Abstract
The ability to smell is governed by the largest gene family in mammalian genomes, the olfactory receptor (OR) genes. Although these genes are well annotated in the finished human and mouse genomes, we still do not understand which receptors bind specific odorants or how they fully function. Previous comparative studies have been taxonomically limited and mostly focused on the percentage of OR pseudogenes within species. No study has investigated the adaptive changes of functional OR gene families across phylogenetically and ecologically diverse mammals. To determine the extent to which OR gene repertoires have been influenced by habitat, sensory specialization, and other ecological traits, to better understand the functional importance of specific OR gene families and thus the odorants they bind, we compared the functional OR gene repertoires from 50 mammalian genomes. We amplified more than 2000 OR genes in aquatic, semi-aquatic, and flying mammals and coupled these data with 48,000 OR genes from mostly terrestrial mammals, extracted from genomic projects. Phylogenomic, Bayesian assignment, and principle component analyses partitioned species by ecotype (aquatic, semi-aquatic, terrestrial, flying) rather than phylogenetic relatedness, and identified OR families important for each habitat. Functional OR gene repertoires were reduced independently in the multiple origins of aquatic mammals and were significantly divergent in bats. We reject recent neutralist views of olfactory subgenome evolution and correlate specific OR gene families with physiological requirements, a preliminary step toward unraveling the relationship between specific odors and respective OR gene families.
Living mammals (∼5400 species) originated ∼217–238 million years ago (Mya), inhabit every biome on earth, and are arguably one of the most phenotypically diverse groups of vertebrates (Wilson and Reeder 2005; Springer and Murphy 2007). From the largest, 170-ton blue whale, to the smallest, ∼2-g flying, echolocating bumblebee bat, the huge diversity and extraordinary adaptive radiations in mammalian form and function have fascinated evolutionary biologists for centuries (Darwin 1859). Increasingly, environmental niche specialization is reflected in animal genomes (Li et al. 2005; Seehausen et al. 2008; Zhao et al. 2009), and studying the molecular mechanisms that are responsible for this vast diversity has allowed some of the greatest insights into the functioning and evolution of our own genome.
Within mammals, olfaction is considered as one of the most valuable modes of sensory perception and provides the basis for the extraordinary sensitivity required to discriminate environmental and sexual cues. Accordingly, olfactory receptor (OR) genes form the largest gene superfamily (Buck and Axel 1991; Niimura and Nei 2007; Zarzo 2007; Keller and Vosshall 2008), accounting for ∼6% of the protein-coding genes in a typical mammalian genome (total OR genes/total number of protein-coding genes in dog ∼1100/19,300) (Lindblad-Toh et al. 2005). Each OR gene is ∼1 kb long, intronless, found in clusters on almost all chromosomes, and codes for a seven transmembrane G-coupled protein receptor that is uniquely expressed in individual olfactory sensory neurons, within the main olfactory epithelium (Buck and Axel 1991; Glusman et al. 2000b; Young and Trask 2002; Kambere and Lane 2007). Olfactory abilities vary among mammals, from vital importance in dogs and rodents, which use smell to navigate, forage, and communicate, to casual use in humans, which rely more on visual and auditory cues. This is reflected in the number of functional OR genes present in these species (functional/total number of OR genes: rat = 1259/1767, human = 388/802) (Nei et al. 2008). Not only is the number of genes considered to reflect the high level of evolutionary plasticity of the OR subgenome, but also the proportion of nonfunctional OR genes (OR pseudogenes: rat = 28%, human = 52%) (Nei et al. 2008). Indeed, the “vision-priority” hypothesis implies that the independent acquisition of trichromatic color vision in primates caused the parallel “death” of functional OR genes through relaxed purifying selection, evidenced by the higher number of OR pseudogenes in the genomes of primates with trichromatic vision compared with all other primates (Gilad et al. 2004; Nei et al. 2008), suggesting that trichromats have a higher reliance on vision than olfaction.
Previous comparative studies of OR diversity have focused on the pseudogene percentage within a species (Gilad et al. 2004; Niimura and Nei 2007; Kishida 2008) without paying particular attention to the actual repertoire of functional OR genes present in each species (Keller and Vosshall 2008). This has led to speculations that “genomic” drift and chance, as opposed to natural selection, play a major role in molding the complement of OR genes. In contrast, no studies have investigated the importance of “birth and death” of various OR gene families.
To investigate if the evolution of the OR gene repertoire has been influenced by habitat, sensory specialization, and other ecological traits; to elucidate if there is a signature of OR natural selection within mammals; and to identify which gene families are important in each ecological niche, we compared the functional OR subgenome repertoire across 50 phylogenetically and ecologically diverse mammals (Supplemental Table 1). We extracted OR gene sequence data from all of the 32 publicly available mammalian genome sequences present in the Ensembl database v52 (Supplemental Table 2) and combined these with PCR-based survey sequencing of the OR subgenome from 18 additional taxa (Supplemental Table 3). Since most genome sequence assemblies sample terrestrial taxa, our PCR-based survey sequencing focused on eco-specialist species that colonized the air and water during the past 65 million years: the bats and aquatic and semi-aquatic mammals (Supplemental Table 3). We uncover spectacular examples of OR gene losses in three independent lineages of aquatic and semi-aquatic mammals, yet convergent, selective retention of similar functional OR families. We show that the importance of olfactory receptor families—and thus the odors they bind—is directly associated with the habitat in which the animal exists. We reject recent neutralist views (Nei et al. 2008) of olfactory subgenome evolution and show that adaptive evolution plays a large role in determining the composition of the largest gene family in our genome. These results show how the mammalian olfactory system has adapted to different ecosystems long hypothesized but never shown before (Touhara and Vosshall 2009) and indicate that different OR gene families are important in different ecological niches.
