Abstract
Background
Genotyping error can increase both type I and II errors. In order to elucidate potential genotyping errors, data quality control often includes testing genotype data for deviations from Hardy-Weinberg Equilibrium (HWE).
Methods
The Hardy-Weinberg Disequilibrium (HWD) coefficient and the ability to reject the null hypothesis of HWE were calculated analytically for genotype data from parents and unaffected siblings of affected probands.
Results
Genotype data from parents and unaffected siblings display deviations from HWE when functional or markers in LD with functional locus are tested. For the parental genotype data all deviations from HWE are negative, indicating an excess of heterozygous genotypes with the strongest deviations from HWE observed for the multiplicative model. In contrast, for affected proband genotype data, there is no deviation from HWE under the multiplicative model and the deviations from HWE for the recessive model are positive. For the unaffected sibling data, patterns of deviation from HWE are similar to those observed in the proband data with the exception of the multiplicative model where the HWD coefficient although close to 0 can be either positive or negative depending on the allele frequency.
Conclusion
Deviations from HWE in parental and unaffected sibling genotype data could be due to an association with the functional locus. However these deviations for genotypic relative risk ≤2.0 are not large and therefore the power to detect them is usually low. Testing for deviations from HWE in parental and unaffected sibling genotype data is still beneficial for quality control even though functional loci, in parental and unaffected sibling genotype data, can produce an association signal.
Key Words: Association studies, Data quality control, Family and population based data, Hardy-Weinberg Equilibrium
Introduction
In the past few years the emphasis in gene mapping has shifted to association studies to map complex traits. Association studies are carried out using either population- or family-based data. Genotyping error can be detrimental for both of these study designs. For population-based studies (e.g. case-control), random genotyping error can increase type II error and thereby decrease power [1,2,3,4]. For family-based data (e.g. trio data), genotyping error can increase both type I and II errors [3, 5]. Therefore it is important to be able to assess SNP marker loci for genotyping error so that problematic markers can either be removed or genotype calls corrected. For population- and family-based data, duplicate samples can be genotyped to determine the rates of genotyping error; however, if a systematic genotyping error has occurred it will not be detected and genotyping error rates will be underestimated. For family data genotyping errors can sometimes be detected by observation of Mendelian inconsistencies. However, a substantial portion of genotyping errors cannot be detected since many errors are compatible with Mendelian inheritance laws [5,6,7]. The ability to detect errors via Mendelian inconsistency depends on the error model (e.g. random, heterozygous to homozygous genotype) and the marker allele frequency. Errors are especially difficult to detect for diallelic markers, with the lowest detection rates for markers with alleles of equal frequency [5, 6]. Although it is easier to identify genotyping errors for markers with multiple alleles, the ability to uncover them can still be low [6, 8]. Families with multiple offspring not only increase the ability to detect Mendelian errors [6, 9], but also aid in uncovering of genotyping errors through the detection of double recombination events over short genetic distances [8,10,11,12,13].
In addition to checking pedigrees for Mendelian inconsistencies, often genotype data from parents or unaffected siblings of affected probands are tested for deviations from Hardy-Weinberg Equilibrium (HWE) in order to detect potential genotyping error. For population based studies, genotype data from all individuals for quantitative trait studies and controls from cases-control studies are analyzed to determine whether there are deviations from HWE [14,15,16]. Genotyping errors can create positive, negative or no deviation from HWE, depending on how the genotyping error occurred. In general testing for deviations from HWE is not a powerful approach to detect genotyping errors [17].
Deviations from HWE are not necessarily due to genotyping error and may be due to chance or genetic factors which include a heterozygous advantage, population admixture/substructure, inbreeding or copy number variants [18,19,20,21,22]. For example, population substructure creates an excess of homozygote genotypes and therefore a positive HWD coefficient. Deviations from HWE which are only observed in genotype data from cases can be due to an association between the trait and either a functional locus or a SNP marker which is in linkage disequilibrium (LD) with a functional locus. The ability to detect deviations from HWE depends on the magnitude of the deviation, sample size and α level. When tests for HWE are performed for genotype quality control there is no consensus on which α level should be used, and the p value criterion used to reject the null hypothesis of HWE varies greatly within the literature. Some studies use a criterion for tests of HWE which is as stringent as those used for genome-wide significance for association studies which is a p value of 1 × 10−7 or lower [23].
In this article it is demonstrated that deviations from HWE observed in the genotype data from parents or unaffected siblings of affected probands can also be due to an association where the tested SNP is either in LD with or is the functional locus. For comparison purposes the deviation from HWE is also examined for affected probands and unrelated controls that are disease free. Depending on the genetic model the pattern, strength and direction of deviations from HWE are different in the parental, unaffected sibling, affected proband and control genotype data. Additionally it is shown that incomplete LD, genotyping error and population admixture/substructure play a role in attenuating or amplifying the deviations from HWE, thus affecting the power to detect a deviation from HWE.
