetics data, a concentration series of Fab in HBS-EP was injected for 2 min at 35 µL/min over all flow cells and dissociation was observed for 3 min. The Fab samples were injected in random order, interspersed with a number of buffer blank injections for double referencing (Myszka, 2000). Flow cells were regenerated between Fab injections with a 0.5 min wash of 2.5 M NaCl at 100 µL/min. Raw data was processed with Scrubber and analyzed with CLAMP using a 1:1 bimolecular interaction model (Myszka and Morton, 1998). KD values were calculated (kd/ka) from the on and off rates determined by CLAMP. Standard free energies of binding were calculated from the KD values (ΔG° = −RT ln(C/KD), R = 1.987 × 10−3 kcal mol−1 K−1, T = 298.15 K, and C = 1 mol L−1).
Results
Selection of a random peptide library against an anti-polyhistidine mAb
The peptide selection experiment, originally designed to target a His6-tagged protein immobilized by an anti-polyhistidine mAb, utilized a random, unconstrained 27-mer peptide library. During PCR and transcription the complexity of the library was maintained by having at least 7 × 1013 sequences at the start of each reaction. The initial mRNA display pool contained at least 1012 unique peptide sequences, estimated from the initial mRNA and methionine concentrations in the translation reaction, out of a maximum complexity of 2027 peptides (~1.3 × 1035).
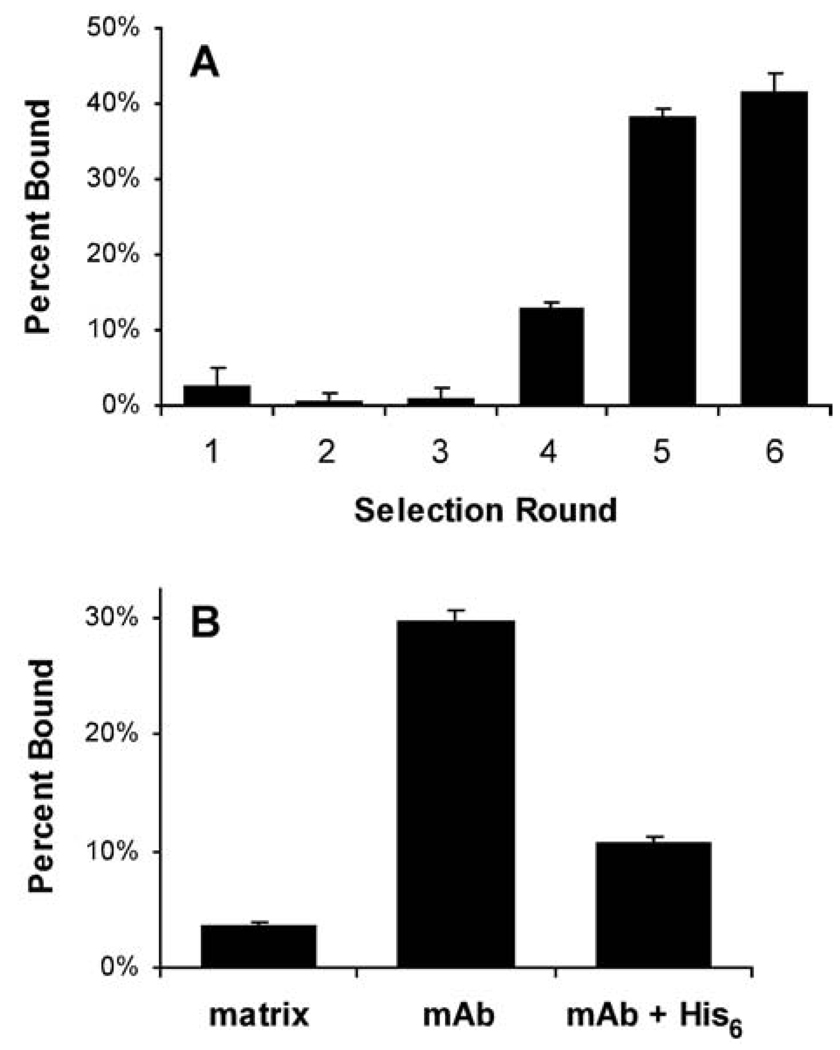
Five rounds of selection were performed on the immobilized anti-polyhistidine mAb, pre-saturated with an N-terminal His6-tagged protein (Figure 2A). Bound RNA-peptide fusions were eluted with acetic acid, which generally recovered >80% of the remaining 35S counts. To determine the progress of the selection, a separate 35S-Met labeled RNA-peptide fusion pool from the 5th round was purified, RNase-treated, and assayed for binding (Figure 2B). This assay revealed specific binding of the peptide pool (now modified only at the C-terminus with puromycin and a short DNA linker) to the antibody rather than to the immobilization matrix (protein G-sepharose) or to the His6-tagged protein. The reduced binding observed when a His6 peptide was added as a competitor further evinced that the selected peptide sequences specifically targeted the antigen-binding region of the mAb. A 6th round of selection, performed with unblocked mAb as the target, demonstrated that the enrichment for active peptides against the mAb was essentially complete (Figure 2A).
Fig. 2.
Selection of peptides against the anti-polyhistidine mAb. (A) the percent binding from each round of selection was determined by scintillation counting of an aliquot of the 35S-Met labeled RNA-peptide fusions before and after affinity selection on the immobilized antibody. (B) binding assay of 5th round mRNA display library. Purified, RNase-treated 35S-labeled fusions from the 5th round pool were assayed on protein G-sepharose matrix with and without immobilized anti-polyhistidine mAb. The addition of 10 mM His6 peptide competitor resulted in reduced binding to the mAb, suggesting that the selected peptides interact specifically with the antigen-binding site.