Results and Discussion
Classification and annotation of OR gene families across mammals
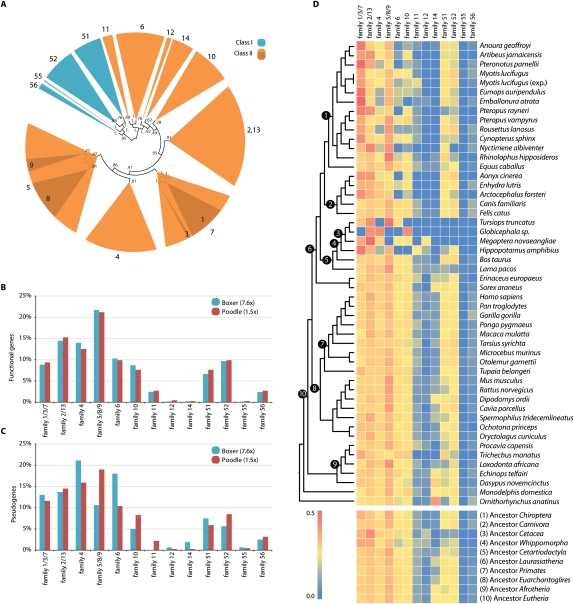
OR genes are traditionally classified into 17 families based on genetic similarity: four Class I, postulated to bind to water-borne molecules, and 14 Class II, hypothesized to bind mainly to airborne molecules (Glusman et al. 2000a; Zhang and Firestein 2002; Olender et al. 2004b; Warren et al. 2008). However, the function of these different families and the range of odorants that they can bind is poorly understood (Nei et al. 2008). Although single ORs can bind multiple odorants and odorants can bind to multiple receptors (Zarzo 2007), statistical analyses of odorant profile databases have classified odors into approximately 17–19 groups (Jeltema and Southwick 1986; Abe et al. 1990). It has been suggested that particular OR families may be important for particular dimensions in odor space (Zarzo 2007). Although simplistic, using this logic, the number of clades of OR genes and the number of genes found in each clade may broadly reflect environmental niche specialization, if this has, indeed, molded OR diversity. However, the monophyly of these 18 families has never been tested using a large sample of mammals. Therefore, we performed a Bayesian phylogenetic analysis of all functional OR genes for 50 mammals (∼24,000 genes). We recovered 13 main OR gene families (Fig. 1A; Supplemental Fig. 1).
Figure 1.
Phylogenomic partitioning of olfactory receptor genes. (A) Bayesian phylogeny of functional mammalian olfactory receptor (OR) genes available in Ensembl v52. OR gene family names are labeled on the termini of each subtree. MCMC posterior probabilities are given for each node. Polyphyletic families are clustered in subsequent analyses. (B) Ratio of functional OR genes per family (number of functional genes per family/total number of functional genes per species). The two C. familiaris (dog) genome assemblies are not significantly different (χ2 = 4.8, degrees of freedom [df] = 10, P ≈ 0.91). (C) Ratio of OR pseudogenes per family (number of pseudogenes per family/total number of pseudogenes per species). The two C. familiaris genome assemblies, boxer (7.6×) and poodle (1.5×), are significantly different (χ2 = 79.0, df = 8, P < 0.0001). (D) Heatmap displaying the relative percentage of functional genes in each OR gene family of different mammalian genomes, mapped onto the consensus phylogenetic tree for mammals. Selected ancestral reconstructions are shown at the bottom.
Markovian machine learning algorithms were used to classify the OR genes into their family groups using the nomenclature present in the HORDE database (Olender et al. 2004a). OR genes were further classified as functional, if they contained no stop codons and had an open reading frame of at least 650 bp, or as nonfunctional (pseudogene), if they contained stop codons and/or were shorter than 650 bp, and hence unable to code for seven potential transmembrane domains. Most traditional families were monophyletic (posterior probabilities [pp] = 0.62–1.00) (Fig. 1A). However, families 2 and 13 were phylogenetically indistinguishable (pp = 0.91) (Fig. 1A); family 7 was not monophyletic but contained the monophyletic families 1 and 3 (pp = 1) (Fig. 1A); family 5 was not monophyletic but contained the monophyletic families 8 and 9, albeit with weak support (pp = 0.47). This suggests that families (2, 13), (1, 3, 7) and (5, 8, 9) may respectively bind similar classes of odor molecules, and all other monophyletic families (56, 55, 52, 11, 6, 12, 14, 10, 4), may bind different classes of odor molecules. These results are consistent with analyses of whole-genome data from five vertebrate taxa (Warren et al. 2008).
Distribution of OR genes across divergent mammalian lineages
A comparison of the number and familial distribution of both functional and nonfunctional OR genes extracted from (1) low (∼2×) and (2) high (∼7×) coverage genome assemblies; and (3) laboratory-generated data, revealed no significant difference in the functional OR gene repertoire between low versus high coverage assemblies, or between laboratory-generated and low coverage genome data (Fig. 1B; Supplemental Fig. 2). However, there was a significant difference between the number of pseudogenes and their distribution between the low and high coverage genome assemblies (Fig. 1C; Supplemental Table 4), which can be attributed to reduced coverage and contiguity in low-coverage genome assemblies. Therefore, when comparing all three types of OR gene data, we only examined the distribution of functional genes.
An examination of the distribution of the familial percentage of OR genes across the mammalian phylogenetic tree revealed a striking difference in levels of variability present across lineages (Fig. 1D; Supplemental Tables 2, 3). Ancestral state reconstructions suggest that most Afrotherians, primates, and rodents appear to have maintained the ancestral mammalian distribution of functional OR genes despite the reported significant differences in the number of pseudogenes among these taxa (Fig. 1D; Nei et al. 2008). Within Laurasiatheria, however, there was much deviation from the eutherian ancestral composition (Fig. 1D, node 6), concentrated within bats, cetaceans, and the semi-aquatic carnivores. This is not an artifact derived from laboratory-generated data, as the whole-genome sequence data from the dolphin (Tursiops truncatus) and two bat species (Myotis lucifugus and Pteropus vampyrus) showed a similar pattern within the same superordinal group (Figs. 1D, 2).