Methods
Testing Proband and Unrelated Control Genotype Data for Deviations from HWE
Calculations are performed for a SNP marker locus with two alleles which is in LD with a functional locus. The two alleles at the functional locus are represented by A1 which has a population allele frequency of p and A2 which has a frequency of q = 1 – p. The two alleles at the SNP marker are B1 and B2 with allele frequency of pm and qm = 1 – pm. Let P11, P12 and P22 denote the frequencies of genotypes G11, G12 and G22 at the functional locus and Q11, Q12 and Q22 denote the frequencies of genotypes M11, M12 and M22 at the SNP marker. Under HWE, P11, P12 and P22 are equal to p2, 2pq, and q2 respectively. Let f11, f12 and f22 denote the penetrances of genotypes G11, G12 and G22, respectively. The genotypic relative risks (RRs) are defined as γ1 = f12/f11 and γ2 = f22/f11. The genotypic RRs satisfy γ2 = γ21 for the multiplicative model, γ2 = 2γ1 − 1 for the additive model, γ2 = γ1 for the dominant model and γ1 = 1 for the recessive model. Given genotype penetrances and allele frequencies, the disease prevalence, PD, can be calculated as PD = p2f11 + 2pqf12 + q2f22. If a sample of N trios is ascertained based on the child's phenotype, using Bayes rule the expected genotype proportions for the probands are PDG11 = f11P11/PD, PDG12 = f12P12/PD and PDG22 = f22P22/PD. For a given strength of LD (e.g. r2) between the functional locus and the SNP marker, the expected genotype frequencies at the maker locus in probands, denoted as Pd(M11), Pd(M12) and Pd(M22), can be calculated assuming allele A1 is positively associated with allele B1 (Appendix). Let pd be the expected allele frequency of B1 at the marker locus, then pd = Pd(M11) + Pd(M12)/2 and the expected HWD coefficient, Dd at the SNP marker is defined as
Dd can range from −0.25 to 0.25 for a locus with two alleles. Under the alternative hypothesis, HWE is false, the power of rejecting HWE is determined by the noncentrality parameter (ncp) of noncentral χ21 distribution [24], which is given by
The power of rejecting HWE in proband genotype data is ηd = Pr(χ21 (vd) ≥χ21,1–α).
In the unrelated unaffected controls the expected genotype proportions at the functional locus are Pc(G11) = (1 – f11)P11/ (1 – PD), Pc(G12) = (1 – f12)P12/(1 – PD) and Pc(G22) = (1 – f22)P22/ (1 – PD). For a given r2 between the marker and the functional locus, the genotype frequencies Pc(M11), Pc(M12) and Pc(M22) at the marker locus are calculated (Appendix) and the frequency of allele B1 in unaffected control genotype data is pc = Pc(M11) + Pc(M12)/2. The HWD coefficient is Dc = Pc(M11) – p2c and the ncp of noncentral χ21 distribution is
where Nc is the number of unaffected controls. The power to reject HWE in unaffected control genotype data is ηc = Pr(χ21(vc) ≥ χ21,1–α).
Testing Parental Genotype Data for Deviations from HWE
The expected genotype frequencies within the parental genotype data are calculated based upon the proband genotype frequencies. There are 3 possible genotypes for each parent and 9 possible mating types for each trio. Each of the 9 mating types has a specific probability of producing an offspring with G11, G12 or G22 genotypes, according to Mendelian law. For example, a father with genotype G11 and a mother with genotype G12 have probability of 0.5, 0.5 and 0 respectively to have a child with genotypes G11, G12 or G22. Given a child's genotype, the expected proportion of each mating type denoted by T is calculated using Bayes rule as
where Gj, j = 1, 2, 3 denotes proband's genotype G11, G12 and G22, and P(Ti) and P(Gj) denote population proportions of mating types and children genotypes, respectively. Then the expected proportion of each mating type in the sample is
where summation is over the 3 genotypes. Parental genotype proportions, denoted as Pp(Gj), j = 0, 1, 2, are calculated as
where If {Gi} and Im {Gi} are indicator functions with value 1 if the father's (If) or mother's (Im) genotype is Gi and 0 otherwise. For a given LD between the marker and the functional locus, the genotype frequencies at the marker locus, denoted as Pp(M11), Pp(M12) and Pp(M22), are calculated similarly as in proband genotype data. The allele frequency of B1 is pp = Pp(M11) + Pp(M12)/2 and the HWD coefficient in parental data is defined as Dp = Pp(M11) – p2p. The ncp of the noncentral χ21 distribution is
and the power to detect the deviation from HWE in parental genotype data is ηp = Pr(χ21(vp) ≥ χ21,1–α).
Testing Unaffected Sibling Genotype Data for Deviations from HWE
The expected genotype frequencies of unaffected siblings of the probands can also be calculated based on the frequencies of each mating type in the ascertained sample. Let P(Gj∣Ti) be the probability of producing a child with genotype Gj when the parents are of mating type Ti under random transmission. The proportion of children's genotype Gj, denoted as Ps(Gj), is given by
The proportion of genotype Gj in the unaffected siblings is given by
where fGj is the penetrance of genotype Gj. Similar to the case for the parents, for a given r2 the expected genotype frequencies Pu(M11), Pu(M12) and Pu(M22) at the SNP marker in unaffected sibling genotype data can also be calculated assuming A1 is positively associated with B1 (Appendix). The expected allele frequency of B1 is pu = Pu(M11) + Pu(M12)/2 and the expected HWD coefficient is Du = Pu(M11) – p2u. The ncp of the noncentral χ21 is
and the power of rejecting HWE in unaffected sibling genotype data is given by ηu = Pr(χ21 (vu) ≥ χ21,1–α).
Genotyping Errors
For genotyping errors, let e be the genotyping error rate. For random genotyping error model, genotyping errors are introduced to either allele of the marker locus independently. The genotype frequencies in parental data with random genotyping errors are PpE1(M11) = Pp(M11)(1 – e)2 + Pp(M12)e(1 – e) + Pp(M22)e2, PpE1(M22) = Pp(M11)e2 + Pp(M12)e(1 – e) + Pp(M22)(1 – e)2 and PpE1(M12) = 1 – PpE1(M11) – PpE1(M22). For homozygote to heterozygote error model, the genotype frequencies with genotyping errors are PpE2(M11) = Pp(M11)(1 – e), PpE2(M22) = Pp(M22)(1 – e) and PpE2(M12) = 1 – PpE2(M11) – PpE2(M22). For heterozygote to homozygote error model the genotype frequencies are PpE3(M11) = Pp(M11) + Pp(M12)e/2, PpE3(M22) = Pp(M22) + Pp(M12)e/2 and PpE3(M12) = 1 – PpE3(M11) – PpE3(M22). For unaffected siblings genotype data the genotype frequencies are calculated similarly.