DNA sequencing of the final 6th round pool revealed a variety of peptides containing 2 to 5 consecutive His residues with no other apparent consensus except a bias for Arg. The His-track was seen in various positions in the random region of the library suggesting that the mAb had little preference for the epitope at either terminus or as an internal binding site. One sequence, peptide C, emerged as the dominant member of the selection (Table I). Further rounds of selection using His6 peptide and/or poly-L-His as competitors in the selection buffer generally resulted in changes in the percentage of peptide C in the pool rather than the emergence of new, beneficial mutations or peptides defining a single consensus (data not shown). Peptide C remained the most prevalent sequence in all subsequent selection rounds, with a collective frequency of 20 out of 53 sequences (Table I).
Table I.
Peptide sequences from anti-polyhistidine mAb selection using a random 27-mer library
| YRTNHHYDVGRFAARGRRD | |
| NGRSSMNWRSQEITRYTSEHHYRMAFL | |
| PEQYDHHHLEARRRASSTRQVRARARR | |
| RAYTPHHHAEGRLVRLEPHPAPYKNRT | |
| YYVKNRLHHHRLARLVAAEHAHRLRVQ | |
| NKRNLSYPWSHHHQVARRTHMRAQHTM | |
| RPTKNFEAEVVRSTGPMHHHDTAKQRY | |
| DFLTYNKSMGGRPTNFRHHHSSVVQSQ | |
| DEPEVVGRVLGERPAGALADHHHMMKW | |
| EVLHGHHHVVARVRASCTGPTRRASCA | (6/53) |
| HVYEKANNRLGHKHHHLAARRRSKSWN | |
| SNKGFSWRKKGMAVTPNRHLHHHMVAH | |
| TNHRHHHGVLERRQDILTGSLIEHKH | |
| ILKRLREQHRHHHAAAHHVRVRRRGRH | |
| NYTTRRAEWNRQDAHRHHHQEARRGAL | A (3/53) |
| SKKDNAVGLQELRLREGHRHHHDVMLT | |
| KKVRGHHRHHHQVALLDAAERGPGRMS | |
| GIHHHHAMAVLAELGMNPMGFALPDMW | |
| AGVHHHHDAARGGTRSRRSTPRSATRR | * |
| TMNWHHHHENGLRARMYDAGRR | |
| KVRRDVMRWHHHHRMARRKANR | B (4/53) |
| RVQDRLGHRAVQPVLHHHHQAARRRVR | |
| AALHHHHHDAGRASAMRRPGTPATSWR | |
| DGHPERHDAGDHHHHHGVRQWRLISTG | C (20/53) |
Only the random domain is shown. Sequences contained between 2 and 5 consecutive histidines and were aligned at the C-terminal end of the His-track. A consensus was not observed except for a strong bias for Arg several residues C-terminal to the His-track. His and Arg residues are shown in bold. The frequency (out of 53) is shown for peptides that appeared more than once from DNA sequencing of individual clones. For these sequences, amino acids that differed between clones are in italics, with the most common residue at that position shown. Several sequences contained multiple deletions that shortened the random domain but left the C-terminal constant region intact and in-frame. The sequence marked with an asterisk contained a 2 bp insertion which resulted in a frame-shift of the C-terminal constant region (not shown). Peptides A, B, and C are named.
Selection for a minimal binding epitope
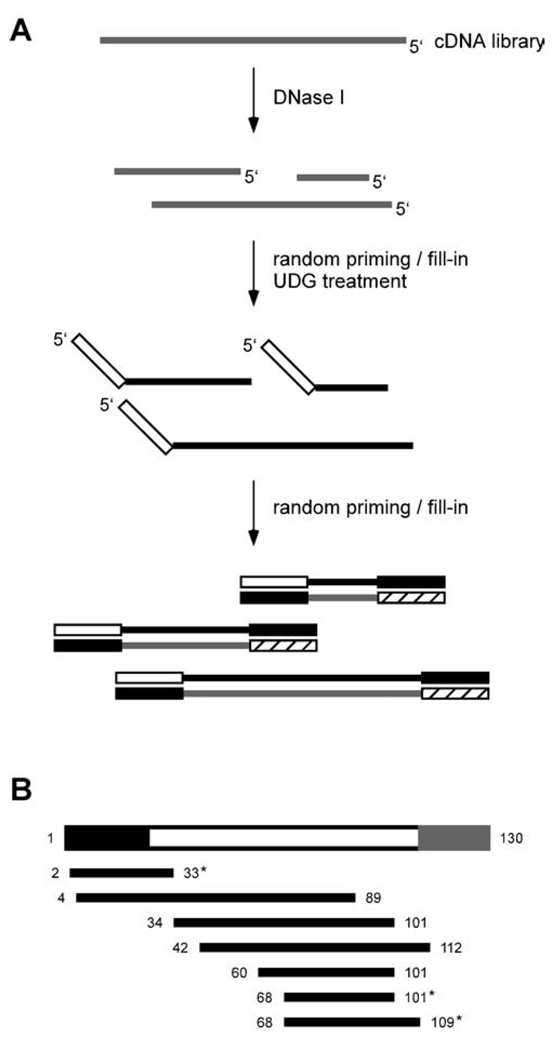
To narrow down the epitope and isolate shorter, high-affinity peptide sequences, a nested deletion library was constructed from the peptide C-dominated library. This library is composed of fragments of DNA that encode shorter stretches of the parent peptides. By using the fragment-library in an mRNA display selection, minimal binding sequences can be identified. Initial attempts to generate nested deletions using random priming on cDNA resulted in nearly full-length sequences (unpublished observations), possibly due to the strand-displacement abilities of the tested polymerases (I.N. Hampson, personal communication; Hamilton et al., 2001). This attribute was exploited in the final fragmentation scheme (Figure 3A). DNase I was used to generate random fragments from the cDNA of a functional library (any pool after the 6th round of selection). Various dilutions of DNase I were used to find the optimal conditions for producing a range of ssDNA products from ~50 to 130 bases (data not shown). Successive random priming and fill-in reactions with a modified T7 polymerase (Sequenase v2.0) and primers containing 3’-random hexamers produced the initial DNA pool. PCR-amplified dsDNA was agarose gel-purified to retain fragments between 150 and 200 bp, corresponding to peptides approximately 10 to 30 amino acids long.