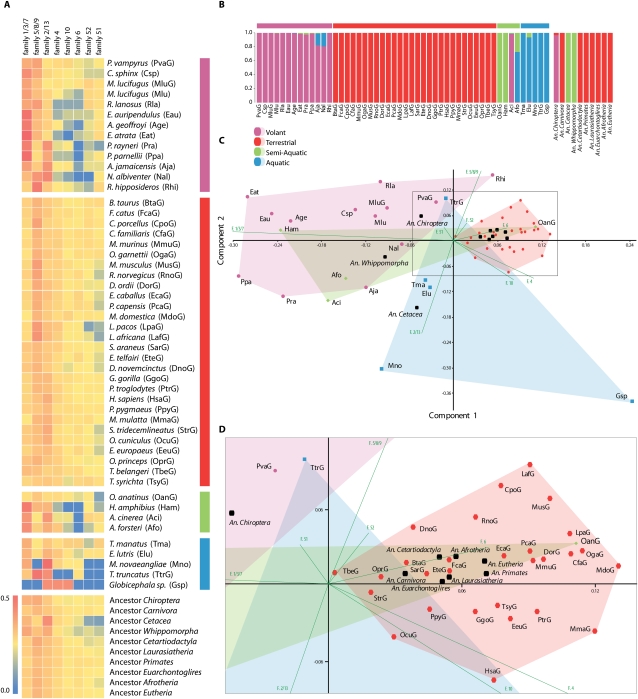
Figure 2.
Ecotype partitioning by olfactory receptor genes. (A) Heatmap of the informative OR gene families partitioned by ecological group (color bars on right follow legend). (B) Naïve Bayes pattern analysis of OR gene repertoires showing species assignment into ecological groups: (pink) volant, (red) terrestrial, (blue) aquatic, or (green) semi-aquatic based on the familial distribution of OR genes. Species abbreviations follow A (color bars on top follow legend). (C) Scatterplots showing the results of PCA analysis on functional OR genes within their respective OR gene families. The first and second axes explain 70% of the variance within the data set (ANOSIM: r = 0.6095, P < 0.001). Colored polygons highlight the different ecological groups of mammals. (Green lines) The contribution of particular families to the first two principal components. Species abbreviations, including ancestors, follow A. (D) Close-up view of the terrestrial polygon in C (boxed).
The distribution of the ratio of functional OR genes ranged from 0% to 45.7% across OR families within mammals. For the majority of mammals examined, families 11, 12, 14, 55, and 56 seem to play a minor role in olfaction, comprising just 0%–6% of each OR subgenome (Fig. 1D; Supplemental Tables 2, 3). Within monotremes there was a higher percentage of family 14 OR genes compared with therian mammals and our results confirm that there was no eutherian specific “OR37” OR genes present in the platypus genome (Hoppe et al. 2006). Class I OR families (51–56) are typically associated with the perception of water-borne odors and have diversified in fish and amphibian lineages compared with mammals (Freitag et al. 1995; Glusman et al. 2000a). As Class I OR genes have not undergone extensive expansion or contraction like the Class II OR genes in mammals, this indicates some ancestral “fish-like” waterborne odorant binding functions have been fixed in the mammalian genome (Kambere and Lane 2007). Intuitively, it might be expected that Class I OR genes should be greatly expanded in aquatic mammal lineages; however, this is not the case. Rather, cetaceans appear to have lost all functional Class I water-borne receptor families, with this reduction starting in the ancestor of hippopotamus and whales (Whippomorpha) (Fig. 1D, node 4; Waddell et al. 1999). This suggests that either (1) Class I ORs perform different functions in mammals versus fish or (2) another Class II OR family is capable of binding waterborne odors in cetaceans, such as family (2, 13), which contains a high percentage of cetacean OR genes; or (3) perhaps olfaction is not important for cetaceans and this is reflected in the small number of OR genes present within these genomes when compared to other mammals (only 26 OR genes were found in the dolphin) (Supplemental Table 2).
Association of OR gene families with divergent ecological niches
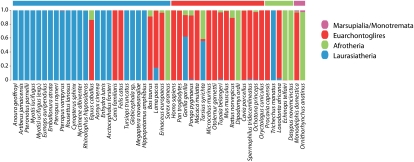
We used principal component analysis (PCA) and Bayesian assignment tests to visualize and identify significant differences in the functional OR gene repertoire between aquatic, semi-aquatic, terrestrial, and flying/volant mammals, and to distinguish which OR families, if any, were driving these differences (Fig. 2; Supplemental Table 5). Based on PCA, ANISOM, and naïve Bayes assignment tests, it was possible to correctly separate and assign most of the taxa into their respective ecogroups (Fig. 2; Supplemental Table 5). There was a significant difference, no overlap, and no misassignment between terrestrial and volant taxa. Similarly, there were significant differences between terrestrial and aquatic taxa, and terrestrial and semi-aquatic taxa (Fig. 2; Supplemental Table 5). However, using PCA analyses, it was not possible to distinguish between semi-aquatic and aquatic taxa, nor between volant and semi-aquatic taxa. Furthermore, Bayesian assignment tests revealed that more than half of the semi-aquatic taxa were incorrectly assigned (Fig. 2). By definition, semi-aquatic taxa inhabit both aquatic and terrestrial habitats for different proportions of their life histories, and the difficulty in differentiating them from fully aquatic and terrestrial ecogroups based on their functional OR repertoire reflects this. It was neither possible to distinguish, nor correctly assign most taxa, on the basis of phylogeny (Fig. 3).
Figure 3.
Naïve Bayes assignment test of OR gene repertoires showing species assignment into phylogenetic groups based on the familial distribution of OR genes. (Pink) Marsupialia/Monotremata, (red) Euarchontoglires, (green) Afrotheria, (blue) Laurasiatheria.