Populations Substructure
For population substructure, assume there are 2 subpopulations and let c denote the proportion of population 1 in the sampled families. Given population-specific allele frequencies and genetic models in population 1 and 2, the genotype frequencies at the marker locus in parental data in population 1, denoted as PpS1(Mij), and in population 2, denoted as PpS2(Mij), can be calculated as described in the section on Testing Parental Genotype Data for Deviations from HWE. The genotype frequencies in the combined populations are PpS(Mij) = cPpS1(Mij) + (1 – c)PpS2(Mij). For unaffected siblings data the genotype frequencies are calculated in the same manner.
Parameters Used for the Calculations
Deviations from HWE at a SNP marker in LD of r2 = 1, r2 = 0.8 and r2 = 0.5 with the functional locus in parental, unaffected sibling, affected proband and unrelated control genotype data were investigated under multiplicative, additive, dominant and recessive genetic models. A sample size of 5,000 pedigrees (i.e. 10,000 parents, 5,000 unaffected siblings and 5,000 probands) and 5,000 unrelated controls were used to calculate the power of rejecting the null hypothesis of HWE at the stringent α level of 1 × 10−7 (χ21 = 28.37). Genotypic RRs of γ1 = 1.5 and γ1 = 2.0 were employed and the population allele frequencies were varied from 0.05 to 0.95. The phenocopy rate, f0 was set to 0.01 and thus the disease prevalence ranged from 0.01 to 0.03. In order to study the effects of genotyping error and population substructure, a genotypic RR γ1 = 1.5 was used with the marker in perfect LD (r2 = 1) with the functional locus. The genotyping error rate was set to 0.01 for all error models. To study the effects of population substructure the sample consisted of two populations with proportion of 0.2 for population 1 and 0.8 for population 2. Three examples were used where the ratio of the allele frequency in the two populations was set to 0.9, 0.8 and 0.6 while keeping all other parameters equal in the two populations.
Results
Parental Genotype Data
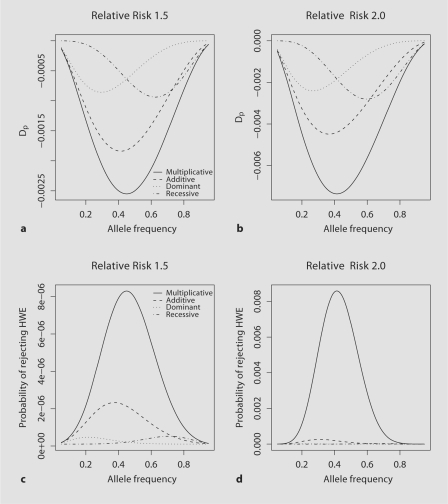
The strength and power of detecting a deviation from HWE at the marker locus which is in perfect LD (r2 = 1) with the functional locus in the parental genotype data are illustrated in figure 1. For the parental data for all genetic models the HWD coefficient Dp is negative, indicating an excess of heterozygous genotypes. As the genotypic RR increases not only does the deviation from HWE increase, but also the population allele frequency at which the maximum deviation occurs declines (fig. 1a, b; table 1). Of the four genetic models, the multiplicative model displays the greatest deviation from HWE, with the additive model presenting with second strongest deviation from HWE. For example, maximum deviations from HWE of −0.00736 and −0.00449 are observed for γ1 = 2.0 under the multiplicative and additive model, respectively (fig. 1b; table 1). For both of these genetic models the deviations are approximately symmetric. The deviations from HWE for the dominant and recessive models are approximately mirror images with the largest HWD coefficient being roughly −0.0009 for γ1 = 1.5 at the population allele frequency of 0.29 for the dominant model and 0.63 for the recessive model (fig. 1a; table 1).
Fig. 1.
For parental genotype data for population allele frequencies ranging from 0.05 to 0.95; the HWD coefficient for a genotypic RR of γ1 = 1.5 (a) and γ1 = 2.0 (b) and the power of rejecting the null hypothesis of HWE for α = 1 × 10−7 forgenotypic RR of γ1 = 1.5 (c) and γ1 = 2.0 (d).
Table 1.