Fig. 3.
Construction of a unidirectional nested deletion library. (A) cDNA library reverse transcribed with dUTP is partially digested with DNase I. A randomly-primed fill-in reaction is performed with degenerate DNA hexamers containing a constant 5’ sequence, resulting in complete second-strand cDNA for each fragment. After uracil-DNA glycosylase (UDG) digestion to remove first-strand cDNA, the anti-sense strand is filled-in again by random priming. The constant region of the 2nd primer encodes a suitable peptide sequence in all 3 frames (lacking stop codons) and serves as the reverse primer site for subsequent PCR. PCR of the resulting dsDNA produces the initial library suitable for in vitro selection. (B) Representation of the peptide C parent DNA sequence in the initial fragment-library. The 5’-UTR, peptide coding region, and 3’-constant region are in black, white, and gray, respectively. The bases spanned by each library member are shown. Sequences marked with an asterisk are in-frame with the 5’-constant region added during the generation of the library. The sequence spanning bases 4 through 89 is also viable assuming translation occurs at the first Met codon.
Because stop codons hinder RNA-peptide fusion formation, the 3’-constant sequence of the fragment-library was chosen such that TAA, TAG, and TGA codons did not exist in any frame. The 5’-constant region added a c-Myc epitope tag and provided a primer site for subsequent PCR amplification (for additional attachment of the T7 promoter and UTR sequence). This method resulted in a unidirectional fragmented pool; all transcribed RNA maintained the sense orientation. DNA sequencing of the initial pool demonstrated reasonable representation of the dominant sequence (peptide C) and confirmed the expected 1/3 fraction of in-frame sequences (Figure 3B). DNA alignments with peptide C derivatives typically contained several mismatches at the beginning and end of the fragment region, most likely due to imperfect annealing of the random hexamer primers.
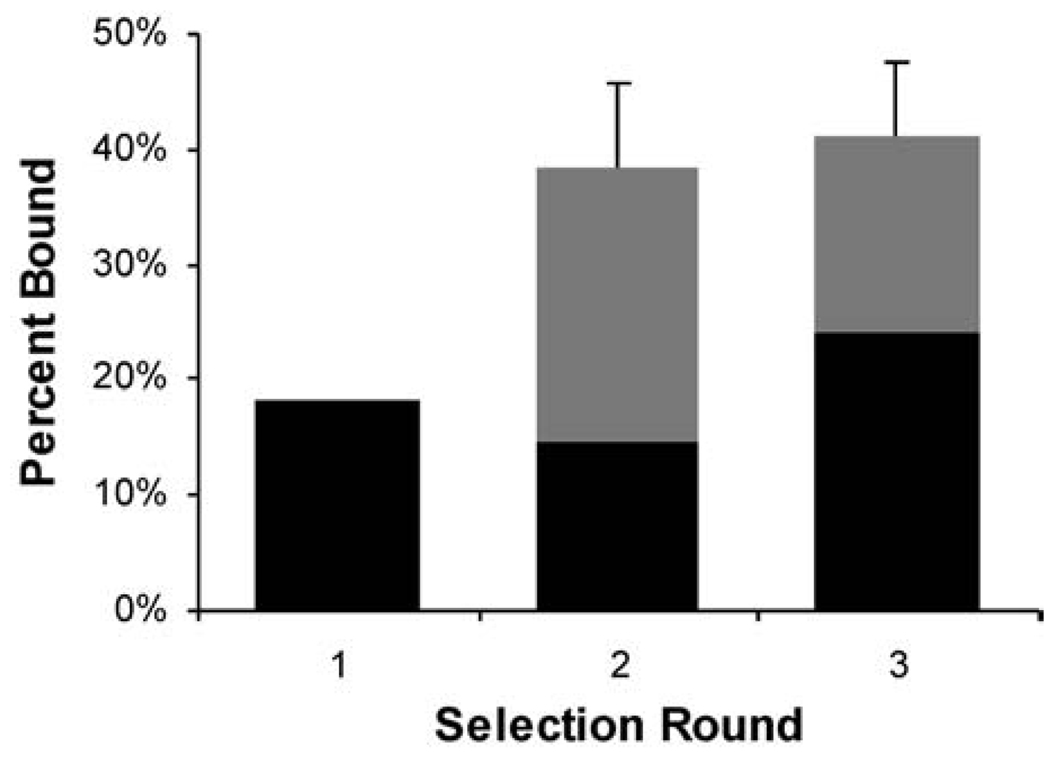
The nested deletion library was used for selection against the anti-polyhistidine mAb (Figure 4). Poly-L-His and His6 peptide were used as competitors in the 2nd and 3rd rounds. Although the binding of the 2nd and 3rd round pools was similar, more RNA-peptide fusions were retained after the stringent, competitive wash in the 3rd round, suggesting that the washes were indeed enriching the pool for the highest affinity peptides. DNA sequencing of the final pool revealed three distinct classes of peptides (Table II). Class 1 sequences were fragments corresponding to N- and C-terminal deletions of peptide C. A sequence alignment of the fragments identified RHDAGDHHHHHGVRQ (peptide Cmin) as a minimal functional sequence for peptide C.
Fig. 4.
