Abstract
We report an evolutionary analysis of the V1R gene family across 37 mammalian genomes. V1Rs comprise one of three chemosensory receptor families expressed in the vomeronasal organ, and contribute to pheromone detection. We first demonstrate that Trace Archive data can be used effectively to determine V1R family sizes and to obtain sequences of most V1R family members. Analyses of V1R sequences from trace data and genome assemblies show that species-specific expansions previously observed in only eight species were prevalent throughout mammalian evolution, resulting in “semi-private” V1R repertoires for most mammals. The largest families are found in mouse and platypus, whose V1R repertoires have been published previously, followed by mouse lemur and rabbit (∼215 and ∼160 intact V1Rs, respectively). In contrast, two bat species and dolphin possess no functional V1Rs, only pseudogenes, and suffered inactivating mutations in the vomeronasal signal transduction gene Trpc2. We show that primate V1R decline happened prior to acquisition of trichromatic vision, earlier during evolution than was previously thought. We also show that it is extremely unlikely that decline of the dog V1R repertoire occurred in response to selective pressures imposed by humans during domestication. Functional repertoire sizes in each species correlate roughly with anatomical observations of vomeronasal organ size and quality; however, no single ecological correlate explains the very diverse fates of this gene family in different mammalian genomes. V1Rs provide one of the most extreme examples observed to date of massive gene duplication in some genomes, with loss of all functional genes in other species.
Pheromones are chemical signals used for intraspecies communication that affect many important mammalian behaviors, including aggression, maternal behavior, courtship, and mating (Wyatt 2003). In rodents, pheromone detection was previously thought to be accomplished solely through the vomeronasal organ (VNO), but it is now recognized that the main olfactory epithelium is also involved in pheromone sensing (Brennan and Zufall 2006). Neurons of the vomeronasal epithelium project to the accessory olfactory bulb (AOB), from which signals are passed on to various other areas of the brain, including some areas that seem to control innate and stereotyped behavioral and endocrine responses (Dulac and Axel 1995).
Several large families of 7-transmembrane G-protein–coupled receptors (GPCRs) are expressed in neurons of the vomeronasal and main olfactory epithelia and recognize odorant and pheromone ligands directly. These GPCR families include olfactory receptors (ORs) and trace-amine associated receptors (TAARs) in the main olfactory epithelium and three families expressed in the vomeronasal organ (V1Rs, V2Rs, and formyl peptide receptor-like proteins [FPRs]) (Dulac and Axel 1995; Mombaerts 2004; Liberles and Buck 2006; Rivière et al. 2009). Based on analysis of a limited number of genome assemblies, the size and composition of these gene families are known to differ between mammalian species (Ache and Young 2005; Grus et al. 2005; Young et al. 2005; Liberles and Buck 2006; Shi and Zhang 2007; Young and Trask 2007; Rivière et al. 2009), with the V1R and V2R families appearing especially variable. There are over 100 functional V1Rs in the rat and mouse genomes, but only five (or fewer) intact V1Rs in the human and chimpanzee genomes along with a large number of pseudogenes (Liman 2006). The platypus possesses the largest V1R repertoire of any mammal studied to date, with more than 270 intact V1Rs (Grus et al. 2007).
Here, we examine V1Rs in a much larger and more diverse set of 37 mammalian genomes to address several questions that were difficult to examine with smaller data sets. Is the variation in V1R family size seen in the eight previously analyzed species a prevalent feature across the mammalian tree? Are the large V1R repertoires seen in platypus, rat, and mouse exceptional among mammals, with most mammalian genomes encoding small functional V1R repertoires like primates and dog? Or do most mammals, like rodents, have large V1R repertoires? Can any correlations between V1R family size and ecological or behavioral observations be seen by studying a much larger range of mammalian species? And, with data from many more primate genomes, we could examine a previous hypothesis that functional decline of the V1R family in humans and other apes and Old World monkeys might reflect a decreased reliance on chemical sensing due to the acquisition of trichromatic visual abilities (Liman and Innan 2003; Zhang and Webb 2003). In order to address these questions, we took advantage of ongoing efforts to generate low-redundancy comparative sequence data on numerous mammalian genomes (Margulies et al. 2005). Such data, although not providing full coverage of the genomes being sequenced, allow both the estimation of the number of V1Rs possessed by various mammals and the generation of partial phylogenies of mammalian V1Rs. We show that the size of the V1R repertoire varies widely among mammals and that most mammals examined appear to have experienced species-specific duplications resulting in “semi-private” functional V1R repertoires.
We also generated additional sequence from V1R family members in wolves to examine the evolutionary timing of the decline of the dog V1R repertoire to only approximately nine apparently intact V1R genes and ∼60 pseudogenes (Grus et al. 2005; Young et al. 2005). Dog also has only pseudogenes in the V2R family (Shi and Zhang 2007; Young and Trask 2007) and possesses a relatively thin vomeronasal epithelium and relatively small AOB (Dennis et al. 2003). Vomeronasal decline is surprising given that dogs and wolves appear to exhibit pheromone-based behaviors (Anisko 1976; Goodwin et al. 1979), such as the well-known effect of a female in heat on males, and urine-marking and examination of urine marks. We and others considered the idea that selective pressures imposed by humans during dog domestication favoring more docile, human-centric animals might have led to a rapid, recent degeneration of the dog V1R repertoire to pseudogenes (Grus et al. 2005; Young et al. 2005). Here, we disprove that hypothesis by sequencing the least degenerated dog V1R pseudogenes (i.e., those most likely to be recently inactivated) from wolf DNA, finding that they are also pseudogenes in wolf. Our result implies that loss of functional V1R genes occurred before dogs were domesticated.
Results and Discussion
Trace Archive data provide good coverage of the V1R family
We have greatly extended previous studies on V1R repertoire sizes and evolution to a much more comprehensive set of mammalian species. Previous analyses examined V1Rs in a total of eight mammals using draft whole-genome assemblies (Grus et al. 2005, 2007; Young et al. 2005; Shi and Zhang 2007). Here, we add another five species for which relatively high-coverage assemblies are available (orangutan, macaque, marmoset, guinea pig, and horse) and take advantage of the rapidly increasing availability of large amounts of low-grade comparative genomic sequence (“Trace Archive” data) to add 24 more species to our study. First, we provide proof-of-principle evidence that reasonable estimates of V1R repertoire size and sequences of most V1R family members can be obtained from Trace Archive data as well as from whole-genome assemblies, and then discuss our findings.