Maximum deviation from HWE (D) and the population allele frequency (freq) at which it occurs for a genotypic RR of γ 1 = 1.5 and γ 1 = 2.0 for parental, unaffected sibling and proband genotype data under an additive, multiplicative, dominant and recessive model
| Models | Parents |
Unaffected siblings |
Probands |
|||
|---|---|---|---|---|---|---|
| Dp | freq | Du | freq | D | freq | |
| Genotypic RR of γ1 = 1.5 | ||||||
| Additive | –0.00184 | 0.41 | –0.00175 | 0.4 | –0.00736 | 0.41 |
| Multiplicative | –0.00255 | 0.45 | 0.00023 | 0.8 | 0 | any |
| Dominant | –0.00086 | 0.29 | –0.00596 | 0.45 | –0.02526 | 0.45 |
| Recessive | –0.00094 | 0.63 | 0.00603 | 0.45 | 0.02526 | 0.45 |
| Genotypic RR of γ1 = 2.0 | ||||||
| Additive | –0.00449 | 0.37 | –0.00423 | 0.35 | –0.01795 | 0.37 |
| Multiplicative | –0.00736 | 0.41 | 0.00101 | 0.78 | 0 | any |
| Dominant | –0.00240 | 0.27 | –0.00996 | 0.41 | –0.04289 | 0.41 |
| Recessive | –0.00280 | 0.60 | 0.01019 | 0.42 | 0.04289 | 0.41 |
Under HWE, the power of detecting a deviation from HWE is α. When Dp ≠ 0 the power of detecting a deviation from HWE increases with increasing sample size. The power of rejecting HWE is greatest for multiplicative model followed by additive model while dominant and recessive models have much lower power (fig. 1c, d). For example for γ1 = 1.5, the maximum power of rejecting HWE is 8.3 × 10−6 for multiplicative model and it is even lower for other models (table 2). For γ1 = 2.0 the power of rejecting HWE increased to 8.58 × 10−3 for multiplicative model (table 2). When the analyzed marker is not in perfect LD (r2 = 1) with the functional locus, the magnitude of HWD coefficients and power of rejecting HWE are reduced (suppl. figure 1; suppl. table 1, 2 (suppl. material see www.karger.com/doi/10.1159/000179558)). For example, the maximum HWD coefficient for the additive model is decreased from −0.00184 to −0.00147 at r2 = 0.8 and to −0.00092 at r2 = 0.5 (suppl. figure 1; suppl. table 1). Corresponding to the attenuation of HWD coefficients, the power of rejecting HWE is also lessened (suppl. figure 1; suppl. table 2).
Table 2.
Maximum power of rejecting HWE and the population allele frequency (freq) at which it occurs for a genotypic RR of γ1 = 1.5 and γ1 = 2.0 for parental, unaffected sibling and proband genotype data under an additive, multiplicative, dominant and recessive model
| Models | Parents |
Unaffected siblings |
Probands |
|||
|---|---|---|---|---|---|---|
| power | freq | power | freq | power | freq | |
| Genotypic RR of γ1 = 1.5 | ||||||
| Additive | 2.32E–06 | 0.37 | 7.21E–07 | 0.36 | 5.86E–04 | 0.41 |
| Multiplicative | 8.30E–06 | 0.45 | 1.55E–07 | 0.95 | 1.0E–07 | any |
| Dominant | 4.67E–07 | 0.21 | 1.41E–04 | 0.42 | 0.97 | 0.45 |
| Recessive | 5.22E–07 | 0.71 | 1.51E–04 | 0.43 | 0.97 | 0.45 |
| Genotypic RR of γ1 = 2.0 | ||||||
| Additive | 2.611E–04 | 0.31 | 2.35E–05 | 0.29 | 0.40 | 0.37 |
| Multiplicative | 8.58E–03 | 0.41 | 3.54E–06 | 0.95 | 1.0E–07 | any |
| Dominant | 1.77E–05 | 0.19 | 6.94E–03 | 0.36 | >0.99 | 0.41 |
| Recessive | 2.83E–05 | 0.68 | 8.07E–03 | 0.38 | >0.99 | 0.41 |
Unaffected Sibling Genotype Data
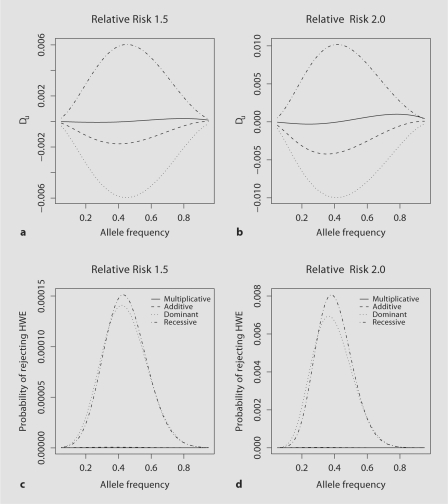
The power of rejecting the null hypothesis and the pattern of deviation from HWE are very different for the parental and the unaffected sibling genotype data (fig. 1, 2; table 1, 2). For the genotype data for the unaffected siblings the HWD coefficient Du is positive for the recessive model indicating an excess of homozygous genotypes, while similar to the parental genotype data the deviation from HWE is negative for the additive and dominant model. For the multiplicative model Du is sigmoidly shaped with negative values for low and positive values for high population allele frequencies, with the position at which Du passes from negative to positive dependent on the genotypic RR (data not shown). Additionally there is a greater departure from Du = 0 for the dominant and recessive model for unaffected sibling genotype data compared to the deviation from HWE observed in the parental genotype data when the genotype RR ≤2.0 (fig. 1a, b; fig. 2a, b; table 1).
Fig. 2.
For unaffected sibling genotype data for population allele frequencies ranging from 0.05 to 0.95; the HWD coefficient for a genotypic RR of γ1 = 1.5 (a) and γ1 = 2.0 (b) and the power of rejecting the null hypothesis of HWE for α = 1 × 10−7 for genotypic RR of γ1 = 1.5 (c) and γ1 = 2.0 (d).
In the unaffected sibling genotype data, the power of rejecting HWE also shows dramatic differences from the parental data (fig. 2c, d; table 2). Dominant and recessive models have the highest power of detecting deviations from HWE while the additive and multiplicative model show much lower power of rejecting HWE (fig. 1c, d). However the power to detect deviations from HWE is not high due to the small magnitude of the deviation. For example, for the dominant model for γ1 = 1.5 the maximum power of rejecting HWE is 1.41 × 10−4, at population allele frequency of ∼0.42, and increases to 6.94 × 10−3 for γ1 = 2.0, at population allele frequency of ∼0.36. The recessive model has similar power of rejecting HWE to the dominant model. For the additive and multiplicative model the maximum power of rejecting HWE is only slightly elevated over the value of α (fig. 2c, d; table 2). When the r2 value between the marker and the functional locus is not equal 1, the HWD coefficients and corresponding power to detect deviations from HWE are both reduced (suppl. figure 1; suppl. table 1, 2).