Selection of the peptide fragment-library on anti-polyhistidine mAb. The percentage of recovered fusions (black) was determined as in Figure 2. In rounds 2 and 3, the competitive washes (gray) removed a portion of the initially bound counts.
Table II.
Peptide sequences from fragment-library selection
| Class 1 | |||
| * | MDGHPERHDAGDHHHHHGVRQ | ||
| ERHDAGDHHHHHGVRQWRLIS | |||
| RHDAGDHHHHHGVRQWRLIS | |||
| Class 2 | |||
| ITNSPGRFRHHHVLARRHALYR | D (6/20) | ||
| MTSAGWTAMHYISARRHAMRSMKFAQ | E (2/20) | ||
| NYTTQRAEWNRQDAHRHHHQEARRGQ | A1 | ||
| * | MKVRRDVMRWHHHHRMARRKANR | B | |
| DHHHHHGAARPVFRRGLYQKRG | F | ||
| DHRHHHGVARVREQMARYV | |||
| Class 3 | |||
| VTMFDVDAYFGLAVWSSGDLRAFQ | |||
| VTMFDVDAYFGLAVW | (2/20) | ||
| * | MFDYDAFYGYNGSAVGSPTLQHVRLQP | ||
| * | MNFDEYLRLLR |
Only the fragment domain of the peptides is shown. Class 1 peptides are derived from peptide C (Table I) and the putative minimal epitope is underlined. Class 2 sequences contain portions of the ARRXA motif. Conserved residues are in bold. Sequences derived from parent peptides A and B, as well as new peptides D, E, and F, are labeled. The C-terminal RGQ in the sequence derived from peptide A is encoded by part of the 3’-constant region. Class 3 peptide sequences were aligned using CLUSTALW (http://npsa-pbil.ibcp.fr) with key residues determined automatically. Clone frequency (out of 20) is shown and differing residues are italicized as described in Table I. Peptide sequences translated from alternate start codons are marked (*).
The majority of fragments recovered after the selection came from parent sequences other than peptide C (Table II, Class 2). An alignment of peptides D and E (which collectively represented 40% of the final, 3rd round selection pool) revealed the consensus motif ARRHA. This exact motif was not seen in the original selection, although three peptide sequences contained ARRXA (X = R, G (peptide A), or K (peptide B)) two residues C-terminal to the His-track (Table I), as in peptide D. Additional N- and C-terminal deletions for peptides D and E were not observed. Hence, these sequences may already represent minimal high affinity binding epitopes. Alternatively, there may have been an insufficient number of clones sequenced to find other corresponding fragments. Other recovered sequences in this peptide class retained at least part of the ARRXA, suggesting that the first few residues of the consensus motif are more critical for high affinity.
Several additional peptides were discovered that encoded a weak consensus sequence non-related to the mAb-binding peptides (Table II, Class 3). Binding assays with a couple of these peptides revealed significantly weaker affinity for the mAb than a His6-containing peptide control (data not shown). These peptides may bind to an alternate interaction site and were consequently enriched when high stringency, competitive washes were introduced for the last rounds of selection. Site-specific, competitive washes (e.g., with poly-L-histidine) would result in the enrichment of peptides with higher affinity for the antigen-binding region, as well as for peptides with affinity for other sites.
Immunoprecipitation of selected peptides
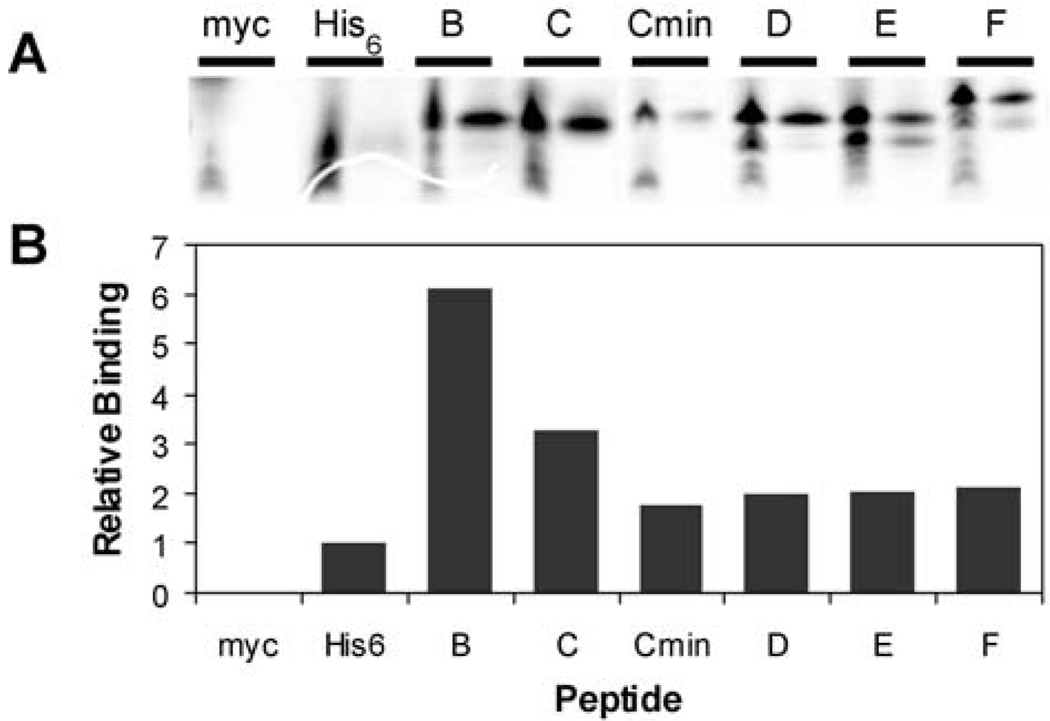
Selected clones were qualitatively assessed for binding by immunoprecipitation with the anti-polyhistidine mAb (Figure 5A). 35S-Met labeled peptides were assayed directly from the in vitro translation reactions. The selected peptides demonstrated significantly increased binding compared with a C-terminal His6-tagged peptide control (Figure 5B). Non-specific binding was shown to be minimal with a c-Myc epitope control peptide. Correct translation of the fragment-selected peptides and the Myc control was confirmed by immunoprecipitation on the 9E10 anti-c-Myc mAb (data not shown).