Trace Archive data consist of millions of individual sequencing traces generated by subcloning random fragments of the entire genome of an organism and sequencing a large number of subclones. Although Trace Archive sequences are short (∼800 bp), contain errors, and do not provide complete genomic coverage, they still yield very useful genomic surveys (Margulies et al. 2005). We modified our series of V1R gene-identification scripts in order to work with Trace Archive data as well as with complete assemblies, incorporating a phredphrap assembly phase into our analysis. V1R genes comprise a single coding exon of ∼900 bp (Dulac and Axel 1995) and therefore are tractable for analysis using Trace Archive data, because assembly of a small number of overlapping reads is enough to cover an entire gene, unlike most intron-containing coding regions.
The V1R family contains many recently duplicated members whose high levels of sequence identity complicate sequence assemblies. Without extremely high sequence coverage and intensive manual finishing efforts such as those performed for the human and mouse genomes, close duplicates in the V1R and other gene families may never be satisfactorily resolved (see Supplemental Table 1). Such intensive finishing work is unlikely ever to happen for other mammalian genomes due to the time and expense involved. Therefore, the analyses we describe here, as well as those in previous publications on assemblies other than human and mouse, must be considered as approximate surveys rather than accurate estimates of gene numbers. A second important caveat to our findings is that, as with any bioinformatic study, we are inferring function based on sequence similarity without experimental evidence. Although it is quite likely that these V1R-like sequences function as receptors for external signals in the vomeronasal organ, it is possible that some of the sequences have taken on a novel function in a different organ in one or more species. For example, some human, goat, and mouse V1Rs have been suggested to function in the main olfactory epithelium rather than the vomeronasal organ (Rodriguez et al. 2000; Wakabayashi et al. 2002; Karunadasa et al. 2006).
In the following analysis, we classify the V1R sequences we find as being either “intact” (for which full-length protein-coding sequence was obtained that does not contain any frameshift or nonsense mutation) or “other,” a category that includes true pseudogene sequences as well as intact V1Rs with sequence errors and V1Rs for which only partial sequences are available so that it is not possible to determine whether or not they are intact.
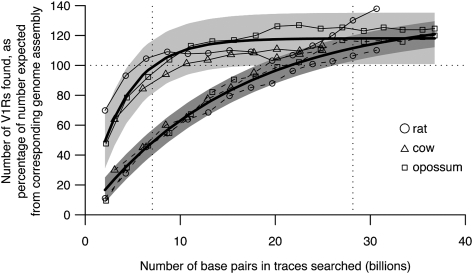
We tested our strategy using trace data from rat, cow, and opossum, where high-coverage draft genome assemblies are also available. Although these genome assemblies are not 100% complete or correct, they provide a useful benchmark against which to test our strategy of mining V1Rs from Trace Archive data. We did not perform tests with mouse and human, because the BAC-based strategies used to sequence these genomes were very different to the random shotgun sequencing approach used for all other species in our analysis. For our tests, we constructed artificial “Trace Archive” databases containing varying numbers of rat, cow, or opossum sequence traces (filtered to include only whole-genome shotgun traces and not, e.g., BAC-derived shotgun sequences) and identified V1Rs in those data sets using the same series of steps as for the other species. We compared the resulting V1R data sets to those identified using the corresponding whole-genome assembly. We found that the total number of V1Rs identified by our method increases with the size of the trace data set that was analyzed, approximately following a Poisson curve (Fig. 1) as predicted from Lander and Waterman's mathematical model (Lander and Waterman 1988). We also found that at ∼2× coverage, the minimum level used in the real data sets we analyzed, we identify at least 90% of the V1Rs we would have found using the whole-genome assemblies; “2× coverage” means that each base pair in the genome is found in an average of two sequence reads. Our independent tests using data sets of conserved sequences (see Methods) also indicate that all the data sets we analyzed contain sequences representing at least 80% of the genome (Supplemental Table 2) and thus should allow identification of the majority of V1R family members in each species. At high coverage (>10 billion bp), we actually found slightly more V1Rs using Trace Archive data than with the whole-genome assembly. In most cases the extra genes appear to be true recent duplicates that had been merged together in the whole-genome assemblies, but were left as separate contigs in our phredphrap reassembly of V1R-like traces because they have noticeable, albeit low, levels of sequence divergence (data not shown). The finding that draft assemblies underestimate numbers of recent gene duplicates is not surprising: Our analyses of several versions of the mouse genome assembly show that, even though earlier drafts were based on reasonably high levels of coverage, many additional V1Rs were found as finishing efforts progressed (Supplemental Table 1). For ease of comparison between species, we decided to adjust trace numbers so that they are comparable to the numbers we find in draft assembled genomes, rather than trying to adjust all numbers to account for falsely merged recent duplicates in draft genome assemblies. This approach might underestimate true V1R numbers.
Figure 1.
Proof-of-principle studies show that Trace Archive data can provide good estimates of V1R repertoire sizes. We used sequence traces from rat (circles), cow (triangles), and opossum (squares) to create artificial Trace Archive data sets of varying sizes. We compared the numbers of V1Rs found using these artificial data sets with the numbers expected based on analysis of draft genome assemblies to determine what proportion was successfully found (expressed as a percentage). As expected, the proportion of total V1Rs found increases as trace data set size increases (solid lines). Likewise, as trace data set size increases, the proportion of intact V1Rs found also increases (dashed lines), but more slowly than does the total V1R number. The thicker lines show curves fitted to these data using Poisson models based on Lander and Waterman's predictions (Lander and Waterman 1988; see Supplemental Methods). The vertical dotted lines show the minimum and maximum trace data set sizes used in our study; within these limits, coverage is reasonably high, and actual numbers follow modeled numbers well. Shaded gray areas show the 95% prediction limits resulting from our modeling.
The number of intact V1Rs identified rises with trace database size, as does the total number of V1Rs (Fig. 1). However, in order to obtain the “correct” number of intact V1Rs, a higher level of coverage is required than is needed to simply count V1Rs (Fig. 1). At lower levels of coverage, frameshifting errors cause some intact genes to be miscalled as pseudogenes, and partial genes are more frequent.