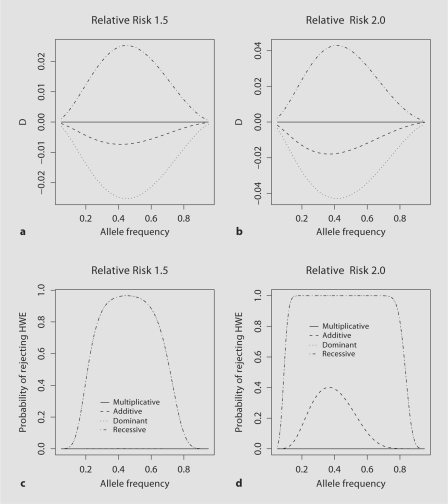
Proband Genotype Data
Deviation from HWE in genotype data from affected individuals was observed and proposed for use in detecting associations in case only studies [25] and various scenarios were more extensively explored by Wittke-Thompson et al. [26]. The strength of deviation for HWE, D is shown in figure 3a, b and table 1 and the power of rejecting the null hypothesis of HWE is displayed in figure 3c, d and table 2. Similar to the genotype data for unaffected siblings deviations from HWE are negative for the additive and dominant model and positive for the recessive model. However the strength of deviations from HWE is greater in the genotype data from probands compared to the unaffected siblings. For γ1 = 1.5 the deviation from HWE is about four times greater in the proband genotype data than in the genotype data from unaffected siblings for the additive, dominant and recessive models. For the multiplicative model there are no deviations from HWE, regardless of the population allele frequency (fig. 3a, b; table 1).
Fig. 3.
For proband genotype data for population allele frequencies ranging from 0.05 to 0.95; the HWD coefficient for a genotypic RR of γ1 = 1.5 (a) and γ1 = 2.0 (b) and the power of rejecting the null hypothesis of HWE for α = 1 × 10−7 for a genotypic RR of γ1 = 1.5 (c) and γ1 = 2.0 (d).
For a fixed genotypic RR and population allele frequency, the power to detect a deviation from HWE is always greater in the proband data compared to the power to detect a deviation from HWE in either the parental or unaffected sibling genotype data with the exception of the multiplicative model, where for the proband genotype data there is no deviation for HWE and the power to detect a deviation from HWE is α. For example for γ1 = 1.5 the maximum power to reject the null hypothesis of HWE is 0.97 for both the dominant and the recessive genetic model in the proband genotype data compared to 4.67 × 10−5 (dominant model) and 5.22 × 10−6 (recessive model) in the parental genotype data and 1.41 × 10−4 (dominant model) and 1.51 × 10−4 (recessive model) in the unaffected sibling genotype data (fig. 3; table 1). When the genotypic RR is increased to 2.0, the maximum power to reject HWE for both the dominant and recessive models is close to 1 for the proband genotype data (fig. 3; table 1). It should be noted that while the comparison is made for equal number of probands and unaffected siblings, since each proband has two parents the evaluation is made for 5,000 probands vs. 10,000 parents. For the additive model although the power to reject the null hypothesis of HWE is greater in the genotype data from probands compared to both the parental and unaffected sibling genotype data the disparity is not as great as observed for the dominant and recessive models. For example, the maximum power to reject HWE in the genotype data from probands is 5.86 × 10−4 for γ1 = 1.5 and 0.4 for γ1 = 2.0 for the additive model, while for the parental genotype data the maximum power of rejecting HWE is 2.32 × 10−6 for γ1 = 1.5 and 2.61 × 10−4 for γ1 = 2.0, and for unaffected sibling data the corresponding power is 7.21 × 10−6 for γ1 = 1.5 and 2.35 × 10−5 for γ1 = 2.0 (table 1).
Genotype Data in Unrelated Unaffected and Cohort Controls
The magnitude of HWD coefficients in the unrelated unaffected individuals are marginally greater than zero and the power to detect deviations from HWE is extremely low. For example, for genotype data from 5,000 unrelated unaffected controls the maximum HWD coefficient is −0.00016 for the multiplicative model at γ1 = 1.5 and the corresponding power is 1.03 × 10−7. When genotypic RR is increased to γ1 = 2.0, the maximum HWD coefficient and power to detect a deviation from HWE increases to −0.00065 and 1.54 × 10−7 respectively for the multiplicative model. For other genetic models the HWD coefficients and power to detect a deviation from HWE are even smaller (data not shown).
Genotyping Errors
Random error model always decreases the magnitude of the HWD coefficients; however, the reduction is not dramatic (suppl. fig. 2; suppl. table 3). For example, in parental genotype data the largest effect is observed for the multiplicative model where the maximum HWD coefficient is reduced from −0.00255 to −0.00245 (suppl. table 3). For the unaffected sibling genotype data the largest effect is seen for the recessive model where the maximum HWD coefficient is reduced to 0.00579 from 0.00603 (suppl. table 3). For other genetic models the reduction in the HWD coefficient is even more marginal (suppl. table 3). Correspondingly the power of rejecting HWE is also reduced for all genetic models in both parental and unaffected sibling genotype data (suppl. table 4).