Fig. 5.
Binding of in vitro translated peptides to anti-polyhistidine mAb. (A) 35S-labeled peptides were assayed for binding directly from the translation reaction. Myc is a peptide encoded by the constant regions of the fragment-library primers with only an arginine residue in between. The His6 sequence encoded a 31-mer peptide with a C-terminal His6 tag. Equivalent aliquots of the translation reactions (left lanes) were analyzed by tricine SDS-PAGE adjacently to immunoprecipitated peptides (right lanes). (B) quantitation of peptide binding in (A). Relative binding is shown as a fold-change versus the His6 sequence. Peptide sequences are given in Table I and Table II.
Kinetics by surface plasmon resonance
Various peptides from the fragment selection were synthesized and purified for kinetics analysis by SPR. In an SPR experiment, one binding partner (ligand) is immobilized on the surface of a sensor chip while the other reactant (analyte) is in solution. Binding of the analyte is seen as a refractive index change on the sensor chip surface and is measured in real-time in resonance units (RU). Peptides were synthesized with a C-terminal biocytin residue for immobilization on streptavidin-coupled surfaces. Full-length peptides B and C were also assayed by expressing the peptides as fusion proteins with a C-terminal MBP and an N-terminal bio-tag, which is biotinylated in vivo by biotin holoenzyme synthetase (BirA). By purifying these proteins via monomeric avidin, they retained their biotin moieties and a homogeneous ligand surface could be produced on the SPR sensor chips.
Rebinding and bivalency effects of mAb interactions with immobilized antigens have previously been shown to offset kinetics measurements considerably, rendering both absolute and relative binding constants unreliable (Nieba et al., 1996). To avoid these problems, Fab fragments were prepared from anti-polyhistidine mAb and used as the analyte. Using the peptides as the immobilized ligands and Fab as the analyte ensured fair comparisons between the kinetics measurements, avoiding bias in protein quantitation, since all Fab concentrations were prepared from a single stock solution. Kinetics parameters were determined using a 1:1 bimolecular interaction model (Table III).
Table III.
Kinetic parameters for peptide interactions with Fab determined by surface plasmon resonance
| Peptide sequence | ka | kd | KD | χ2 | ΔG° | |
|---|---|---|---|---|---|---|
| M−1 s−1 (× 104) | s−1 (× 10−2) | nM | kcal/mol | |||
| HHHHHH | 9.9 | 5.78 | 580 | 0.72 | −8.5 | |
| HHHHHH-protein | 7.3 | 23.82 | 3260 | 0.80 | −7.5 | |
| HHHHHHHHHH | 62.4 | 6.56 | 105 | 1.19 | −9.5 | |
| MDGHPERHDAGDHHHHHGVRQ | 11.8 | 0.85 | 72 | 1.28 | −9.7 | |
| Cmin | RHDAGDHHHHHGVRQ | 21.5 | 0.82 | 38 | 1.19 | −10.1 |
| C | MDGHPERHDAGDHHHHHGVRQWRLISTG-MBP | 52.5 | 0.97 | 18.5 | 1.48 | −10.5 |
| B | MKVRRDVMRWHHHHRMARRKANR-MBP | 40.9 | 0.31 | 7.6 | 1.28 | −11.1 |
| D | NSPGRFRHHHVLARRHALYR | 17.4 | 0.27 | 15.5 | 0.77 | −10.7 |
SPR experiments monitored binding between immobilized peptides and purified Fab fragments. On and off rates were determined by global fit analysis on CLAMP using a 1:1 bimolecular interaction model (Myszka and Morton, 1998). KD values were calculated from kd/ka.
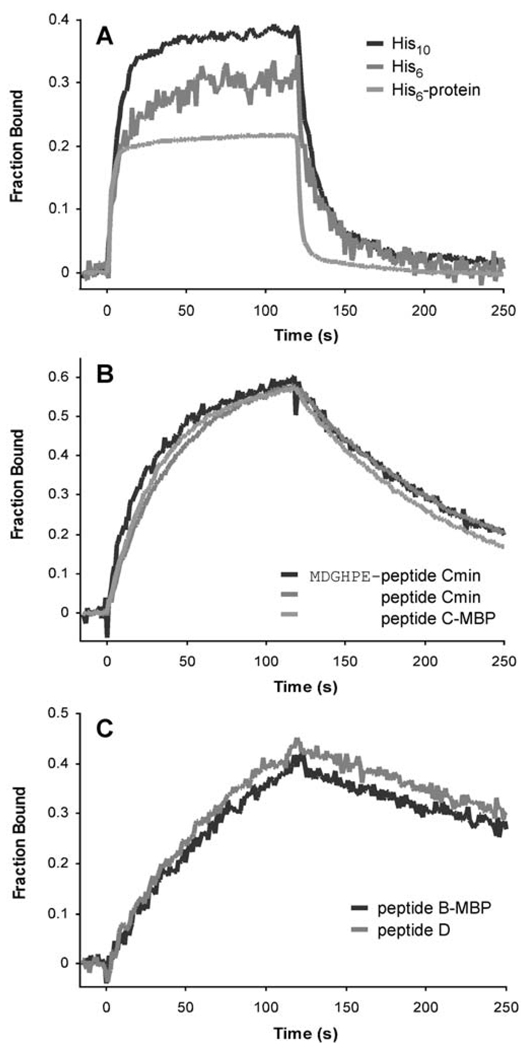
The assayed peptides could be categorized by their dissociation rates from the Fab (Figure 6). The cited epitope, His6, bound weakest to the Fab; the His6 peptide and the His6-tagged protein used in the original selection exhibited dissociation constants of 0.6 and 3 µM, respectively. Additional His residues (His10 peptide) increased the association rate 6-fold without changing the dissociation rate significantly. Peptides from the selection demonstrated dissociation constants less than 75 nM, approximately 10- to 75-fold better than the control His6 sequence, with increased affinities as a result of faster association (up to 5-fold) and considerably slower (6- to 21-fold) dissociation rates (Table III). Class 2 peptides with the ARRXA motif demonstrated the highest affinities, with ~3-fold slower dissociation rates compared to sequences derived from peptide C (Figure 6C). While the flanking residues on the minimized peptide C contribute at least 1.6 kcal/mol to the binding free energy compared with the His6 peptide, sequences with the ARRXA motif demonstrate 2.6 (peptide B) and 2.2 (peptide D) kcal/mol improvements. The contributions from these flanking residues is likely even greater, as these calculations do not account for any loss of binding free energy from having shorter (<6) stretches of His residues in the core site.