Given that our proof-of-principle studies show that V1Rs can be effectively mined from Trace Archive data, we identified V1R gene sequences from any mammalian species for which whole-genome shotgun sequence data of at least ∼2× coverage was available (Methods; Supplemental Table 2). We used the most recent versions of high coverage (>∼5×) genome assemblies, where available, and used Trace Archive data sets for the remaining species. The three species studied in our proof-of-principle experiments exhibit similar relationships between the numbers of V1Rs found and the trace data set size (Fig. 1). We therefore fit curves to the coverage measures according to Lander and Waterman's model (Fig. 1) and used them to adjust the numbers of intact and total V1Rs to account for the varying sizes of the trace data sets searched (Methods; Supplemental Methods), so that numbers we obtain from Trace Archive data sets are approximately comparable to those we obtain from whole-genome assemblies. The slight differences we observe among the three test species in their relationships between data set size and coverage are likely due to genome size differences, to variation in sequence read quality, and/or to possible differences in the history of V1R evolution between species (for example, it might be more difficult to estimate V1R numbers in species that have experienced more recent V1R duplications than others). However, the differences between the species’ relationships are small and are well covered by the confidence intervals of our predictions, enabling us to use a single modeled relationship to adjust V1R numbers for all species studied. A caveat to our analyses is that our model might not perform well for any species with a particularly unusual genome size compared to other mammals (see Supplemental Text).
Functional V1R repertoire size varies widely among mammals
We identified a total of 6853 V1R-like sequences in the 37 species studied, of which 1809 V1Rs are intact and the rest are either incomplete sequences or contain inactivating mutations/sequence errors. Adjusting these numbers to account for the fact that we used low-coverage trace data, we estimate a total of ∼6580 V1Rs in the 37 genomes, of which ∼2280 are intact (about one-third).
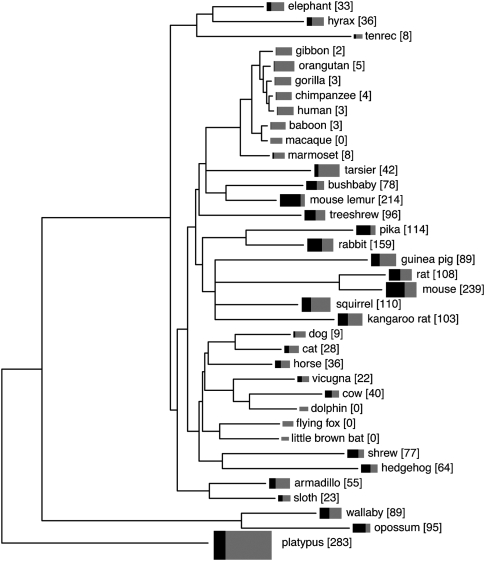
The size of the V1R repertoire varies widely among mammalian species, as does the proportion of V1Rs that appear functional (Fig. 2). Variation had been noted in previous studies based on only eight whole-genome assemblies (Grus et al. 2005, 2007; Young et al. 2005). Our analysis of a much more diverse set of 37 species shows that V1R family evolution has been extraordinarily dynamic across the entire mammalian family and illuminates the biology of many additional species, as discussed below. Where previously published V1R repertoire sizes are available, our numbers compare well (Grus et al. 2005, 2007; Young et al. 2005), with the exception of the mouse and platypus repertoires. The discrepancy for mouse stems from the use of a greatly improved genome assembly (Supplemental Table 1). For platypus, we deliberately took a less conservative approach toward counting possible recent duplicates/allelic variants than did Grus et al. (2007), and thus we report a greater number of platypus V1R pseudogenes.
Figure 2.
Numbers of intact V1Rs and V1R pseudogenes we estimate to be present in various mammalian genomes. The tree depicts mammalian species relationships and is adapted from supporting Figure 6E of Margulies et al. (2005) (for details, see Supplemental Methods). Black and gray rectangles indicate the size of the V1R repertoire in each species—the total area (black plus gray) is proportional to total V1R repertoire size, and the relative size of the black area reflects the proportion of V1Rs that are intact. Numbers in square brackets after species names are the numbers of intact V1Rs. The estimates derived from analyses of Trace Archive data have been adjusted using the fitted curves shown in Figure 1 so that they are comparable to numbers obtained from draft-level whole-genome assemblies. The human and mouse genome assemblies are exceptions in that they are “finished” and likely deal better with very recent duplicates than do any of the other assemblies. Numbers for all other species likely represent underestimates of true V1R numbers.
Of the 37 species studied, platypus (∼280 intact V1Rs) (Grus et al. 2007), mouse (∼240 intact V1Rs), mouse lemur (∼210 intact V1Rs), and rabbit (∼160 intact V1Rs) have the largest V1R repertoires (Fig. 2). A large group of species have ∼60 to ∼110 intact V1Rs, including all remaining members of the glires (rodents and lagomorphs) as well as bushbaby, treeshrew, shrew, hedgehog, and the two marsupials. Almost all of the species with large V1R repertoires have well-developed vomeronasal organs and/or AOBs, where anatomy has been studied (Evans and Schilling 1995; Meisami and Bhatnagar 1998; Malz et al. 2000; Takami 2002; Smith et al. 2005; Schneider et al. 2008). The sole possible exception is the shrew, which appears to have a small AOB (Meisami and Bhatnagar 1998) despite its fairly large intact V1R repertoire (∼80).
On the opposite end of the spectrum, the genomes of dolphin, little brown bat, and flying fox (a megabat) appear to contain no intact V1Rs. This lack of V1Rs might be expected given the absence of a vomeronasal organ in these species (in the case of the flying fox, whose vomeronasal anatomy has not been described, we assume the organ is absent because all eight other members of the same genus that have been examined lack a VNO) (Oelschlager 1989; Bhatnagar and Meisami 1998). In addition, our analyses of the Trpc2 gene in each of these genomes suggest that the gene has acquired inactivating mutations in all three species (Supplemental Fig. 1). Trpc2 is a signaling molecule necessary for neuronal signaling in the vomeronasal organ (although a recent study suggests that some vomeronasal function may be Trpc2-independent, likely through V2R-expressing neurons) (Kelliher et al. 2006). Trpc2 is also a pseudogene in apes and Old World monkeys (Liman and Innan 2003; Zhang and Webb 2003), species that have almost no intact V1Rs (Fig. 2). The adaptation of the dolphin's ancestor to a fully aquatic lifestyle likely rendered its chemosensory apparatus ineffective and removed selective pressure maintaining function of its chemosensory components. Paralleling V1R loss in dolphin, most OR sequences identified in other cetaceans are pseudogenes (Kishida et al. 2007).