The error model which converts homozygote to heterozygote genotypes causes an excess of heterozygotes which pulls the HWD coefficient in a negative direction. For example, the HWD coefficient became more negative and changed from −0.00184 to −0.00431 for the additive model in parental genotype data (suppl. fig. 2; suppl. table 3) and has similar effects for other genetic models (suppl. table 3). This type of genotyping error exacerbates HWD and increases the power of rejecting HWE in parental genotype data (suppl. table 4). On the other hand, the effects of this error model on the genotype data in unaffected siblings is dependent on the genetic model; for the dominant and additive model the HWD coefficient is more negative, for the recessive model the HWD coefficient is less positive and for the multiplicative model the HWD coefficient can either decrease or increase depending upon its original value (suppl. table 3). Genotyping error which converts heterozygote to homozygote genotypes creates an excess of homozygote genotypes and pushes the deviation from HWE in a positive direction. The effects of this error model on both parental and unaffected sibling genotype data are in the opposite direction compared to the homozygote to heterozygote genotyping error model (suppl. fig. 2; suppl. table 3, 4).
Population Substructure
For all genetic models in both parental and unaffected sibling genotype data population substructure pushes the HWD coefficients in the positive direction. When the allele frequency ratio between the two populations is 0.9 the effect is not dramatic (suppl. fig. 3; suppl. table 5, 6). However, when the ratio is decreased to 0.8, the substructure effect subjugated the genetic effect and the maximum deviation for the HWD coefficient went from negative to positive values, with the exception of the recessive model for the unaffected sibling genotype data where the HWD coefficient was already positive (suppl. fig. 3; suppl. table 5, 6). For example, for a dominant model in the parental genotype data the maximum deviation from HWE changed from −0.00086 to 0.00535 for an allele frequency ratio of 0.8 and the maximum power to detect a deviation from HWE increased from 4.67 × 10−7 to 0.93 (suppl. table 5, 6). For allele frequency ratio of 0.6 and allele frequencies >0.8 in population 2, the population substructure effect is so large that the power is close to 1 to detect a deviation from HWE (suppl. table 6). The HWD coefficients and the maximum power of rejecting HWE for other genetic models in both parental and unaffected sibling genotype data in the presence of population substructure are shown in supplemental tables 5 and 6.
Discussion
The family-based association study design is popular, since it allows for control of population admixture/substructure by using the non-transmitted parental alleles as control alleles. For family-based studies, genotype data from unrelated individuals are usually unavailable to evaluate deviations from HWE. Therefore it is common practice to carry out genotype quality control by testing for deviations from HWE using the parental or unaffected sibling genotype data. When trio data are used in association studies, parents which are included in the analysis are not phenotyped and can be either unaffected or affected for the trait understudy. Even when parents of affected probands are truly unaffected they have a higher probability than the general population of being susceptibility loci carriers. For fixed trait prevalence this probability increases with increasing genotypic RR. For family-based studies unaffected siblings are especially useful when parental data are missing. For case-control studies unaffected siblings are not commonly used as controls due to the reduction in power compared to when unrelated controls are analyzed. An exception is in the study of dizygote twins, where the unaffected co-twin is employed as a control. The advantage of this design is that the cases and controls are matched on environmental factors, since co-twins share many environmental and intrauterine exposures.
For most current genome-wide association studies, a large sample and a small α value are used (i.e. ≤1 × 10−7) to have adequate power to detect associations and guard against false positive results due to multiple testing. However even for studies with thousands of study subjects for low genotypic RR (≤1.2) these studies are often underpowered for genome-wide significance levels. In this study we used a small α value i.e. 1 × 10−7 and a large sample size i.e. 5,000 trios. This sample size was selected for sufficient power to detect an association for a large variety of genotypic RRs and allele frequencies.
Although testing for deviations from HWE in genotype data from controls or unaffected family members is often used as quality control to detect markers with genotyping error, deviation from HWE can be also be caused by other factors. In this study it is demonstrated that family ascertainment can also cause deviations from HWE in the genotype data of parents and unaffected siblings at the disease/trait susceptibility locus. Two measures are calculated: the HWD coefficientand the power to reject the null hypothesis of HWE. It is shown that detection of deviation from HWE due to a true association is negligible for a sample of 5,000 trios at α level of 1 × 10−7. The power will vary depending on sample size and α levels for a specific HWD coefficient and allele frequency. The genotypic RR also plays an important role in the strength of deviation from HWE, with higher genotypic RRs causing larger deviations of the HWD coefficient from 0. For the parental genotype data for 5,000 trios, under a multiplicative model for an allele frequency of 0.45 and a genotypic RR of 1.5, the power is 8.3 × 10−6 for an α level of 1 × 10−7 and increases to 0.175 for an α level of 0.05. Likewise it can be seen that the sample size has an effect on power for the same example using an α level of 1 × 10−7; the power is 5.6 × 10−7 for 1,000 trios and increases to 5.15 × 10−5 for 10,000 trios.
The phenomenon of deviations from HWE at the functional locus does not only occur because of ascertainment through families. When individuals are excluded from the control group due to having the phenotype understudy, deviations from HWE are also observed in the control genotype data at the disease/trait susceptibility locus. In this situation the HWD coefficient is negative for all genetic models except the dominant model for which the HWD coefficient is positive. Although the largest deviation from HWD is observed for the multiplicative model the magnitude of deviation is only marginally greater than 0. For a fixed genotypic RR the HWD disequilibrium coefficient increases with increasing disease prevalence. If the controls are collected from the general population without any exclusion criteria and the laws of HWE are not violated, no deviation from HWE will be observed in the genotype data.