Fig. 6.
Representative sensorgrams from SPR experiments. Purified anti-polyhistidine Fab fragments at concentrations corresponding to ~0.5 KD were injected over immobilized peptides or peptide-MBP fusions. Peptides fell into three categories describing (A) weak (His6, His10, and His6-tagged blocking protein), (B) intermediate (peptide C-derived sequences), and (C) strong (sequences containing the ARRXA motif) binding for the Fab fragments. For comparison, sensorgrams were divided by the computed maximum signal.
Discussion
During an in vitro selection experiment against a target protein immobilized using an anti-polyhistidine antibody, mAb-binding peptides were inadvertently enriched. The weak affinity of the His6-tagged fusion protein for the mAb and the existence of alternative peptide motifs that confer significantly higher affinity are the likely causes for the inability to enrich for peptides that bind the original target protein. A preclearing step that included the mAb may not have been totally effective in preventing the selection of antibody-specific peptides, as even the final selection round resulted in an incomplete, ~40% pull-down of the RNA-peptide fusions. Although the cited mAb epitope is hexahistidine, the recovered peptides surprisingly each contained a shorter (≤5) stretch of consecutive His residues and a bias for Arg.
To better characterize the mAb epitope and demonstrate the feasibility of gene fragment mRNA display, a nested deletion library was constructed from the final selection pool. A previously described protocol—directional random oligonucleotide primed (DROP) synthesis of cDNA (Hampson et al., 1996)—was modified to maintain as many viable library fragments as possible. Due to the difficulty in obtaining a broad size distribution of sequences with degenerate oligos, DNase I was used for the random fragmentation of cDNA. DROP-synthesis using a highly processive DNA polymerase, capable of potent strand-displacement, yielded intact copies of the cDNA fragments while maintaining the sense strand (Figure 3A).
In vitro selection with the fragment-library resulted in the identification of a 15-mer functional sequence derived from the full-length 35-mer, peptide C. Because the initial fragment-library was produced from a pool dominated by peptide C, we expected to recover and identify numerous overlapping peptides that defined a minimal epitope for this sequence. Surprisingly, the majority of recovered sequences came from unknown parents. The enrichment of these peptides implies that these fragments were more highly favored after truncation. The flanking regions of the original peptides may have hindered access to the epitope by the mAb, suggesting that peptide length may be an important attribute in the fine-tuning of affinity and/or function. Alternatively, these particular sequences may have been negatively biased by the constant C-terminal peptide used in the original random peptide library. The 3-frame constant sequence used in the fragment-library construction increases the sensitivity of the selection when one of the translation frames causes negative bias. Additionally, a random distribution between the three translation frames would indicate that the constant region does not affect selectability. The 6 independent clones of peptide D, for example, had all 3 frames represented in the 3’ constant region (Table II and data not shown).
Based on the selected peptide sequences, two major protein interaction motifs were identified: a core epitope consisting of at least three consecutive His residues and a 2nd interaction site encoded by the consensus motif, ARRXA. SPR experiments demonstrated a significant increase in the association rate of His10 compared with His6, suggesting that additional His residues present a more accessible core interaction, rather than slow dissociation by enhancing rebinding from multivalency effects. Only additional contacts, made by the addition of interacting residues such as the ARRXA motif, result in significantly slower dissociation rates. These flanking residues can contribute significantly to the binding free energy—at least 2.6 kcal/mol in the case of peptide B in comparison with His6, which assumes the loss of 2 out of 6 histidines in the core has no effect. The two interaction cassettes we have identified here are likely juxtaposed sites from the fusion protein used as the original antigen, a proprietary sequence (Sigma-Aldrich Corp., technical specifications for unconjugated mouse anti-polyhistidine mAb).
Our results also highlight the importance of flanking residues outside of the two consensus motifs and their contribution to binding affinity with antibodies. Residues adjoining core amino acids in an epitope can substantially influence antibody binding, the effects of which can only be assessed through quantitative affinity measurements (Choulier et al., 2001; Coley et al., 2001). This is demonstrated in our experiments, where the rank order of binding in the immunoprecipitation assay did not entirely correspond with quantitative kinetics measurements. Epitope tags are often appended to proteins and used as molecular handles for detection, isolation, and analysis of protein-protein interactions. Their functionality in this context, however, is highly variable. Tandem repeats of tags (e.g., the popular c-Myc or FLAG epitopes) have been used to ensure robust affinity and recognition by antisera (Nakajima and Yaoita, 1997; Hernan et al., 2000). By identifying longer functional peptides with appropriate flanking residues, high affinity can be maintained with less variability depending on the linker region and the protein to which the epitope is attached.