Eight OR gene families were responsible for most of the variance observed in the entire data set (Fig. 2). PCA analyses indicate that for bats, families (1, 3, 7) and (5, 8, 9) appear to be important. For aquatic mammals, family (2, 13) appears to play a large role in their differentiation, whereas families 6, 4, and 10 reflect the main diversity in OR genes present in terrestrial mammals. Interestingly Tupaia belangeri (tree-shrew), which is considered to have maintained the ancestral mammalian state (Emmons 2000), is placed at the center of the PCA analyses. All reconstructed ancestors for each major lineage were also correctly assigned to their hypothesized ecogroups (Fig. 2). As it is possible to assign taxa to their correct ecogroup based on their functional OR gene repertoire rather than phylogenetic relatedness, these results suggest that natural selection occurring through environmental niche specialization plays a large role in molding the OR gene repertoire in mammals, rather than shared evolutionary history and chance.
OR adaptation
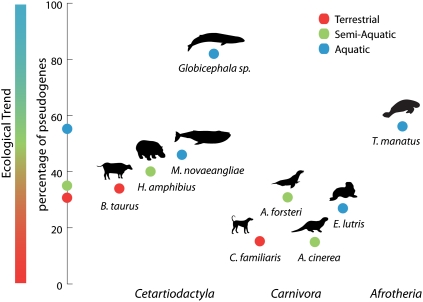
What are the selective forces that could have driven these changes? Comparison of the percentage of OR pseudogenes (using high-coverage genomic and laboratory generated data only) in terrestrial, semi-aquatic, and aquatic taxa in three orders—Carnivora, Cetartiodactyla, and Sirenia—revealed an independent gradual loss of functional OR genes (Fig. 4), consistent with the anatomical reduction and loss of the olfactory bulb in the transition to a fully aquatic lifestyle (McGowen et al. 2008). Within primates the loss of function of OR genes has been mainly attributed to a “trade-off” between vision and olfaction (Gilad et al. 2004). In the aquatic and semi-aquatic cetacean and pinniped lineages, this cannot be the case, as they are L cone visual-monochromats and have lost the function of their SWS opsin gene (Peichl 2005). Consequently, vision and olfaction appear not to provide the predominant sensory inputs; therefore, sound and taste most likely play a larger role in their sensory perception, as has been previously hypothesized (Macdonald 2006). However, these results suggest that the OR gene family 2/13 is highly important for survival in an aquatic environment as this gene family is found in the highest proportion (median, 33%) in aquatic mammals versus other eco-groups (volant, 13%; terrestrial, 17%; semi-aquatic, 21%). It appears that this family has selectively been retained in aquatic lineages because the loss appears nonrandomly across the OR gene families. This suggests that family 2/13 may be important for the perception of waterborne odors or, if olfaction is not important for aquatic mammals, that this OR gene family may have acquired a nonolfactory role.
Figure 4.
Ecological specialization influences OR gene repertoires. Illustrated scatter plot showing the global percentage of pseudogenes within Cetartiodactyla, Carnivora, and Sirenia. The median values of the percentage pseudogenes from high coverage genomic and laboratory generated data for terrestrial, aquatic, and semi-aquatic taxa are depicted along the y-axis. The genomic pseudogenes are considered at 650 bp, which is similar to the length of the PCR product that we amplify. Low-coverage assemblies were excluded.
Bats are highly derived mammals, uniquely capable of both flight and echolocation since the Eocene (Teeling et al. 2005). The majority of bats rely heavily on echolocation for sensory perception, and the acquisition of this unique sensory capability may explain their divergent OR repertoires when compared to other mammals. Despite this unique sensory capability, we rejected the “echolocation priority” hypothesis, which would support the loss of olfactory capabilities with the acquisition of echolocation, as there was no significant difference in the percentage of pseudogenes across bats regardless of echolocation capabilities (Supplemental Fig. 3).
Although most bat OR repertoires are derived from the ancestral mammalian state, Rhinolophus hipposideros (lesser horseshoe bat) appears to have maintained the ancestral mammalian OR composition (Fig. 1D, node 10) and also has the lowest percentage of nonfunctional OR genes (10%) when compared to other bats (10%–36%) (Supplemental Fig. 3). This result is surprising as Rhinolophus sp. exhibit the smallest olfactory bulb-to-forebrain ratio of all bat species examined (Neuweiler 2000). All Rhinolophus sp. are capable of a unique type of advanced Doppler-shift echolocation (Jones and Teeling 2006) but have drastically reduced visual capabilities (nonfunctional SWS1 opsin gene), compared to most other bat species (Zhao et al. 2009). Rhinolophus sp. do not possess a functional vomeronasal organ (Bhatnagar and Meisami 1998), and coupled with limited visual capacities, perhaps olfaction likely plays a larger role in intra-specific chemosensory communication within this family.
Although genomic drift may play a role in driving the diversity of the OR system, our findings indicate that the “volcanic” birth and death of OR gene families (Kambere and Lane 2007) are indeed driven by natural selection owing to spectacular niche-specific adaptation. Particular OR gene families are associated with different ecotypes; therefore, ongoing comparative investigations into the molecular motifs involved in odorant recognition should target these OR gene families. Finally, our results suggest that additional comparative and population genomic studies of OR genes in ecologically diverse taxa will more precisely determine the molecular mechanisms that drive the evolution of sensory perception.
Methods
Laboratory methods
OR gene sampling
OR genes from genomic DNA of 11 chiropterans, three carnivores, two cetaceans, an artiodactyl, and a sirenian (Supplemental Table 3), were amplified by PCR. GPC1 and GPC2 primers (modified from Gilad et al. 2004 by the addition of a guanine on the 5′-end) are described below. These PCR products were visualized on a 1% agarose gel using SYBR Safe DNA gel stain (Invitrogen Corporation). The 12 PCR products per primer pair, per species from the initial gradient PCR were pooled and concentrated using Millipore MultiScreen PCR plates then gel-purified using a Montage purification kit (Millipore) to remove any smaller fragments than the OR genes (∼700 bp). The pooled OR gene PCR products were cloned into Escherichia coli using a TOPO TA cloning kit to isolate the individual OR genes. GenBank accession numbers are detailed in Supplemental Table 3.
PCR screening
To faithfully represent the OR genome, we estimated that we needed to amplify ∼50% of potentially amplifiable (using our primers) OR genes present. This 50% threshold was validated by comparing laboratory-generated data with whole-genome sequence data for Myotis lucifugus (little brown bat) (Supplemental Fig. 2) and using the Gazey and Staley algorithm as described below to estimate how many OR genes were most likely amplifiable (using our primers) in each species (Supplemental Table 3). Initially, 192 colonies were picked, but if this did not yield enough genes, more colonies were sampled (Supplemental Table 6). These clones were PCR-screened using M13 vector primers and the PCR conditions described below. PCR products were visualized on a 1% agarose gel using SYBR Safe DNA gel stain. Clones containing an insert of correct length were then subjected to another round of PCR using M13 primers and Platinum grade Taq. PCR products were purified using 1 U of Exonuclease I and 1 U of shrimp alkaline phosphatase, and they were sequenced with M13 primers using Sanger sequencing methods in both forward and reverse directions.
Primers
GPC1F, 5′-GCTSCAYSARCCCATGTWYHWYTTBCT-3′; GPC1R, 3′-GGTYYYSAYDCHRTARAYRAYRGGGTT-3′; GPC2F, 5′-GYTNCAYWCHCCHATGTAYTTYTTBCT-3′;GPC2R, 3′-GTTYCTNARGSTRTAGATNANDGGRTT-3′; M13F, 5′-GTAAAACGACGGCCAG-3′;M13R, 5′-CAGGAAACAGCTATGAC-3′.
PCR conditions
The initial PCRs were performed in a total volume of 25 μL containing: 50 μmol/L deoxynucleotides, 0.4 μmol/L of each primer, 1.5 mmol/L MgCl2, 1× PCR buffer, 1 unit of Platinum Taq (Invitrogen Corporation), and 25 ng of DNA. Conditions for initial PCR were modified from Gilad et al. (2004). A first step of denaturation for 10 min at 94°C followed by 35 cycles of denaturation for 15 sec, annealing for 30 sec at a temperature gradient of 38°C to 50°C, and an extension for 1 min at 72°C. The final step was an extension for 10 min at 72°C.
The screening colony PCRs were performed in a total volume of 11 μL containing 80 μmol/L deoxynucleotides, 0.2 μmol/L of each primer, 1 mmol/L MgCl2, 1× Green GoTaq Flexi Buffer, 1 unit of GoTaq DNA polymerase (Promega Corporation), and 2 μL of bacteria (a picked colony, twirled in 10 μL of water). Conditions for the clone screening PCR were as follows: A first step of denaturation for 10 min at 95°C followed by 30 cycles of denaturation for 15 sec at 95°C, annealing for 30 sec at 58°C, and extension for 1 min at 72°C.
A third PCR was carried out using high-grade Taq of screened colonies that contained inserts. These were performed in a total volume of 25 μL containing 50 μmol/L deoxynucleotides, 0.4 μmol/L of each primer, 1.5 mM MgCl2, 1× PCR buffer, 1 unit of Platinum Taq (Invitrogen Corporation), and 25 ng of DNA. Conditions for this PCR were as follows: A first step of denaturation for 10 min at 95°C followed by 30 cycles of denaturation for 15 sec at 95°C, annealing for 30 sec at 58°C, and extension for 1 min at 72°C.
Plasmid extraction
Clones that contained inserts of ∼700 bp were then grown on Luria-Bertani agar with 0.02 g/L kanamycin. Plasmid DNA was purified using a Millipore Plasmid Miniprep Kit. Plasmids were sequenced using both forward and reverse M13 primers.
Sequence analysis
Between 192 and 667 colonies were picked for each species (see Gazey and Staley algorithm below), and between 27 and 288 OR genes were sequenced from each study species (see Supplemental Table 6 for numbers of sequences and genes). The nucleotide sequences of the OR genes were examined using Sequencher v4.7 (Gene Codes Corporation). Forward and reverse sequences were aligned and checked for ambiguities. Assembly of the consensus sequences was carried out at a 98% similarity level to allow for Taq-generated mutations that may have been sequenced in individual clones. Each consensus sequence was counted as one gene.
Statistical validation
Gazey and Staley algorithm
The Gazey and Staley Bayesian algorithm (Gazey and Staley 1986) was used in R (Ihaka and Gentleman 1996) to estimate how many OR genes could possibly be amplified using the GPC1 and GPC2 primers (described above) for each species. This algorithm is typically used to estimate the size of animal populations by analyzing “capture-mark-re-capture” data from field studies. The algorithm assumes that the population is closed and that its size will not change between captures and that there is no bias in the capture method. Subsequently, as the number of captured and marked individuals caught reaches the maximum population size, the number of new individuals captured will significantly decrease per capture session. Using this method, we examined what percentage of the probable amount of genes amplifiable with our primers had been sequenced per species given our sampling effort (i.e., number of sequences). If we generated 500 DNA sequences and this resulted in only 10 OR genes, we would have almost sequenced all genes in the OR repertoire. However, if having generated 500 DNA sequences we obtained 475 OR genes, therefore, we have only “captured” a smaller proportion of the OR repertoire with our PCR primers and total clones sequenced (Supplemental Tables 3, 6). For example, we generated 260 DNA sequences for Emballonura atrata (Peters's sheath-tailed bat), and this resulted in 137 OR genes (Supplemental Table 6). Using the Gazey and Staley Bayesian algorithm, we estimated that we could amplify 180 genes with our primers; therefore, we have amplified 76% of the amplifiable OR repertoire (Supplemental Table 3).
The total number of OR genes per species was considered as the maximum number of individuals contained in a closed population. The number of DNA sequences generated per species was considered as the number of individuals caught, and the number of genes obtained from those sequences was considered as the number of individuals “marked and re-captured.” We tested that there was no bias of “capture method,” which in our case was primer amplification bias, using a homogeneity test as described below (Supplemental Fig. 5). As the population was closed and there was no bias in “capture method,” we were able to use this method. These data described above were input into the algorithm, and a posterior probability distribution of the total number of OR genes (i.e., maximum population size) was generated. A 95% probability for the maximum, minimum, and mode of the number of OR genes potentially amplifiable using our primer pairs was obtained. We compared the number of genes that we had amplified with this probable distribution of total gene number to assess what percentage of the OR genes had been amplified given our sequencing efforts per species (Supplemental Fig. 4).
Homogeneity test
A homogeneity test developed by Puechmaille and Petit (2007) was used in R (Ihaka and Gentleman 1996) to investigate if the primer sets preferentially amplified any gene. This test simulated the sampling of OR genes by assuming a homogeneous capture probability (amplification per primer set) and compared the expected (obtained from Gazey and Staley test above) with the observed captures per individual (gene). This algorithm took into account n, the number of DNA sequences amplified, N, the expected number of genes that could be amplified with our primers and the number of genes that were an aggregate of one DNA sequence only, two DNA sequences, three DNA sequences, and so forth (for details, see Puechmaille and Petit 2007). Simulations were conducted using R (Ihaka and Gentleman 1996). The sampling simulation was repeated 1000 times to compute the average boundaries of the 95% CI of the number of DNA sequences per gene. These data were not considered biased if the frequency of observed values lay inside the simulated 95% CI (Supplemental Fig. 5).
In silico methods
OR family assignment and classification of retrieved and amplified sequences
A unique probabilistic genomic search algorithm was developed to identify the OR genes, assign their class and family, and distinguish between functional and nonfunctional genes. The olfactory receptor family assigner (ORA) performs HMMER (Eddy 1998) searches, using profile hidden Markov models (profile HMM) established on alignments of OR gene family sequences. Profile HMM was used to perform a sensitive database search using statistical descriptions of a gene family's consensus sequence. Non-OR genes were eliminated, and the remaining sequences were classified based on their similarity to a profile OR gene family HMM. The sequence was then translated and the functionality checked (ORA Perl code is available from the Comprehensive Perl Archive Network, CPAN).
To design the profile HMM for each gene family, a collection of protein sequences from HORDE OLFR database #42 (Safran et al. 2003) was used as the training set. The Homo sapiens (human), Pan troglodytes (chimpanzee), Canis familiaris (dog), Monodelphis domestica (gray short-tailed opossum), and Ornithorhynchus anatinus (platypus) complete sets of annotated OR genes were extracted and aligned using ClustalW v2.0 (Larkin et al. 2007), and one profile HMM was designed for each of the original 17 OR gene families. These species acted as the training set. The relative weight of each profile was adjusted to allow 97% of the training set to be reassigned correctly. Only 3% were misassigned, and when they were manually checked, all corresponded to sequence fragments of <150 nt, annotated as OR genes in HORDE. As the accuracy of such short fragments was dubious, we had confidence in our assignment.
To evaluate ORA's prediction sensitivity, sequences of completed human, chimpanzee, dog, opossum, and platypus genomes were downloaded from the Ensembl v52. ORA detected all the annotated OR genes consistently. To evaluate ORA's prediction selectivity, a random sequence database (totaling 21 Gb) was generated by a fifth-order Markov chain based on 6-mer frequencies of each genomic sequence. ORA did not detect any OR gene sequence in this database. We also ensured that ORA's predicted numbers of genes and pseudogenes were comparable with the recently published annotations (Niimura and Nei 2007). Only data for Mus musculus (mouse) were not comparable; in this case, a newer genome assembly was used by ORA, considerably reducing the number of false pseudogenes (Supplemental Table 2). To generate comparable data sets between the genome sequence-derived data sets and the laboratory-generated OR fragments (700 to 750 bp), we fixed a threshold of 650 bp, which corresponds to the seven transmembrane domains, to be the minimal size of an open reading frame to be considered as functional (which would underestimate the actual number of pseudogenes) (see Supplemental Table 2).
Phylogenetic analyses of the OR gene families
The amino acid sequences of all functional OR genes assigned from the genome sequence assemblies (32 species) were collected into one file (family 1: 1419 genes; family 2: 2839; family 3: 115; family 4: 2750; family 5: 2916; family 6: 1878; family 7: 1377; family 8: 1662; family 9: 547; family 10: 1794; family 11: 433; family 12: 147; family 13: 874; family 14: 772; family 51: 1231; family 52: 1556; family 55: 38; family 56: 232. Total number of sequences: 22,580). Class I and class II genes were aligned separately using GramAlign v1.17 (Russell et al. 2008). The resulting alignments were analyzed using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003) under the relaxed model of sequence evolution for 10,000,000 cycles, with a tree sampled every 1000 generations once the Markov chain reached stationary (determined by empirical checking of likelihood values). The consensus tree of the stable models (GTR + Γ) was then established. For visualization purposes, nodes were collapsed when they supported the same OR family members. To further reduce the total number of visualized branches, we allowed a 1% mis-assignment threshold when collapsing the branches (i.e., if there were 100 leaves and one did not belong to the same OR family, they were still collapsed and treated as the same OR family). In the final tree we merged the separate class I and II subtrees in the same graphic, with polyphyletic families (i.e., families 2, 5, 7, and 13) clustered together (Fig. 1). These analyses were also repeated by including the shorter sequence laboratory generated data (∼700 bp) included (family 1: 1594 genes; family 2: 3027; family 3: 134; family 4: 2848; family 5: 3128; family 6: 1912; family 7: 1668; family 8: 1705; family 9: 572; family 10: 1863; family 11: 459; family 12: 150; family 13: 917; family 14: 774; family 51: 1349; family 52: 1646; family 55: 38; family 56: 247. Total number of sequences: 24,085). The nodes recovered were the same, but the posterior probabilities were slightly lower (Supplemental Fig. 1).
Comparison of the wet-laboratory data generated with the publicly available whole-genome data at both low (∼2×) and high (∼7×) coverage
To ascertain if there was any significant difference between our data sets (experimentally sampled OR genes vs. low-coverage whole-genome sequences and high-coverage whole-genome sequences), the distribution of OR genes into gene families was compared (1) between the M. lucifugus laboratory generated OR gene distribution data versus the 2× coverage M. lucifugus genomic sequences and (2) the 1.5× genomic poodle dog sequence versus the 7.6× coverage boxer dog genome sequence. A χ2 test was carried out to ascertain if there was any difference in the distribution of genes into OR gene families between the different data sets for all genes and both functional and nonfunctional genes independently in the following manner: (1) We pooled the families with small effective values. (2) We established the “Expected” number of genes per family as the ratio of the number of genes found in a particular OR gene family obtained from the genomic sequence divided by the total number of OR genes found in the genomic sequence multiplied by the total number of genes found experimentally (e.g., M. lucifugus, functional family 1/3/7: (105/381) × 131 = 36.102). (3) We ran a χ2 test between the “Expected” values and the experimental PCR values.
There was no significant difference between low-coverage genomic data (∼2×) versus the laboratory-generated data (Supplemental Fig. 2), when the functional repertoire was compared as a whole. However, when individual families are tested alone, it appears that families 6 and 2/13 could be slightly biased in the laboratory-generated data. Despite this, PCA and naïve Bayes analyses yield very similar results even if each of these families are removed, so possible bias in these families does not affect the overall conclusions of this study (data not shown).
There was no significant difference between the distribution of the functional OR genes between low- (∼2×) versus high- (∼7×) coverage genomic sequence (Fig. 1B). However, there was a large and significant difference in both the number of pseudogenes and the percentage distribution of pseudogenes between low- versus high-coverage data (Fig. 1C). This indicates that it is not meaningful to compare low versus high genome coverage at the pseudogene level.
Aquatic versus terrestrial versus semi-aquatic versus volant mammals
Classification of taxa into ecogroup
Each species was assigned to an ecological group: terrestrial, aquatic, semi-aquatic, or volant (Supplemental Table 1). This classification was based on the amount of time each mammal spends in a particular habitat. For example, Aonyx cinerea (oriental small clawed otter) was classified semi-aquatic because it occupies an aquatic habitat for feeding but breeds and spends a considerable amount of time on land, whereas Enhydra lutris (sea otter), classified as aquatic, occupies an aquatic habitat for feeding, mating, and sleeping, and rarely ventures on to shore (Nowak 1999). Bats were considered different from other terrestrial mammals because of their unique ability for flight (volant).
Principal component analysis (PCA) and analysis of similarities (ANOSIM)
A PCA was performed on all functional genes using the program PAST v1.89 (Hammer et al. 2001) to explore the overall variation in the distribution of functional OR genes into OR gene families across mammals occupying different ecological niches as described above. The PCA algorithm used was the covariance matrix of the data. An analysis of similarities, ANOSIM (Clarke 1993), was then performed using the program PAST v1.89 (Hammer et al. 2001) to test the plausibility of the above groupings. The ANOSIM procedure is a nonparametric test based on the rank-ordering of the values of a distance matrix among all observations (in our case, the Euclidean distance among species) and the derivation of an R statistic, which expresses the difference between the mean rank of between-group (Rb) and within-group (Rw) distances. To test for the significance of positive values of R, the observed value is compared to the 95% confidence interval of a simulated distribution.
Bayesian assignment test
The proportion of OR genes in each OR gene family for each mammal examined along with ecological information for that mammal (e.g., volant, terrestrial, aquatic, and semi-aquatic) were entered into the WEKA package (Whitten and Frank 2005), which contains a variety of machine-learning algorithms, including naïve Bayes, a classifier with independent assumptions that requires a low amount of data for training to estimate parameters. Naïve Bayes is an assignment test that learns using the maximum, minimum, and mode of OR gene percentages for each OR gene family in a particular group (volant, terrestrial, aquatic, or semi-aquatic) within all species studied. Naïve Bayes then classifies each species into a particular group based on the levels of OR genes in each OR gene family. Permutatively, one species is removed from the training set, and subsequently its ecogroup is then assigned by the algorithm. The probability score for the assignment of each species into each of the ecogroups is obtained along with various error scores and a k statistic (a measure of how different our assignment is from random) for the overall assignment. The significance of the obtained k values was inferred by comparing them to the distributions of 10,000 simulated values from randomly assigned data sets (respecting the original distribution frequency). The k statistics for each random assignment were gathered and plotted in a graph to ensure that the k statistic for our assignment was distanced from the curve of random k statistics (Supplemental Fig. 6). When the taxa were assigned to their correct ecogroups, the error statistic was significantly better than random, giving us support for the presence of a unique eco-signature in the ORs. When the semi-aquatic ecogroup was excluded from the initial training set and then reassigned, they were classified as follows: A. cinerea (oriental small clawed otter) and Hippopotamus amphibius (hippopotamus) were classified as volant; Arctocephalus forsteri (New Zealand fur seal) was classified as aquatic; and O. anatinus (platypus) was classified as terrestrial. These results correspond with their positions in the PCA analyses (Fig. 2B). This was also repeated with phylogenetic groupings.
Ancestral state reconstruction
We used Mesquite v2.0 (Maddison and Maddison 2007) to reconstruct the ancestral-state OR repertoire for various key nodes using the consensus mammalian phylogenetic tree (Murphy et al. 2007). We used parsimony with the continuous character option to trace the ancestral states of each OR gene family distribution (e.g., Supplemental Fig. 7). The ancestral states of OR gene distribution were obtained for the major phylogenetic groupings within the data set, and also Whippomorpha (Waddell et al. 1999). These ancestral states were input into the PCA and naïve Bayes analysis to elucidate if these ancestors were classified within the ecological niches of the species that preceded them (Supplemental Fig. 2).
Acknowledgments
This study was supported by a Science Foundation Ireland PIYRA 06/YI3/B932 award to ECT and Embark IRCSET PhD scholarship awarded to ECT and SH. WJM is supported by NSF grant EF0629849. Computational resources were supplied by the Irish Centre for High-End Computing. We thank E. Baerwald for samples, A. Goodbla for technical assistance, and J. Yearsely for statistical advice.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. GQ215701–GQ217490.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099416.109.
References
- Abe H, Kanaya S, Komukai T, Takahashi Y, Sasaki S-I. Systemization of semantic descriptions of odors. Anal Chim Acta. 1990;239:73–85. [Google Scholar]
- Bhatnagar KP, Meisami E. Vomeronasal organ in bats and primates: Extremes of structural variability and its phylogenetic implications. Microsc Res Tech. 1998;43:465–475. doi: 10.1002/(SICI)1097-0029(19981215)43:6<465::AID-JEMT1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors—a molecular-basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analysis of changes in community structure. Austral Ecol. 1993;18:117–143. [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. J. Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Emmons LH. Tupai. A field study of Bornean treeshrews. University of California Press; Berkeley: 2000. [Google Scholar]
- Freitag J, Krieger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Gazey WJ, Staley MJ. Population estimation from markrecapture experiments using a sequential Bayes algorithm. Ecology. 1986;67:941–951. [Google Scholar]
- Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:120–125. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm Genome. 2000a;11:1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]
- Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics. 2000b;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST. Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electronica. 2001;4 http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- Hoppe R, Breer H, Strotmann J. Promoter motifs of olfactory receptor genes expressed in distinct topographic patterns. Genomics. 2006;87:711–723. doi: 10.1016/j.ygeno.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
- Jeltema MA, Southwick EW. Evaluations and application of odor profiling. J Sens Stud. 1986;1:123–136. [Google Scholar]
- Jones G, Teeling EC. The evolution of echolocation in bats. Trends Ecol Evol. 2006;21:149–156. doi: 10.1016/j.tree.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kambere MB, Lane RP. Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci. 2007;8:S2. doi: 10.1186/1471-2202-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Vosshall LB. Better smelling through genetics: Mammalian odor perception. Curr Opin Neurobiol. 2008;18:364–369. doi: 10.1016/j.conb.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T. Pattern of the divergence of olfactory receptor genes during tetrapod evolution. PLoS One. 2008;3:e2385. doi: 10.1371/journal.pone.0002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li X, Li W, Wang H, Cao J, Maehashi K, Huang L, Bachmanov AA, Reed DR, Legrand-Defretin V, Beauchamp GK, et al. Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Macdonald D. The encyclopedia of mammals. The Brown Reference Group; London: 2006. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis. Version 2.6. 2007. http://mesquiteproject.org.
- McGowen MR, Clark C, Gatesy J. The vestigial olfactory receptor subgenome of odontocete whales: Phylogenetic congruence between gene-tree reconciliation and supermatrix methods. Syst Biol. 2008;57:574–590. doi: 10.1080/10635150802304787. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Neuweiler G. Biology of bats. Oxford University Press; New York: 2000. [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak RM. Walker's mammals of the world. Johns Hopkins University Press; Baltimore, MD: 1999. [Google Scholar]
- Olender T, Feldmesser E, Atarot T, Eisenstein M, Lancet D. The olfactory receptor universe—from whole genome analysis to structure and evolution. Genet Mol Res. 2004a;3:545–553. [PubMed] [Google Scholar]
- Olender T, Fuchs T, Linhart C, Shamir R, Adams M, Kalush F, Khen M, Lancet D. The canine olfactory subgenome. Genomics. 2004b;83:361–372. doi: 10.1016/j.ygeno.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Peichl L. Diversity of mammalian photoreceptor properties: Adaptations to habitat and lifestyle? Anat Rec A Discov Mol Cell Evol Biol. 2005;287:1001–1012. doi: 10.1002/ar.a.20262. [DOI] [PubMed] [Google Scholar]
- Puechmaille SJ, Petit EJ. Empirical evaluation of non-invasive capture-mark-recapture estimation of population size based on a single sampling session. J Appl Ecol. 2007;44:843–852. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Russell DJ, Otu HH, Sayood K. Grammar-based distance in progressive multiple sequence alignment. BMC Bioinformatics. 2008;9:306. doi: 10.1186/1471-2105-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, Shmoish M, Peter Y, Glusman G, Feldmesser E, et al. Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HD, Miyagi R, van der Sluijs I, Schneider MV, Maan ME, Tachida H, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ. Mammalian evolution and biomedicine: New views from phylogeny. Biol Rev Camb Philos Soc. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- Waddell PJ, Okada N, Hasegawa M. Towards resolving the interordinal relationships of placental mammals. Syst Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten IH, Frank E. Data mining: Practical machine learning tools and techniques. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]
- Wilson DE, Reeder DM. Mammal species of the world: A taxonomic and geographic reference. The Johns Hopkins University Press; Baltimore: 2005. [Google Scholar]
- Young JM, Trask BJ. The sense of smell: Genomics of vertebrate odorant receptors. Hum Mol Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- Zarzo M. The sense of smell: Molecular basis of odorant recognition. Biol Rev Camb Philos Soc. 2007;82:455–479. doi: 10.1111/j.1469-185X.2007.00019.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S. The evolution of color vision in nocturnal mammals. Proc Natl Acad Sci. 2009;106:8980–8985. doi: 10.1073/pnas.0813201106. [DOI] [PMC free article] [PubMed] [Google Scholar]