The HWD coefficient reflects the difference between observed homozygote frequency and the corresponding expected frequency under HWE. Negative values indicate an excess of heterozygous genotypes and a deficiency of homozygous genotypes while positive HWD coefficients indicate the opposite. Negative HWD coefficients are indicative of gentoyping error under a random error model and when homozygous genotypes are incorrectly called as heterozygous genotypes [17]. Under all genetic models considered, HWD coefficients are negative for the parental genotype data (fig. 1a, b). For the additive and dominant model, HWD coefficients are also negative in the unaffected sibling genotype data (fig. 2a, b). Therefore, it is important not to make the assumption that negative HWD coefficients indicate genotyping errors when observed in parental and unaffected sibling genotype data.
The deviation from HWE caused by a true association can be further compounded by genotyping error and population substructure. The influence of genotyping error on the HWD coefficients is dependent on the error model. Genotyping error can create either an excess of homozygote or heterozygote genotypes depending on the underlying genotyping error model [17]. Genotyping error usually does not have a large effect on HWD coefficients unless the genotyping error rate is high. Genotyping error at the disease/trait susceptibility locus in the parental and unaffected sibling genotype data can either attenuate or amplify the deviation of HWD coefficient from 0; in turn this will affect the power to detect a deviation from HWE. The absolute power will be dependent on genetic model, genotypic RR, type of genotyping error, frequency of genotyping error, allele frequency, sample size and α value. Population substructure always creates an excess of homozygotes when the subpopulations have different allele frequencies. When the allele frequency difference is large in the two populations the deviation from HWE can be dominated by population substructure and the HWD coefficients shift from negative to positive or become more positive.
All calculations are based upon pedigrees with one affected proband. If calculations were carried out for kindreds with multiple offspring, the genotype probabilities for the 9 parental mating types would be modified. With increasing number of affected offspring the probability would increase that the parents are susceptibility allele carriers, since the probability that affected offspring are phenocopies is reduced with increasing number of affected offspring. For the unaffected siblings calculations are carried out conditional on their parents having one affected offspring. Based upon the probability of each possible mating type, the probability for all three possible genotypes is then calculated conditional on the offspring being unaffected.
The similarity between probands and unaffected siblings is due to low penetrance of susceptibility loci since unaffected siblings and probands can share a large proportion of high risk genotypes. When the penetrances were raised to high values, the patterns of HWD in unaffected siblings showed dramatic differences from probands (data not shown) since the probability that the unaffected sibling is a susceptibility allele carrier is greatly diminished.
For both the deviation from HWE and the power of rejecting the null hypothesis of HWE the results are shown for population allele frequencies which range from 0.05 to 0.95. Although it is unlikely that a disease susceptibility locus will have high allele frequencies (e.g. ≥0.5) it is not unlikely to observe such high allele frequencies for variants which are involved in human variation.
Unless genotype data for probands, parents or unaffected siblings are genotyped in different batches it is expected that the type of genotyping error and error rates should be consistent. Therefore, potentially different patterns of deviations from HWE in proband data compared to patterns observed in parental or unaffected sibling genotype data could be an indication that the deviation is due to an association and not genotyping errors. It can be observed that for the recessive and multiplicative model the pattern of deviation from HWE is different in the parental genotype data compared to proband genotype data. For the proband genotype data there is no deviation from HWE for the multiplicative model, and the deviation from HWE is positive for the recessive model, while for the parental genotype data the deviation from HWE is negative for both the multiplicative and recessive model. However, even though for the parental genotype data the HWD coefficients are negative the divergence from 0 is not large, especially under the recessive model. For unaffected sibling genotype data the strength of deviation from HWE is less than for the proband genotype data for the same genetic model and there is no difference in the direction, with the exception of the multiplicative model where D = 0 for the proband genotype data. In most circumstances differences in the deviation in HWE in the genotype data between the proband and either parental or unaffected sibling genotype data are difficult to distinguish from random variability.
In family-based studies, erroneous genotypes could bias linkage or association study. Mendelian inconsistency is usually used to detect errors in family-based studies. Errors which include wrong pedigree structure and sample mix-ups will usually cause a large portion of markers to display Mendelian inconsistency and therefore are easily detected. However, genotyping errors which often dependent on genotyping methods are more difficult to detect, since genotyping errors are often compatible with Mendelian inheritance. Undetected genotype errors can increase type I and II errors. Detection of genotyping error via deviation from HWE is often carried out in unrelated controls from case-control association study [16, 17]. However deviation from HWE is not necessarily caused by genotyping errors and may be due to chance, population admixture/stratification, inbreeding, selection or copy number variants [18,19,20,21,22]. In this article it is demonstrated that in genotype data obtained from parents and unaffected siblings of probands the deviation from HWE at the trait locus could be due to probands’ ascertainment. However the deviations are not large and at a genome-wide association study α value the power of detecting deviations from HWE at the functional locus is low. Deviations from HWE in either parental or unaffected sibling genotyping data can be used to flag markers for potential genotyping error. For these markers, cluster quality score should be examined for potential problems. Information on duplicate samples and Mendelian inconsistencies may give further evidence of genotyping error. Genotypes can also be confirmed by obtaining genotyping results from another platform. Additionally, markers with high rates of missing genotype data (e.g. >0.05) may also be indicative of problems with genotyping error.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgements
The work was funded by NIH grant R01-DC03594.
Appendix
The following procedure calculates genotype frequencies at a SNP marker given genotype frequencies at the functional locus in a specific sample and the LD between them.