The ability to access high complexity libraries is a great advantage for mRNA display over other selection systems. Library construction methods that involve PCR and DNA reassembly are better suited for the mRNA display format, thereby avoiding cloning steps that are required in techniques such as phage display. A comparative study on epitope mapping using random 6-mer and 15-mer peptide phage display libraries successfully identified consensus motifs for only 2 of the 4 mAbs examined (Fack et al., 1997). For one of the mapped mAbs, the random peptide selection succeeded only with the 6-mer library, identifying a short consensus motif that was not discovered with the 15-mer library, which the authors attributed to a statistical lack of representation. Previously, mRNA display with a random 27-mer library revealed epitope-like consensus motifs for the trypsin active site and the anti-c-Myc antibody, 9E10 (Baggio et al., 2002). These experiments achieved relatively fine-mapping of the epitopes, uncovering the core residues as well as some of the allowed flanking amino acids. By utilizing high complexity, long peptide libraries, mRNA display selections can identify rare sequences of high affinity and determine linear or discontinuous epitopes. The full-length consensus peptide, Hm-X2-ARRXA, for example, may not have been identified with more traditional X6 or X10 phage display libraries.
One of the difficulties noticed in the fragment selection was the disproportionate number of peptides that did not contain an N-terminal deletion. Because of the 5’-UTR on the mRNA used to make the fragment-library, more fragments containing the first start codon (with varying lengths of UTR sequence) were probably present in the initial fragment pool. 5’-UTR and/or promoter sequences most likely do not hinder the fragment selection process, as ribosome scanning can initiate translation at the correct start codon, regardless of which frame was amplified. This was seen in several of the selected fragment sequences (Table II). This property increases the number of viable (i.e. translatable) templates, but introduces some bias favoring intact N-terminal sequences.
Although not utilized in this experiment, the c-Myc tag introduced in the fragmentation library can be used to generate and purify a fragment-library enriched with in-frame sequences. Although the tag is at the N-terminus of the library, in general RNA-peptide fusions will form only when the ribosome can translate most of the sequence and reach the end of the mRNA (unpublished results). Hence, only sequences that lack stop codons (and therefore are most likely in-frame) will form fusions and be purified and amplified after a Myc-epitope pre-selection. Another improvement to the protocol includes using Exonuclease I to remove excess degenerate primers during DROP-synthesis, preventing the amplification of sequences without “inserts,” as DNA size fractionation by agarose gel is not completely effective in removing these smaller fragments (data not shown).
Due to the higher efficiency of synthesizing the nested deletion library completely in vitro, the fragment-library construction described here maintains a higher number of unique sequences, in contrast to DNA libraries produced by enzymatic ligation and cloning, which are limited by in vivo transformation efficiencies. Additionally, the DROP-synthesis is unidirectional for all amplified sequences so that the sense orientation is maintained and only the minimal 2/3 of the fragments are non-viable due to frame shifts. This protocol produces a well-distributed library and is technically less challenging as the random oligonucleotide priming is used only to “copy” the cDNA fragments produced by DNase digestion, and need not be optimized for generating a fragment distribution. mRNA display with fragment-libraries combine the ease and versatility of working with cDNA in vitro with the benefits of expression cloning. The method permits the minimization of functional domains, as well as the isolation of optimal binding contexts through the removal of negative-acting flanking regions. Although the technique may not be sufficiently processive for the fine-mapping of short peptide sequences, it should be highly applicable for constructing cDNA or tissue-specific expression-libraries and the subsequent determination of minimal binding domains and novel protein-protein interactions.
Acknowledgments
We thank Dr. David S. Waugh (National Cancer Institute at Frederick) for the pDW363 in vivo biotinylation vector, Prof. Pamela J. Bjorkman and Anthony M. Giannetti for time and support on the Biacore 2000, William Hunter (Biacore, Inc., Piscataway, NJ) for technical advice on SPR, Cindy I. Chen and Christopher T. Balmaseda for preparative and technical assistance on library construction and protein purification, and Prof. David G. Myszka (University of Utah) for generously providing the kinetics analysis software, Scrubber and CLAMP. We greatly appreciate Dr. Yuri Peterson (Duke University) for suggestions on the paper. We are indebted to Dr. Ian N. Hampson (St. Mary’s Hospital, Manchester, UK) for valuable discussions and technical expertise on random priming and the synthesis of the fragment-library. This work was supported by grants from the NIH (RO160416) and Beckman Foundation to R.W.R. W.W.J. was supported in part by a DOD National Defense and Engineering Graduate Fellowship. R.W.R. is an Alfred P. Sloan Foundation Research Fellow.
References
- Baggio R, Burgstaller P, Hale SP, Putney AR, Lane M, Lipovsek D, Wright MC, Roberts RW, Liu R, Szostak JW, et al. J. Mol. Recognit. 2002;15:126–134. doi: 10.1002/jmr.567. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Biotechnol. Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- Choulier L, Laune D, Orfanoudakis G, Wlad H, Janson J-C, Granier C, Altschuh D. J. Immunol. Methods. 2001;249:253–264. doi: 10.1016/s0022-1759(00)00358-6. [DOI] [PubMed] [Google Scholar]
- Christmann A, Wentzel A, Meyer C, Meyers G, Kolmar H. J. Immunol. Methods. 2001;257:163–173. doi: 10.1016/s0022-1759(01)00461-6. [DOI] [PubMed] [Google Scholar]
- Coley AM, Campanale NV, Casey JL, Hodder AN, Crewther PE, Anders RF, Tilley LM, Foley M. Protein Eng. 2001;14:691–698. doi: 10.1093/protein/14.9.691. [DOI] [PubMed] [Google Scholar]
- du Plessis DH, Wang L-F, Jordaan FA, Eaton BT. Virology. 1994;198:346–349. doi: 10.1006/viro.1994.1039. [DOI] [PubMed] [Google Scholar]
- Fack F, Hügle-Dörr B, Song D, Queitsch I, Petersen G, Bautz EKF. J. Immunol. Methods. 1997;206:43–52. doi: 10.1016/s0022-1759(97)00083-5. [DOI] [PubMed] [Google Scholar]
- Frank R. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- Frank R, Overwin H. Methods Mol. Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- Frohlich MW, Parker DS. Biotechniques. 2001;30:264–266. doi: 10.2144/01302bm05. [DOI] [PubMed] [Google Scholar]
- Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Curtiss R., 3rd Nat. Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- Gupta S, Arora K, Sampath A, Khurana S, Singh SS, Gupta A, Chaudhary VK. Biotechniques. 1999;27:328–330. doi: 10.2144/99272st04. 332–334. [DOI] [PubMed] [Google Scholar]
- Hamilton SC, Farchaus JW, Davis MC. Biotechniques. 2001;31:370–376. doi: 10.2144/01312rv01. 378–380, 382–383. [DOI] [PubMed] [Google Scholar]
- Hammond PW, Alpin J, Rise CE, Wright M, Kreider BL. J. Biol. Chem. 2001;276:20898–20906. doi: 10.1074/jbc.M011641200. [DOI] [PubMed] [Google Scholar]
- Hampson IN, Hampson L, Dexter TM. Nucleic Acids Res. 1996;24:4832–4835. doi: 10.1093/nar/24.23.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes J, Plückthun A. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Taussig MJ. Nucleic Acids Res. 1997;25:5132–5134. doi: 10.1093/nar/25.24.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hernan R, Heuermann K, Brizzard B. Biotechniques. 2000;28:789–793. doi: 10.2144/00284pf01. [DOI] [PubMed] [Google Scholar]
- Irving MB, Pan O, Scott JK. Curr. Opin. Chem. Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Reineke U, Dong L, Hoffmann B, Hoffmüller U, Winkler D, Volkmer-Engert R, Schneider-Mergener J. J. Pept. Res. 1999;54:319–327. doi: 10.1034/j.1399-3011.1999.00108.x. [DOI] [PubMed] [Google Scholar]
- Kurz M, Gu K, Lohse PA. Nucleic Acids Res. 2000;28:e83. doi: 10.1093/nar/28.18.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara I, Maruyama H, Kamisue S, Shima M, Yoshioka A, Maruyama IN. J. Immunol. Methods. 1999;224:89–99. doi: 10.1016/s0022-1759(99)00012-5. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Lenstra JA, Kusters JG, van der Zeijst BA. Arch. Virol. 1990;110:1–24. doi: 10.1007/BF01310699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Barrick JE, Szostak JW, Roberts RW. Methods Enzymol. 2000;318:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- Mattheakis LC, Bhatt RR, Dower WJ. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson M, Yang Y, Hammond PW, Kreider BL. Chem. Biol. 2002;9:691–698. doi: 10.1016/s1074-5521(02)00148-5. [DOI] [PubMed] [Google Scholar]
- Miceli RM, DeGraaf ME, Fischer HD. J. Immunol. Methods. 1994;167:279–287. doi: 10.1016/0022-1759(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Milavetz B. Nucleic Acids Res. 1992;20:3529–3530. doi: 10.1093/nar/20.13.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BP, Pallavicini MG, Marks JD. Infect. Immun. 2001;69:6511–6514. doi: 10.1128/IAI.69.10.6511-6514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KK, Shen S-H, Banville D. Biochem. Biophys. Res. Commun. 1998;248:69–74. doi: 10.1006/bbrc.1998.8912. [DOI] [PubMed] [Google Scholar]
- Myszka DG. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- Myszka DG, Morton TA. Trends Biochem. Sci. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Nucleic Acids Res. 1997;25:2231–2232. doi: 10.1093/nar/25.11.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieba L, Krebber A, Plückthun A. Anal. Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- Parhami-Seren B, Keel T, Reed GL. J. Mol. Biol. 1997;271:333–341. doi: 10.1006/jmbi.1997.1174. [DOI] [PubMed] [Google Scholar]
- Pues H, Holz B, Weinhold E. Nucleic Acids Res. 1997;25:1303–1304. doi: 10.1093/nar/25.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke U, Kramer A, Schneider-Mergener J. Curr. Top. Microbiol. Immunol. 1999;243:23–36. doi: 10.1007/978-3-642-60142-2_2. [DOI] [PubMed] [Google Scholar]
- Roberts RW, Szostak JW. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi E, Capone S, Mennuni C, Lahm A, Tramontano A, Luzzago A, Nicosia A. J. Mol. Biol. 2000;296:497–508. doi: 10.1006/jmbi.1999.3471. [DOI] [PubMed] [Google Scholar]
- Scopes RK. Anal. Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Takahashi TT, Austin RJ, Roberts RW. Trends Biochem. Sci. 2003;28:159–165. doi: 10.1016/S0968-0004(03)00036-7. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BAJ, Tunnacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Tsao K-L, DeBarbieri B, Michel H, Waugh DS. Gene. 1996;169:59–64. doi: 10.1016/0378-1119(95)00762-8. [DOI] [PubMed] [Google Scholar]
- Whitcomb JM, Rashtchian A, Hughes SH. Nucleic Acids Res. 1993;21:4143–4146. doi: 10.1093/nar/21.17.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Byrne CD. Biochem. J. 1999;337:231–241. [PMC free article] [PubMed] [Google Scholar]