V1R/Trpc2 loss in bats is more difficult to explain. Flying fox and little brown bat are representatives of the two deeply diverged bat suborders, Megachiroptera and Microchiroptera, respectively. Because some of the little brown bat's microchiropteran relatives possess well-developed VNOs (Bhatnagar and Meisami 1998), it appears that the flying fox and little brown bat independently lost functional VNOs (and thus V1Rs). Reduced importance of chemical signaling in bats due to acquisition of echolocation and enhanced auditory function might at first appear an attractive hypothesis. However, this hypothesis is not supported by the fact that other bat species with well-developed VNOs also use echolocation (Bhatnagar and Meisami 1998). Furthermore, flying fox and other present-day megabat species do not appear to echolocate, yet flying fox still has no functional V1Rs. Thus, echolocation is not a prerequisite for V1R loss, although it is possible that an ancestor of flying fox might have acquired echolocation abilities, with subsequent loss in all present-day Megachiroptera (Teeling et al. 2000). It would be interesting to perform a more comprehensive study of V1Rs, V2Rs, FPRs and their downstream partners such as Trpc2 across the bat superfamily to determine whether bats with a more well developed VNO retain functional versions of these genes.
We find that all Old World monkey and ape V1R repertoires also consist primarily of pseudogenes, but that marmoset has eight apparently intact V1Rs. In contrast, their relative the tarsier has ∼40 intact V1Rs, and the two prosimian species (mouse lemur and bushbaby) have very large intact V1R repertoires (∼210 and ∼80 intact V1Rs, respectively). Previous primate V1R studies had examined the complete repertoires of only human and chimpanzee (Rodriguez and Mombaerts 2002; Young et al. 2005). Even the few remaining potentially intact human V1Rs and their ape orthologs appear to be evolving neutrally (Zhang and Webb 2003). Our new data show that V1R loss must have begun in the common ancestor of New and Old World primates and/or occurred independently in the New World monkey and Old World lineages. This finding refutes an earlier hypothesis that large-scale vomeronasal degeneration in primates was temporally correlated with the acquisition of trichromatic vision in the common ancestor of Old World monkeys and apes, after divergence from New World monkeys (Liman and Innan 2003; Zhang and Webb 2003). Although marmoset has a very small repertoire of intact V1Rs, its functional repertoire appears larger than that of other primates. This result suggests that some V1R-mediated signaling is still important in marmoset behavior, consistent with the presence of an apparently functional vomeronasal organ, albeit a less well-developed organ than that of rodents (Dennis et al. 2004). Primate V1R repertoire sizes correlate well with anatomical observations: The vomeronasal system is well-developed in bushbabies and lemurs (Evans and Schilling 1995; Smith et al. 2005) and is present in marmosets and neonatal tarsiers (Dennis et al. 2004), but is absent in apes and Old World monkeys (Liman 2006).
Thus, V1R repertoire sizes across the mammalian tree correlate approximately with anatomical observations of VNO and/or AOB size and quality, supporting previous observations based on fewer species (Grus et al. 2005, 2007). We referred above to the well-developed vomeronasal system anatomy and large V1R repertoires of rodents, rabbit, platypus, and mouse lemur and the poorly developed or absent vomeronasal structures of apes, Old World monkeys, some bats, dolphins, and dogs. Among species with intermediately sized V1R repertoires, the elephant (∼30 intact V1Rs) appears to have a well-developed VNO (Johnson and Rasmussen 2002), as do the cat (∼30 intact V1Rs) (Salazar et al. 1996) and armadillo (∼60 intact V1Rs) (Carmanchahi et al. 1999), albeit not the same armadillo species from which DNA sequences are available. We have not attempted any quantitative comparisons here due to the difficulty of comparing anatomical observations between different studies, especially when the species studied have vastly differing body and relative brain sizes.
Most mammalian genomes experienced species-specific V1R gene family expansions
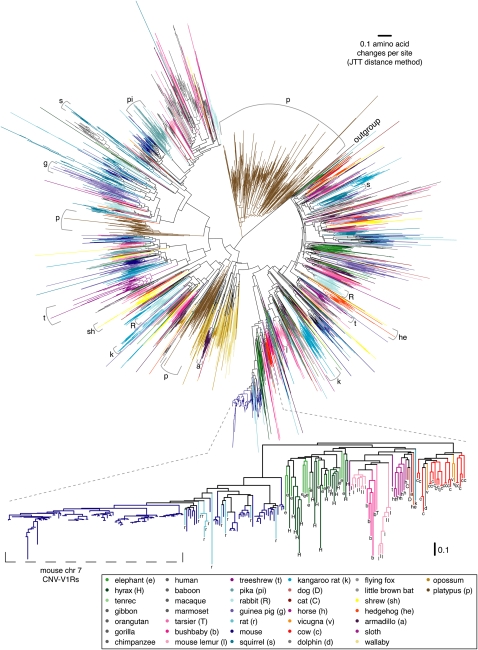
Phylogenetic trees of the V1R sequences we identified (Fig. 3; Supplemental Figs. 2, 3) contain a large number of species-specific clades that likely arose due to post-speciation duplication. The homogenizing effects of post-speciation gene-conversion events might also have contributed to the semi-private nature of these V1R repertoires. Studies of eight mammalian genomes had detected the phenomenon of species-specific V1R clades (Rodriguez and Mombaerts 2002; Grus and Zhang 2004; Grus et al. 2005; Young et al. 2005); here we show that it is widespread across the entire mammalian tree. We constructed the trees after aligning predicted V1R protein sequences (artificially correcting frameshifting mutations/sequence errors where necessary) and excluding sequences that were too short to allow effective phylogenetic inference.
Figure 3.
V1R phylogenetic tree showing that species-specific gene duplications have resulted in “semi-private” V1R repertoires. The tree shows the relationships between predicted protein sequences of 4954 of the V1Rs we identified, including intact genes, sequences that contain inactivating mutations/sequence errors (frameshifts corrected where necessary for alignment), and some genes for which full-length sequence is not available, but excluding sequences too short for phylogenetic analysis (see Methods). We also included zebrafish V1R1 (GenBank ABL01523) as an outgroup sequence to root the tree. We used the neighbor-joining method with Jones-Taylor-Thornton amino acid distances to create a tree. Terminal branches are colored according to species. Internal branches share the species color if all descendent terminal taxa are from the same species, or black if descendents are from two or more species. Thus, clusters of same-colored branches reflect species-specific duplications (and possibly some post-speciation gene-conversion events). Internal black branches help distinguish species-specific clades from clades of mixed species especially when the colors used to represent those species are similar. Due to the difficulties of displaying such a large phylogenetic tree and finding enough colors to distinguish so many species, we also provide subtrees constructed from only glires (rodents and lagomorphs) or primate V1Rs in Supplemental Figures 2 and 3. The lower part of the figure provides a higher resolution view of a selected part of the tree and uses letters (see key) to label non-mouse V1Rs to help distinguish species. All species-specific clades containing more than 30 members are indicated in the large tree using brackets with letters to indicate species (see key), except for a clade containing mouse V1Rs that are found in a large copy-number variable (CNV) region (see text), which is instead labeled in the higher-resolution subtree in the lower half of the figure.
Our trees show that species-specific V1R subfamilies are common throughout the entire mammalian phylogeny and are especially apparent in species with large V1R repertoires. Most V1Rs we identify are the product of duplication since divergence from any other species in our study. In fact, ∼80% of all the V1Rs we found have a more similar match in the same genome than in any other species’ genome in our study (i.e., paralogs have higher amino acid identity than orthologs in BLASTP comparisons of all ∼6850 V1Rs found, see Methods). This finding also holds true if we use a more conservative subset of the data (Supplemental Table 2) that includes only one member of any group of sequences having at least 98% nucleotide identity in order to avoid possible inflation of species-specific duplication estimates caused by incomplete assembly of highly similar sequences, such as atypically divergent allelic pairs. In this more conservative set, 71% of the V1Rs have a more similar paralog than ortholog. Many of the remaining 29% V1Rs are from ape or Old World monkey species, a mammalian clade where draft-level sequence is available from an unusually dense sampling of species; if we exclude all ape/Old World monkey sequences, we find that only 19% of the V1Rs in this conservative subset have a closer ortholog than paralog.
We observe several particularly large species-specific V1R clades in our tree, including clades with 30 or more members in platypus, mouse, treeshrew, pika, guinea pig, kangaroo rat, rabbit, hedgehog, armadillo, lemur, squirrel, and shrew (Fig. 3, brackets). One of the largest species-specific clades comprises a subfamily of ∼90 mostly intact mouse V1Rs, almost all of which are clustered on chromosome 7. This cluster appears to have arisen by a series of local duplication events since mouse diverged from its closest sequenced relative, the rat. Interestingly, this cluster shows extensive copy-number variation: Many mouse strains appear to have fewer genomic copies of this region of chromosome 7 than does the reference strain, C57BL/6J (Cutler et al. 2007; Graubert et al. 2007), suggesting that at least some of the duplications that expanded this subfamily in C57BL/6J happened very recently indeed or that some duplicated copies were deleted in other mouse strains. It would be interesting to see whether behavioral differences exist between these mouse strains. Knockout mice lacking 16 V1Rs on chromosome 6 show altered male sexual behavior and maternal aggression (Del Punta et al. 2002), but the phenotypic effect of having additional, slightly divergent V1R copies is unknown.
Close examination of our phylogenetic trees (e.g., Supplemental Fig. 3) reveals many examples of V1Rs that appear to have duplicated after losing function (or, where an inactivating mutation spread to several related V1Rs by gene conversion). These duplications occur in many of the species we analyzed and are evident as clades in which several V1R pseudogenes from the same species share the same inactivating mutation. These cases are likely to be examples of neutral changes being fixed by genetic drift, as it is difficult to imagine any selective advantage to these duplications/gene-conversion events.
Dog V1R deterioration is not correlated with domestication
We explored the idea that domestication might have imposed selective pressures favoring the acquisition of inactivating mutations in dog V1Rs. Over time, humans might have selected animals that were least responsive to pheromonal cues and therefore perhaps less distracted by conspecifics. Our analysis of Trace Archive sequence data shows that another carnivore, cat, has ∼30 intact V1Rs, indicating that the dog lineage indeed suffered substantial V1R deterioration after canines and felines diverged. However, the wolf DNA sequences we obtained indicate that this deterioration likely occurred before wolf and dog diverged (i.e., before domestication).
In detail, we selected seven dog V1R pseudogenes with fewer disruptions than most others, each containing one to three inactivating mutations (in-frame stop codons, frameshifts, or insertion of interspersed repeats). We reasoned that, given their small number of inactivating mutations, these V1Rs likely became pseudogenes relatively recently in evolutionary time and thus are the best candidates for genes that might have experienced domestication-related selection for loss of function, if such selection had occurred. We designed oligonucleotides from the sequences of these seven V1R pseudogenes (Supplemental Table 3) along with one intact V1R and used these primers to amplify and sequence DNA from a dog and two unrelated wolves, one from Spain and one from Alaska.
We found that all seven dog pseudogenes are also pseudogenes in wolf. The wolf genes contain the same inactivating mutations as their dog counterparts (Supplemental Table 4). These inactivating mutations must therefore have been acquired before dog domestication, making it very unlikely that domestication drove V1R loss. The intact dog gene also appeared intact in both wolves. The remaining possibility that a set of V1R genes was entirely deleted from the dog genome during domestication seems remote given that dog retains V1R pseudogenes in most V1R families (Young et al. 2005). A definitive answer to the question of whether dog domestication was associated with any V1R loss could be obtained by determining sequences of the full wolf V1R repertoire. However, without whole-genome sequence, this might be impossible: The enormous sequence diversity of the V1R family makes an exhaustive PCR-based V1R survey exceedingly difficult—we and others have tried and failed to develop universal degenerate V1R primers (Mombaerts 2004; data not shown).
Concluding remarks
We have shown that variation in V1R repertoire size and species-specific V1R subfamilies are widespread phenomena across the mammalian tree. An approximate correlation of V1R repertoire size with anatomical observations (Grus et al. 2005) is also widespread among mammals: Species with many intact V1Rs tend to have well-developed vomeronasal organs and AOBs, but these structures are absent or poorly developed in species with few V1Rs. However, we see no obvious correlation across all mammals of V1R repertoire size with ecological factors such as body size, nocturnality, diet, sociality, or mating system. Our addition of V1R data from marmoset argues against one previous suggestion that degeneration of primate V1Rs occurred when trichromatic vision was acquired. We also show that the decline of the dog V1R repertoire almost certainly did not occur in response to selective pressures imposed during domestication.
The large number of genetic and ecological changes seen along all branches of the mammalian tree makes it very difficult to correlate changes in V1R family composition with ecological factors, especially given that several ecological factors vary simultaneously in the species studied. We also note that effective population sizes and demographic histories likely differ dramatically between the species we have studied, meaning that there will be differences between species in the efficiency of selection and likelihood that genetic drift could fix selectively neutral changes. Population genetics might, in some cases, impact V1R gene family dynamics as much as ecology and behavior. It would be informative to compare V1R repertoires of several closely related species in subfamilies such as lemurs, rodents, bats, or shrews, where only a few ecological factors vary between species, effective population sizes are roughly similar, and less evolutionary time has elapsed since their divergence. With sets of closely related species, more sophisticated methods of comparing V1R repertoires with ecological factors could be explored, such as the use of phylogenetically independent contrasts (Felsenstein 1985).
Almost all of the mammals we studied possess species-specific V1R subfamilies. We and others previously suggested that species-specificity in the V1R repertoire might arise in response to the need to reinforce, or possibly even establish, mating barriers during speciation (Lane et al. 2002; Grus and Zhang 2004). The species in our study are all separated by millions of years of evolution. In order to elucidate whether changes in the V1R repertoire are involved in speciation, it would be interesting to examine the V1R repertoires of subspecies pairs currently undergoing speciation or to test whether or not V1R copy-number polymorphisms observed among laboratory mouse strains impact their mating preferences.
Given that there are two other families of vomeronasal receptors, the V2Rs and FPRs, and increasing evidence that the main olfactory epithelium in rodents can detect pheromones via TAARs and perhaps also ORs, the data we present here form just one part of the story of the highly variable importance of chemical communication among the amazingly diverse mammals of the world.
Methods
V1R identification from low-redundancy comparative sequence data
We obtained Trace Archive sequences from NCBI (http://www.ncbi.nlm.nih.gov) and genome assemblies from the UCSC Genome Bioinformatics site (http://genome.ucsc.edu). We also performed pilot studies on a small number of ∼2× coverage genome assemblies from the Broad Institute but found that we obtained less fragmented V1R data sets and greater numbers of distinct intact V1Rs by using raw sequence traces (data not shown). In addition, Trace Archive data sets offered an opportunity to search recently generated sequence traces that were not included in assemblies; for example, Ensembl reports that the Broad Institute bushbaby assembly has coverage of ∼1.5×, whereas the Trace Archive contains >15 million bushbaby traces, coverage of ∼4.5× (Supplemental Table 2). Download dates, numbers of sequences analyzed, and other characteristics of the data sets we used are given in Supplemental Table 2.
Our V1R identification method is described in full in a previous publication (Young et al. 2005). We performed an initial round of sensitive TBLASTN searches (Altschul et al. 1997), using local copies of sequence databases for each species, to identify any Trace Archive sequence or genome assembly segment that had even very weak V1R homology. Our queries for this round of TBLASTN searches were 49 divergent V1Rs (Young et al. 2005), and we considered any match with E-value < 10 in our subsequent analysis, filtering out non-V1R sequences at a later stage. For genome assemblies, we extracted sequences of matching regions along with 1 kb of flanking sequence on each side.
For Trace Archive data sets, we used NCBI's query_tracedb interface to download chromatogram files corresponding to any matching sequences and used phredphrap (http://www.phrap.org) to determine DNA sequences from those chromatograms. Interspersed repeat sequences were masked from the resulting FASTA files of individual sequence reads using RepeatMasker (AFA Smit, R Hubley, and P Green, 1996–2004. RepeatMasker Open-3.0; http://www.repeatmasker.org), and masked sequences were used to BLAST the appropriate Trace Archive database once again to obtain overlapping traces, this time using MEGABLAST (Zhang et al. 2000). Chromatograms were obtained for any sequences matching for at least 100 bp with at least 95% identity, and phredphrap was run again to assemble overlapping sequences into contigs where possible. The “contigs” and “singletons” files produced by phredphrap were concatenated (singletons are traces that failed to assemble with others into a contig), and a second round of TBLASTN searches (using the same 49 V1R queries and E-value threshold) was used to determine which of these contigs and singletons contain candidate V1R sequences.
The resulting ∼3-kb sequences (from genome assemblies) and contigs/singletons files (from Trace Archive data sets) were further analyzed for V1R content, using slightly modified versions of our V1R-identification scripts (Young et al. 2005). A brief description of the scripts and full details of the modifications are provided as Supplemental Methods.
Modeling numbers of V1Rs found in Trace Archive data sets of different coverage levels
In order to compare the numbers of intact and total V1Rs we obtained from Trace Archive databases (which have varying levels of coverage) with one another and with numbers obtained from draft genome assemblies, we used the following method to adjust gene numbers. We performed tests of our strategy using artificial trace data sets of varying sizes (increasing in increments of 2.5 million traces) from rat, cow, and opossum (using only traces with the tag “trace_type_code = WGS”, to avoid BAC shotgun sequences, EST traces, etc.). We compared the numbers of intact and total V1Rs obtained from those trace data sets with the numbers we obtained from the corresponding genome assemblies (see Fig. 1 and legend) and found that the proportion of V1Rs found varied with trace data set size in a predictable way that could be modeled. We then used those modeling results to adjust Trace Archive V1R numbers according to the trace data set size, so that they would be comparable to numbers obtained from draft genome assemblies of typical coverage. Further details of the modeling process are given as Supplemental Methods.
Independent estimates of proportion of genome present in Trace Archive data sets
We also wanted to obtain a measure of the proportion of the genome present in each Trace Archive database, so that we did not have to rely wholly on the number of traces searched (e.g., if a large number of poor-quality traces were present, the true coverage might be lower than that expected from the number of traces searched). We therefore collected two data sets of highly conserved sequences that should be present and easily identified in all mammalian genomes, blasted those sequences against genome assemblies/Trace Archive data sets, and determined how many of the conserved sequences were found in each database (see Supplemental Methods). We were also concerned that species mislabeling might be a problem for some Trace Archive sequences—our analyses indicate that this is a very infrequent issue (see Supplemental Methods).
Multiple alignments and phylogenetics
In order to create a multiple sequence alignment of the V1R gene and pseudogene sequences we identified, we first aligned all intact V1Rs to a hidden Markov model (HMM) representing V1R proteins. We then grouped the other V1Rs (pseudogenes, partial V1Rs, and V1Rs with sequencing errors) by sequence similarity and, for each group, chose a best-matching intact V1R, taking the match with highest amino acid identity to any sequence in the group. We then aligned each V1R in the group to that best-matching intact V1R and used that pairwise alignment to add each sequence to the aligned intact V1Rs. Details of this procedure are provided in the Supplemental Methods. Prior to constructing trees, we removed any sequence with length <60% of typical full-length V1Rs to ensure at least some overlap between all sequences in the alignment. Phylogenetic trees were obtained using the neighbor-joining method (Saitou and Nei 1987) as implemented in the neighbor algorithm of the PHYLIP package (J. Felsenstein, University of Washington, Seattle), using protein distances calculated using protdist (PHYLIP) and the Jones-Taylor-Thornton method. Trees were displayed and colored using a custom R script that uses the ape package (Paradis et al. 2004).
Other sequence analyses
In order to determine the proportion of V1Rs that appear to be the product of duplication since divergence from any other species in our study, we compared all V1Rs to each other using BLASTP (Altschul et al. 1997) and determined whether the non-self match with the highest amino acid identity was from the genome of the same or a different species (only considering matches of at least 30% identity over at least 30 amino acids, with E-value ≤ 10−5). Using BLAST-based counts for this analysis allowed us to include a larger fraction of genes than if we had examined the structure of the phylogenetic tree, because the tree omits the ∼30% of V1Rs found that were too short for phylogenetic analysis.
We also performed the analysis on a more conservative subset of the data (where we included only one member of any group of sequences with at least 98% nucleotide identity) to avoid possible inflation of species-specific duplication estimates caused by incomplete assembly of highly similar sequences such as unusually divergent allelic pairs. To create this conservative subset, we used BLASTN (Altschul et al. 1997) to compare all V1R nucleotide sequences (RepeatMasked using the appropriate species settings for each sequence, because some V1R pseudogenes are interrupted by internal repeat sequences) with one another, and a custom Perl script to perform single linkage clustering of groups of sequences with ≥98% pairwise nucleotide identity over ≥200 bp. We then retained the longest sequence in each subset in our conservative data set.
PCR and sequencing
DNA samples from two wolves from Alaska and Spain were kindly provided by R. Wayne and M. Gray (Department of Ecology and Evolutionary Biology, University of California, Los Angeles), and dog DNA was obtained from Clontech. These two wolf samples should represent a true predomestication outgroup to dogs: Other genotype data obtained from the same samples show no evidence of dog introgression (M Gray and R Wayne, pers. comm.). Furthermore, recent genome-wide SNP analyses show that, apart from some Italian animals that are clearly wolf-dog hybrids, wolf genomes do not contain significant genetic contributions from domesticated dogs, except at a positively selected locus for melanism (Anderson et al. 2009); this locus is unlinked to any of the V1Rs we study here.
V1Rs were PCR amplified using standard conditions (available on request). Amplified products were purified using Sephacryl S-300 (Amersham Biosciences) and subjected to big-dye sequencing (Applied Biosystems) using standard protocols. Sequencing primers included those used for PCR as well as additional internal primers. All primer sequences are provided in Supplemental Table 3. One pseudogene consistently failed to amplify in the Spanish wolf but was successfully amplified from dog and Alaskan wolf (Supplemental Table 4). We did not determine whether this failure was due to a SNP in the primer site or to a whole or partial deletion of this gene in that animal.
The wolf and dog sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ713544–FJ713565. All V1R sequences identified from genome assemblies and Trace Archive sequence data are provided in Supplemental Datafiles 1 and 2; 3532 of them are also in RefSeq, and accession numbers are provided in Supplemental Table 6.
Acknowledgments
We thank the many sequencing centers who generated and released the data we analyzed. We also thank Bob Wayne and Melissa Gray for providing wolf DNA, Elaine Ostrander for useful discussions and the opportunity to carry out dog and wolf work, Sean Parghi and Fred Hutchinson Cancer Research Center's Genomics Resource for sequencing assistance, and Elliott Margulies for providing species trees. We thank Wendy Grus for comments on the manuscript and helpful suggestions and other members of the Trask laboratory for useful discussion. We also thank Dirck Bradt and Deborah Reed of the Mouse Genome Informatics sequencing group (Jackson Laboratory) for their assistance with mouse V1R nomenclature. This work was supported by the National Institutes of Health grants DC004209, CA092167, and AG14358.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ713544–FJ713565.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.098913.109.
References
- Ache BW, Young JM. Olfaction: Diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, Padhukasahasram B, Randi E, Leonard JA, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisko JJ. Communication by chemical signals in Canidae. In: Doty RL, editor. Mammalian olfaction, reproductive processes, and behavior. Academic Press; New York: 1976. pp. 283–293. [Google Scholar]
- Bhatnagar KP, Meisami E. Vomeronasal organ in bats and primates: Extremes of structural variability and its phylogenetic implications. Microsc Res Tech. 1998;43:465–475. doi: 10.1002/(SICI)1097-0029(19981215)43:6<465::AID-JEMT1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Brennan PA, Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- Carmanchahi PD, Aldana Marcos HJ, Ferrari CC, Affanni JM. The vomeronasal organ of the South American armadillo Chaetophractus villosus (Xenarthra, Mammalia): Anatomy, histology and ultrastructure. J Anat. 1999;195:587–604. doi: 10.1046/j.1469-7580.1999.19540587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- Dennis JC, Allgier JG, Desouza LS, Eward WC, Morrison EE. Immunohistochemistry of the canine vomeronasal organ. J Anat. 2003;203:329–338. doi: 10.1046/j.1469-7580.2003.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JC, Smith TD, Bhatnagar KP, Bonar CJ, Burrows AM, Morrison EE. Expression of neuron-specific markers by the vomeronasal neuroepithelium in six species of primates. Anat Rec A Discov Mol Cell Evol Biol. 2004;281:1190–1200. doi: 10.1002/ar.a.20124. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Evans C, Schilling A. The accessory (vomeronasal) chemoreceptor system in some prosimians. In: Alterman L, et al., editors. Creatures of the dark: The nocturnal prosimians. Plenum Press; New York: 1995. pp. 393–411. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Goodwin M, Gooding KM, Regnier F. Sex pheromone in the dog. Science. 1979;203:559–561. doi: 10.1126/science.569903. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Rapid turnover and species-specificity of vomeronasal pheromone receptor genes in mice and rats. Gene. 2004;340:303–312. doi: 10.1016/j.gene.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Grus WE, Shi P, Zhang YP, Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc Natl Acad Sci. 2005;102:5767–5772. doi: 10.1073/pnas.0501589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus WE, Shi P, Zhang J. Largest vertebrate vomeronasal type 1 receptor gene repertoire in the semiaquatic platypus. Mol Biol Evol. 2007;24:2153–2157. doi: 10.1093/molbev/msm157. [DOI] [PubMed] [Google Scholar]
- Johnson EW, Rasmussen L. Morphological characteristics of the vomeronasal organ of the newborn Asian elephant (Elephas maximus) Anat Rec. 2002;267:252–259. doi: 10.1002/ar.10112. [DOI] [PubMed] [Google Scholar]
- Karunadasa DK, Chapman C, Bicknell RJ. Expression of pheromone receptor gene families during olfactory development in the mouse: Expression of a V1 receptor in the main olfactory epithelium. Eur J Neurosci. 2006;23:2563–2572. doi: 10.1111/j.1460-9568.2006.04795.x. [DOI] [PubMed] [Google Scholar]
- Kelliher KR, Spehr M, Li XH, Zufall F, Leinders-Zufall T. Pheromonal recognition memory induced by TRPC2-independent vomeronasal sensing. Eur J Neurosci. 2006;23:3385–3390. doi: 10.1111/j.1460-9568.2006.04866.x. [DOI] [PubMed] [Google Scholar]
- Kishida T, Kubota S, Shirayama Y, Fukami H. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: Evidence for reduction of the functional proportions in cetaceans. Biol Lett. 2007;3:428–430. doi: 10.1098/rsbl.2007.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Axel R, Hood L, Trask BJ. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc Natl Acad Sci. 2002;99:291–296. doi: 10.1073/pnas.012608399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Liman ER. Use it or lose it: Molecular evolution of sensory signaling in primates. Pflugers Arch. 2006;453:125–131. doi: 10.1007/s00424-006-0120-3. [DOI] [PubMed] [Google Scholar]
- Liman ER, Innan H. Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc Natl Acad Sci. 2003;100:3328–3332. doi: 10.1073/pnas.0636123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malz CR, Knabe W, Kuhn HJ. Pattern of calretinin immunoreactivity in the main olfactory system and the vomeronasal system of the tree shrew, Tupaia belangeri. J Comp Neurol. 2000;420:428–436. doi: 10.1002/(sici)1096-9861(20000515)420:4<428::aid-cne2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Margulies EH, Vinson JP, Miller W, Jaffe DB, Lindblad-Toh K, Chang JL, Green ED, Lander ES, Mullikin JC, Clamp M. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc Natl Acad Sci. 2005;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisami E, Bhatnagar KP. Structure and diversity in mammalian accessory olfactory bulb. Microsc Res Tech. 1998;43:476–499. doi: 10.1002/(SICI)1097-0029(19981215)43:6<476::AID-JEMT2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Oelschlager HA. Early development of the olfactory and terminalis systems in baleen whales. Brain Behav Evol. 1989;34:171–183. doi: 10.1159/000116503. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Mombaerts P. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr Biol. 2002;12:R409–R411. doi: 10.1016/s0960-9822(02)00909-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Greer CA, Mok MY, Mombaerts P. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat Genet. 2000;26:18–19. doi: 10.1038/79124. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salazar I, Sanchez Quinteiro P, Cifuentes JM, Garcia Caballero T. The vomeronasal organ of the cat. J Anat. 1996;188:445–454. [PMC free article] [PubMed] [Google Scholar]
- Schneider NY, Fletcher TP, Shaw G, Renfree MB. The vomeronasal organ of the tammar wallaby. J Anat. 2008;213:93–105. doi: 10.1111/j.1469-7580.2008.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TD, Bhatnagar KP, Burrows AM, Shimp KL, Dennis JC, Smith MA, Maico-Tan L, Morrison EE. The vomeronasal organ of greater bushbabies (Otolemur spp.): Species, sex, and age differences. J Neurocytol. 2005;34:135–147. doi: 10.1007/s11068-005-5053-9. [DOI] [PubMed] [Google Scholar]
- Takami S. Recent progress in the neurobiology of the vomeronasal organ. Microsc Res Tech. 2002;58:228–250. doi: 10.1002/jemt.10094. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Scally M, Kao DJ, Romagnoli ML, Springer MS, Stanhope MJ. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Mori Y, Ichikawa M, Yazaki K, Hagino-Yamagishi K. A putative pheromone receptor gene is expressed in two distinct olfactory organs in goats. Chem Senses. 2002;27:207–213. doi: 10.1093/chemse/27.3.207. [DOI] [PubMed] [Google Scholar]
- Wyatt TD. Pheromones and animal behavior. Cambridge University Press; Cambridge, UK: 2003. [Google Scholar]
- Young JM, Trask BJ. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23:212–215. doi: 10.1016/j.tig.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Young JM, Kambere M, Trask BJ, Lane RP. Divergent V1R repertoires in five species: Amplification in rodents, decimation in primates, and a surprisingly small repertoire in dogs. Genome Res. 2005;15:231–240. doi: 10.1101/gr.3339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]