Let p1 and q1 = 1 – p1 denote the frequencies of allele A1 and A2 at the functional locus and p2 and q2 = 1 – p2 denote the frequencies of allele M1 and M2 at a SNP marker locus which is in LD with the functional locus. Let h11, h12, h21 and h22 denote the four haplotypes at the two markers. Define LD between the two loci as
where it is assumed that A1 and M1 at the two loci are positively associated. Then the frequencies of the four haplotypes are
Assuming the population is under HWE, the joint distribution of the frequency of the 9 two-locus genotypes GijMkl is
where i, j, {1,2}, i ≤ j and k, l, {1,2}, k ≤ l. The marginal distribution of genotype Gij at the functional locus is given by PGij = Σk,lPGijMkl. Then given genotype frequencies Ps11, Ps12 and Ps22 at the functional locus in a specific sample, the genotype frequencies at the marker locus are
Another commonly used measure of LD for association studies is r2 which is defined as
If an r2 value instead of a δ value is given, the above calculation can be proceeded by replacing δ with r SPACEŽp1q1p2q2.
References
- 1.Boss I. Misclassification in 2 × 2 tables. Biometrics. 1954;10:487. [Google Scholar]
- 2.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: Application to single nucleotide polymorphisms. Hum Hered. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 3.Gordon D, Heath SC, Liu X, Ott J. A transmission/disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am J Hum Genet. 2001;69:371–380. doi: 10.1086/321981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon D, Levenstien MA, Finch SJ, Ott J. Errors and linkage disequilibrium interact multiplicatively when computing sample sizes for genetic case-control association studies. Pac Symp Biocomput. 2003:490–501. doi: 10.1142/9789812776303_0046. [DOI] [PubMed] [Google Scholar]
- 5.Gordon D, Heath SC, Ott J. True pedigree errors more frequent than apparent errors for single nucleotide polymorphisms. Hum Hered. 1999;49:65–70. doi: 10.1159/000022846. [DOI] [PubMed] [Google Scholar]
- 6.Douglas JA, Skol AD, Boehnke M. Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data. Am J Hum Genet. 2002;70:487–495. doi: 10.1086/338919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geller F, Ziegler A. Detection rates for genotyping errors in snps using the trio design. Hum Hered. 2002;54:111–117. doi: 10.1159/000068836. [DOI] [PubMed] [Google Scholar]
- 8.Douglas JA, Boehnke M, Lange K. A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet. 2000;66:1287–1297. doi: 10.1086/302861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon D, Leal SM, Heath SC, Ott J. An analytic solution to single nucleotide polymorphism error-detection rates in nuclear families: Implications for study design. Pac Symp Biocomput. 2000:663–674. doi: 10.1142/9789814447331_0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzustowicz LM, Merette C, Xie X, Townsend L, Gilliam TC, Ott J. Molecular and statistical approaches to the detection and correction of errors in genotype databases. Am J Hum Genet. 1993;53:1137–1145. [PMC free article] [PubMed] [Google Scholar]
- 11.Ehm MG, Kimmel M, Cottingham RW., Jr Error detection for genetic data, using likelihood methods. Am J Hum Genet. 1996;58:225–234. [PMC free article] [PubMed] [Google Scholar]
- 12.Lincoln SE, Lander ES. Systematic detection of errors in genetic linkage data. Genomics. 1992;14:604–610. doi: 10.1016/s0888-7543(05)80158-2. [DOI] [PubMed] [Google Scholar]
- 13.Stringham HM, Boehnke M. Identifying marker typing incompatibilities in linkage analysis. Am J Hum Genet. 1996;59:946–950. [PMC free article] [PubMed] [Google Scholar]
- 14.Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, Bansal A, Riley J, Purvis I, Xu CF. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet. 2004;12:395–399. doi: 10.1038/sj.ejhg.5201164. [DOI] [PubMed] [Google Scholar]
- 15.Tiret L, Cambien F. Departure from Hardy-Weinberg equilibrium should be systematically tested in studies of association between genetic markers and disease. Circulation. 1995;92:3364–3365. [PubMed] [Google Scholar]
- 16.Xu J, Turner A, Little J, Bleecker ER, Meyers DA. Positive results in association studies are associated with departure from Hardy-Weinberg equilibrium: Hint for genotyping error? Hum Genet. 2002;111:573–574. doi: 10.1007/s00439-002-0819-y. [DOI] [PubMed] [Google Scholar]
- 17.Leal SM. Detection of genotyping errors and pseudo-snps via deviations from Hardy-Weinberg equilibrium. Genet Epidemiol. 2005;29:204–214. doi: 10.1002/gepi.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cockerham CC. Group inbreeding and coancestry. Genetics. 1967;56:89–104. doi: 10.1093/genetics/56.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cockerham CC. Variance of gene frequencies. Evolution. 1969;23:72–78. doi: 10.1111/j.1558-5646.1969.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 20.Crow JKM. An Introduction to Population Genetics Theory. New York: Harper and Row; 1970. [Google Scholar]
- 21.Deng HW, Chen WM, Recker RR. Population admixture: Detection by Hardy-Weinberg test and its quantitative effects on linkage-disequilibrium methods for localizing genes underlying complex traits. Genetics. 2001;157:885–897. doi: 10.1093/genetics/157.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weir BS, Hill WG, Cardon LR. Allelic association patterns for a dense snp map. Genet Epidemiol. 2004;27:442–450. doi: 10.1002/gepi.20038. [DOI] [PubMed] [Google Scholar]
- 23.Dudbridge F, Gusnanto A: Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008, in press. [DOI] [PMC free article] [PubMed]
- 24.Agresti A. Categorical Data Analysis. Hoboken, New Jersey: John Wiley & Sons; 2002. [Google Scholar]
- 25.Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet. 1998;63:1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables