Abstract
Organisms of the third domain of life, the Archaea, share molecular characteristics both with Bacteria and Eukarya. These organisms attract scientific attention as research models for regulation and evolution of processes such as transcription, translation, and RNA processing. We have reconstructed the primary transcriptome of Sulfolobus solfataricus P2, one of the most widely studied model archaeal organisms. Analysis of 625 million bases of sequenced cDNAs yielded a single-base-pair resolution map of transcription start sites and operon structures for more than 1000 transcriptional units. The analysis led to the discovery of 310 expressed noncoding RNAs, with an extensive expression of overlapping cis-antisense transcripts to a level unprecedented in any bacteria or archaea but resembling that of eukaryotes. As opposed to bacterial transcripts, most Sulfolobus transcripts completely lack 5′-UTR sequences, suggesting that mRNA/ncRNA interactions differ between Bacteria and Archaea. The data also reveal internal hotspots for transcript cleavage linked to RNA degradation and predict sequence motifs that promote RNA destabilization. This study highlights transcriptome sequencing as a key tool for understanding the mechanisms and extent of RNA-based regulation in Bacteria and Archaea.
The Archaea are one of the three domains of life. They lack a nucleus and have an overall genome organization similar to that of Bacteria, but many of their molecular systems, including the transcription, translation, and DNA packaging apparatuses, are similar to those of eukaryotes (Bell and Jackson 1998). These organisms thus became an axis of interest in questions concerning the evolution of basic molecular machineries. Over the last three decades, Sulfolobus solfataricus, a sulfur-metabolizing aerobic archaeon that grows optimally at 80°C and pH 2–3 (Brock et al. 1972) has been one of the most widely studied archaeal organisms. Sulfolobus serves as a research model for mechanisms of transcription, translation, DNA damage, DNA replication, cell cycle, and RNA processing (She et al. 2001).
Deep transcriptome sequencing (RNA-seq) has recently emerged as a method enabling the study of RNA-based regulatory mechanisms in a genome-wide manner (Wang et al. 2009). In eukaryotes, RNA-seq has been used for the discovery of splice variants, RNA editing sites, and new microRNAs (Glazov et al. 2008; Sultan et al. 2008; Li et al. 2009). Recent studies have utilized RNA-seq for bacterial transcriptome research and demonstrated its effectiveness in accurate operon definition, discovery of noncoding RNAs, and correction of gene annotation (Passalacqua et al. 2009; Perkins et al. 2009; Yoder-Himes et al. 2009).
To date, there has been no systematic characterization of an archaeal transcriptome by deep sequencing. The largest number of transcription start sites (TSSs) characterized for any archaeal organism is 40 (Brenneis et al. 2007), and no complete archaeal operon map has been experimentally determined. Directed efforts have been conducted to detect small noncoding RNAs (sRNAs) in S. solfataricus (Tang et al. 2005; Zago et al. 2005), but since these efforts were based on cloning of size-selected individual transcripts, they had limited capacity and yielded only partially overlapping results.
Using a combination of whole-transcript sequencing and strand-sensitive 5′-end determination, we have generated a map of the S. solfataricus transcriptome to a single-nucleotide resolution. The exact TSS and operon structures were determined for more than 1000 transcripts, and more than 300 noncoding RNAs were detected. This approach also revealed 80 new protein-coding genes and resulted in the correction of annotation for >5% of all genes in the genome.
Results
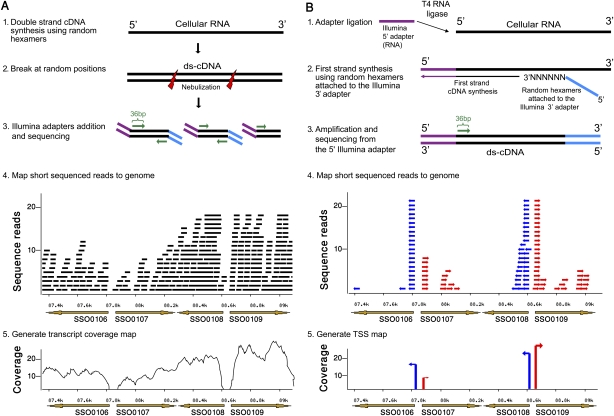
To generate a single-nucleotide resolution map of the S. solfataricus P2 transcriptome, we combined two independent transcriptome sequencing approaches: whole-transcript sequencing and 5′-end sequencing (Fig. 1). In the whole-transcript approach, double-stranded cDNA is prepared using random-hexamer priming on total RNA and then broken in random positions. Resulting cDNA fragments are sequenced using the Illumina Genome Analyzer, yielding strand-insensitive sequence reads that cover the entire lengths of cellular transcripts. In parallel, a 5′-end sequencing protocol is used to identify the exact 5′-end of each transcript and determine the strand of transcription. In this protocol, Illumina 5′ adapters are ligated to the exposed 5′-ends of total RNAs prior to reverse transcription (Fig. 1; Methods).
Figure 1.
Transcriptome mapping using massive cDNA sequencing. (A) Whole-transcript sequencing. S. solfataricus ds-cDNA libraries are prepared from total RNA using random hexamers priming (stage 1). Resulting cDNA is sheared in random positions, and Illumina adapters are ligated (stages 2–3). Sequenced short reads are mapped to the genome (stage 4) to generate a transcript coverage map (stage 5). Shown is a 1.7-kb window of the genome, containing (brown arrows) four genes (right- and left-pointing arrows correspond to genes encoded on the forward and reverse strands, respectively). (Black bars) Sequenced reads, mapped to their respective positions on the genome. (B) 5′-End sequencing. An RNA oligonucleotide corresponding to the Illumina 5′ adapter is attached to exposed 5′ ends of total S. solfataricus RNAs (stage 1). cDNA is then synthesized using random DNA hexamers flanked by the Illumina 3′ adapters (stage 2). Transcripts are primed from the 5′ adapter, thereby generating strand-specific sequences matching 5′-ends of cellular RNAs (stage 3). Sequenced short reads are mapped to the genome (stage 4) to generate a transcription start sites (TSS) map (stage 5). Shown is the same region as in panel A. (Small arrows) Short reads mapped to their respective positions on the genome; (red) forward strand; (blue) reverse. Reads in the middle of genes might represent degraded/fragmented RNAs that present an exposed 5′-end.
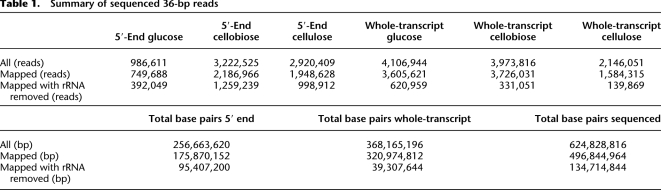
These sequencing protocols were applied on three separate samples of S. solfataricus independently grown on three different carbon sources (Table 1; Methods). The resulting 36-bp reads were mapped to the genome allowing up to two mismatches, and reads that mapped equally well to more than one position on the genome, as well as reads mapped to ribosomal RNAs, were discarded (Methods). This yielded a total of 2.65 million 5′-end reads and 1.09 million whole-transcript reads mapped to the genome (Table 1). In general, 89.5% of the genome was transcribed in at least one growth condition, and 45% of the genome showed transcription in all three conditions. Genes involved in information processing tended to be highly expressed (Supplemental Fig. S1). As detailed below, the mapped transcriptome data were used for a careful automated and manual annotation of the transcriptome structure.
Table 1.
Summary of sequenced 36-bp reads
Transcription start sites and comprehensive operon map
The sequences derived from the 5′-end approach may correspond either to real beginnings of transcripts existing in the native cell or to break points of degraded RNA molecules. To detect reproducible 5′ ends of transcripts, we further considered only positions represented by at least two reads in each of the three sequenced samples, and by more than 10 reads cumulatively (Methods). There were 1.83 million reads that corresponded to such positions, with 53% of them mapped up to 50 bases upstream of an annotated beginning of a gene (and having the strand match the predicted transcription direction). This represents a 10-fold enrichment for 5′ ends of transcripts (P < 10−30). Such enrichment was not detected in the whole-transcript sequencing data. These results indicate that our 5′ sequencing approach largely maps actual beginnings of RNAs in the cell (Fig. 1B).
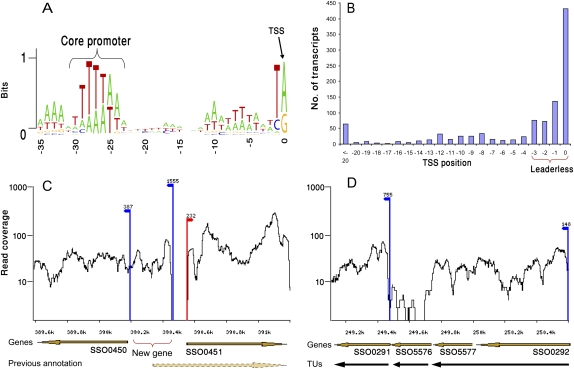
We defined the dominant TSS of annotated genes as the upstream site supported by the highest number of 5′-end reads (Fig. 1B). To verify that the defined sites represent genuine TSSs, we examined the nucleotide composition upstream to these sites. Most archaeal genes are known to be preceded by a TATA-like conserved core promoter motif, centered ∼26 bp upstream of the TSS (Zillig et al. 1988; Reiter et al. 1990); indeed, the vast majority of our defined TSSs were preceded by this core promoter, with a BRE element (Soppa 1999) appearing upstream (Fig. 2A). In addition, our data show that transcription in S. solfataricus invariably begins with a purine, with a pyrimidine at position −1 (Fig. 2A). These results indicate that our TSS mapping is accurate to single-nucleotide resolution.
Figure 2.
The structure of the S. solfataricus transcriptome. (A) Core promoter. Positions relative to TSS are marked below the sequences. The height of each letter corresponds to its frequency at that position. The core promoter motif in Archaea is indicated. The plot was prepared using the WebLogo software tool (Crooks et al. 2004). (B) Distribution of mapped TSS positions relative to the ORF ATG codon (n = 1040). Position 0 depicts transcripts beginning exactly at the adenine of the ATG. (C) Example for correction of gene annotations. (Dashed arrow) Marks the previous annotation of gene SSO0450 (conserved protein implicated in secretion) that was computationally predicted to begin in position 390,329. Transcriptome data indicate that it actually begins 228 bp downstream. A new protein-coding gene, having homology with a small heat-shock protein Hsp20 in other archaea, is identified on the reverse strand. Read coverage is in log scale. (Red arrows) TSS on the forward strands; (blue arrows) TSS on the reverse strands. The number above TSS indicates the number of reads supporting transcript beginning at that position. (D) Definition of transcriptional units (TUs). Operon annotation is refined based on continuous expression and TSS existence. Four closely spaced genes might be predicted to appear in the same operon based on the distance between them. Transcriptome data show at least two separate TUs. A third TU, corresponding to gene SSO5576, does not show expression and might either represent an unexpressed gene or a spurious gene prediction.
We further examined the distance of the identified TSS from the annotated open reading frame (ORF). In the majority of the protein-coding transcripts (69%), the transcription began exactly at the “A” of the ATG translational start site or 1–3 bases upstream of the ATG, leaving no room for a 5′ Shine-Dalgarno ribosomal binding site leader (Fig. 2B). Therefore, these transcripts lack 5′ untranslated regions (UTRs), which, in bacteria, frequently have regulatory capacity. Such “leaderless” transcripts have been previously detected in Sulfolobus and other archaea, both in experimental and bioinformatics analyses (Tolstrup et al. 2000; Moll et al. 2002; Benelli et al. 2003; Brenneis et al. 2007); in these cases, the translation is presumed to be initiated by an initiator f-Met-tRNA bound to the AUG at the beginning of the mRNA (Benelli et al. 2003). Our results suggest that leaderless translation is the preferred strategy for translation initiation of operon-beginning genes in the Crenarchaeon S. solfataricus P2. This is in agreement with results obtained on 40 transcripts from Euryarchaeota (Brenneis et al. 2007), suggesting that the preference for leaderless transcripts might be an Archaea-wide trait.
Since leaderless transcripts in S. solfataricus are the rule rather than the exception, we tested whether 5′-UTR-containing transcripts shared unique features. We found that genes involved in protein translation were twofold over-represented among the genes with 5′ UTRs (P < 0.0008, corrected for multiple testing) (Supplemental Table S2). For example, 18/25 (72%) of expressed ribosomal protein genes that begin an operon have a 5′ UTR 8 bp or larger, suggesting a role for 5′ UTRs in the regulation of these genes.
In the vast majority of sequenced bacterial and archaeal genomes, genes are annotated using automated, error-prone gene prediction software (McHardy et al. 2004). Our data enabled the correction of 162 misannotated gene structures (Fig. 2C). In most corrected cases, the N terminus of the gene was shortened, reflecting the tendency of automated gene prediction to select the largest possible ORF. The large number of corrected gene predictions (5.3% of all annotated genes) suggests that transcriptome sequencing would be beneficial for enhancing the annotation in many sequenced bacterial and archaeal genomes.
Genes in microbial genomes are often arranged in operon structures, with one polycistronic mRNA encompassing several genes. We defined transcriptional units (TUs) in S. solfataricus as regions presenting a continuous coverage of whole-transcript reads, and that had a mapped upstream TSS (Fig. 2D; Methods). Overall, 1040 such TUs were defined, encompassing 1478 (48%) of the protein-coding genes in S. solfataricus. Most (72%) TUs were monocistronic, with only 80 TUs (8%) containing three genes or more. The largest number of genes in a single operon was 14 (ribosomal proteins operon, 601,227–607,782), and the largest TU was 8.7 kb long (purine biosynthesis operon, 542,822–551,545). This is the first experimentally determined operon map for an archaeon.
Noncoding RNAs and cis-antisense transcripts
The availability of a transcriptome map for S. solfataricus now allows the detection of new transcripts that were previously not annotated in the genome. Such transcripts include noncoding RNAs (ncRNAs), which were shown in other microorganisms to regulate important processes such as pathogenesis, iron metabolism, and quorum sensing (Bejerano-Sagie and Xavier 2007; Masse et al. 2007; Toledo-Arana et al. 2007). We have recently developed an algorithm that uses RNA-seq data to detect novel ncRNAs in bacteria, and experimentally demonstrated using Northern blots and microarrays that the predicted ncRNAs are indeed expressed in the bacterial cell (Yoder-Himes et al. 2009).
We searched evidence for transcribed ncRNAs in the transcriptome of S. solfataricus and found 390 transcriptional units not overlapping any annotated coding sequence (CDS). Of these, 80 probably represented unannotated protein-coding genes (Fig. 2C; Supplemental Table S1; Methods), as their downstream sequences matched proteins previously annotated in other genomes (75 cases), or because they preceded an ORF (five cases). Presumably, these ORFs were not previously predicted owing to their relatively short sizes (average 67 amino acids). The remaining 310 transcripts probably represent ncRNAs transcribed in S. solfataricus.
Two previous studies have used large-scale shotgun cloning of small transcripts to identify ncRNAs in S. solfataricus, and detected 57 and 45 ncRNAs, respectively (Tang et al. 2005; Zago et al. 2005). All these previously identified ncRNAs, as well as all previously characterized sno-like RNAs, were detected in our transcriptome data (although some were expressed only in one of the growth conditions) (Methods), further suggesting that our transcriptome map indeed represents the vast majority of the primary S. solfataricus transcriptome. We have also detected the RNase P ncRNA, which was previously missing from the S. solfataricus genome annotation. The ncRNAs detected here were generally highly conserved (P < 0.0005) in six closely related Sulfolobus islandicus genomes (Reno et al. 2009), further establishing them as possibly functional transcripts (Methods). Still, it remains to be determined which of these ncRNAs is functionally important, and under what conditions.
Dominant groups among the S. solfataricus ncRNAs included transposon-associated ncRNAs (28 cases), which putatively regulate transposition (Tang et al. 2005); CRISPR-associated small RNAs (18 cases), a group of ncRNAs that provide protection against phage and plasmid infection (Sorek et al. 2008); and C/D-box RNAs, which guide methylation sites in rRNAs or tRNAs (13 cases) (Supplemental Table S3; Zago et al. 2005). Some guide RNAs were found in repetitive genome regions, where reads could not be uniquely mapped; such cases are absent from our annotations.
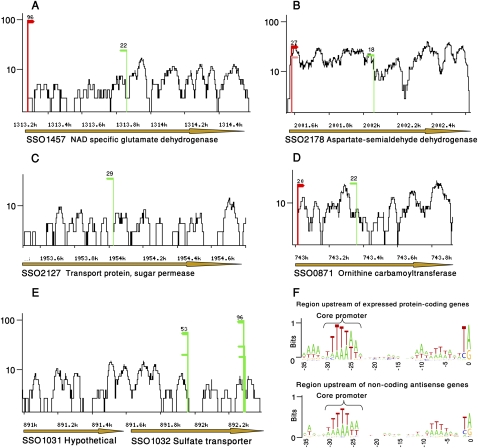
Interestingly, the majority of ncRNAs we discovered (185 cases, 60%) corresponded to cis-encoded antisense RNA transcripts (Fig. 3; Supplemental Table S1). These transcripts reside on the strand opposite to another gene, and are thus fully complementary with their targets. A handful of chromosomally encoded cis-antisense transcripts were documented in bacteria and were shown to regulate translation, mRNA stability, and mRNA degradation (Brantl 2007; Toledo-Arana 2009). We examined whether the antisense transcripts in S. solfataricus preferentially overlap specific types of genes. Genes involved in ion transport and metabolism were found to be threefold over-represented than expected among genes having overlapping antisense transcripts (P < 0.02, corrected for multiple testing), suggesting that antisense transcription might be a common regulatory mechanism for such genes in S. solfataricus (Supplemental Table S1; Methods). Membrane transporters were previously documented to be regulated by cis-antisense in bacteria (Chen and Crosa 1996).
Figure 3.
Cis-antisense transcription in S. solfataricus. Green TSS positions represent transcripts encoded from the strand opposite to the protein-coding gene. Read coverage is in log scale. (A) Antisense transcription in the SSO1457 locus, encoding NAD-specific glutamate dehydrognase. (B) Antisense transcription in the SSO2178 locus, encoding aspartate semialdehyde dehydrognase. (C) Antisense transcription in the SSO2127 locus, encoding a sugar permease (sense transcription not observed). (D) Antisense transcription in the SSO0871 locus, encoding ornithine carbamoyltransferase. (E) Antisense transcription in the SSO1032 locus, encoding a sulfate ABC transporter (sense transcription not observed). (F) Antisense transcripts have promoters embedded within the protein-coding genes they overlap. Sequence motifs upstream of protein-coding genes (upper panel) compared to motifs upstream of noncoding antisense transcripts (lower panel). Positions relative to TSS are marked below the sequences. The height of each letter corresponds to its frequency at that position. The core promoter motif in Archaea is indicated. The plot was prepared using the WebLogo software tool (Crooks et al. 2004).
Our results show that the regions upstream of the TSS of antisense transcripts contain an archaeal core promoter in positions −29 to −23, similar to protein-coding sense genes (Fig. 3F). Interestingly, the promoters of most (75%) of the antisense transcripts were embedded in the protein-coding region of the sense gene (Fig. 3), imposing an evolutionary constraint on the protein sequence. If, indeed, the antisense transcripts hold a regulatory capacity, then our data suggest that these sense genes “carry” their antisense regulation within their own sequence.
We note that the number of 185 antisenses we detected in S. solfataricus is probably very conservative, as it only represented transcripts constitutively expressed in all three growth conditions examined, and were supported by more than 10 sequence reads. Additional, condition-specific antisense transcripts might be detected if thresholds are relaxed. Still, cis-antisense transcription has never been observed in this scale for any Bacteria or Archaea to date but has been documented in many eukaryotes including Arabidopsis, Drosophila, and human (Yamada et al. 2003; Yelin et al. 2003; Numata et al. 2007; see Discussion).
RNA degradation
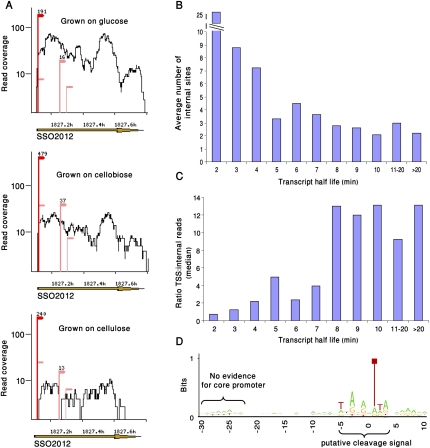
Despite the general concentration of sequenced 5′-ends near annotated beginnings of genes, additional internal sites were found within most transcribed RNAs (Fig. 4A; Supplemental Table S1; Methods). We hypothesized that these sites correspond to positions where the RNAs are cleaved by endoribonucleases to promote RNA turnover/degradation. To test this hypothesis, we compared our data with a transcriptome-wide map of S. solfataricus RNA half-lives collected through microarray analysis (Andersson et al. 2006). We detected an inverse correlation between RNA half-life and the number of internal sites (Spearman correlation, ρ = −0.9; P < 0.00023), that is, genes with shorter half-lives tended to have a larger number of internal sites (Fig. 4B). Similarly, the ratio between the number of reads in the TSS and the number of reads in the internal sites strongly correlated with half-life (Spearman correlation, ρ = 0.9; P < 0.00015) (Fig. 4C). This correlation was not dependent on transcript size. These results suggest that the internal sites we observe are hotspots for transcript cleavage in the process of RNA destabilization.
Figure 4.
Sequencing the 5′ ends of RNA degradation products. (A) Internal sites in RNAs. Shown are the transcriptome data on gene SSO2012. (Red) TSS; (pink) internal sites. The three different sequencing samples are presented to show the reproducibility of internal sites between samples. Numbers above dominant sites represent the number of reads supporting the site. (B) Negative correlation between the observed number of internal sites in our data and transcript half-life as measured in a global survey of S. solfataricus RNA stability (Andersson et al. 2006). Transcripts with shorter half-lives tend to contain a higher number of internal sites (Spearman correlation, ρ = −0.9). (C) Ratio between the number of reads in TSS and number of internal reads is positively correlated with transcript half-life (Spearman correlation, ρ = 0.9). (D) A sequence motif at the position of internal sites. Nucleotide frequency at internal sites (n = 1240) was plotted using the WebLogo tool (Crooks et al. 2004). Position 0 represents the first sequenced nucleotide, that is, the first nucleotide 3′ to the RNA cleavage position. The core promoter is largely absent from the region upstream to this motif.
The reproducible sequencing of the same internal sites across several growth conditions implies that there might be a preference for cleavage at specific sites. By calculating the base distribution surrounding 1240 dominant internal sites, we detected the position-specific motif TNRNR|NTDR (where | marks the cleavage position, R = A/G, D = A/G/T), which was 10-fold over-represented at cleavage positions than at random positions taken from the same genes (Fig. 4D). This motif might, therefore, be a preferential recognition site for cleavage in the RNA, thereby beginning the cascade leading to RNA degradation. Our results therefore suggest that RNA degradation in S. solfataricus possibly includes cleavage at specific positions within the transcript. These results also show that 5′-end RNA sequencing is a useful tool for documenting RNA degradation in Bacteria and Archaea.
In Gram-negative bacteria, RNase E binds RNAs that are marked for degradation and generates internal cleavages in AU-rich sites, resulting in RNA fragments that are degraded in the 3′–5′ direction by the exosome (Evguenieva-Hackenberg and Klug 2009). In Archaea, no endoribonuclease was identified to be responsible for internal mRNA cleavage, and RNA degradation is thought to be driven mainly by the exosome from the 3′ end. However, some S. solfataricus proteins presented endoribonucleolytic activity in vitro (Evguenieva-Hackenberg and Klug 2009). It might be hypothesized that one or more of these endoribonucleases is responsible for the preferential cleavage we had observed.
Discussion
Most sequenced bacterial and archaeal genomes are annotated almost solely based on gene prediction software that can detect CDS and a limited set of ncRNAs. Such annotations are error-prone, fail to predict small protein-coding genes, cannot reliably determine operon structures, and leave most ncRNAs and UTRs undetected. In S. solfataricus, our whole-transcriptome sequencing approach enabled correction of 162 gene annotations, defined 80 new expressed ORFs, detected more than 300 ncRNAs (including the conserved RNase P gene), and defined the operon structures of more than 1000 transcriptional units. The complete extended annotation can be viewed in an interactive browser at http://www.weizmann.ac.il/molgen/Sorek/Sulfolobus_solfataricus_transcriptome/index.html.
Although our approach accurately maps the exposed 5′ ends of transcripts in the cell, it has several limitations. First, the 3′ ends of transcripts are inferred from the whole-transcript coverage only and are thus not accurately determined. Second, our approach detects transcripts constitutively expressed in all three growth conditions examined and hence defines the “primary” transcriptome. Regulated transcripts, expressed in only one of the conditions (or in a condition not tested), have not been annotated here. Indeed, only 48% of the defined operons in this study showed constitutive expression under the growth conditions examined. To map the expression of every gene in the genome, one must grow the organism under a large array of conditions that would allow expression of every gene. This, however, was out of the scope of this study.
A recent study has documented a large number of internal TSSs within operons of the archaeaon Halobacterium salinarum NRC-1 (Koide et al. 2009). Although some of the internal sites we identified might represent sites where transcription was initiated, the lack of core promoter signal upstream of these sites indicates that the majority of internal sites do not represent internal TSSs (Fig. 4). Furthermore, the strong correlation of these sites with RNA degradation data, and the motif we detected at the suggested cleavage positions, suggest that most of these sites are cleavage products rather than sites where new transcription was initiated.
In Bacteria, regulation by noncoding RNAs is mostly mediated by base-pairing with the 5′ UTR of target mRNAs (Waters and Storz 2009). The general lack of 5′ UTRs in the archaeal transcriptome points to a different mode of ncRNA action. Based on the observation that archaeal mRNAs have 3′ UTRs of significant sizes (Brenneis et al. 2007), it is conceivable that ncRNA regulation is mediated by interaction with the 3′ UTR, as in eukaryotes. Indeed, Tang et al. (2005) have demonstrated long regions of complementarity between S. solfataricus ncRNAs and 3′ UTRs of protein-coding mRNAs.
The results of this study indicate that at least 8% of S. solfataricus operons are overlapped by antisense transcripts. Cis-encoded antisense transcripts have been previously documented in Archaea (Tang et al. 2005; Straub et al. 2009) and in Bacteria (Brantl 2007), but not to this extent of abundance. However, such antisense transcripts were documented in many eukaryotic species, including mammals, plants, and flies (Yamada et al. 2003; Yelin et al. 2003; Numata et al. 2007), where such transcripts have been implicated in genomic imprinting, RNA interference, translational regulation, alternative splicing, X-inactivation, and RNA editing (Lavorgna et al. 2004). In human, where antisense transcription was extensively characterized experimentally, it is estimated that 5%–25% of transcripts have a cis-antisense counterpart (Yelin et al. 2003; He et al. 2008). The rareness of cis-antisense documentation in Bacteria might reflect the fact that very few strand-sensitive whole-transcriptome sequencing efforts were done in Bacteria to date; alternatively, this mechanism might be unique to Archaea and eukaryotes. Whether the S. solfataricus antisense transcripts are functionally important, and the mechanism by which these transcripts might regulate their sense counterparts, remains to be determined.
Methods
Growth of S. solfataricus
All salts and reagents used were reagent grade and obtained from Sigma. S. solfataricus (DSMZ 1617) was grown in defined modified Brock's mineral medium containing (final concentrations) 0.25 g/L CaCl2·2H2O, 0.25 g/L KH2PO4, 1.3 g/L (NH4)2SO4, 0.28 g/L MgSO4·7H2O, 0.02 g/L FeCl3·6H2O, and trace mineral solution (100×) containing 10 nM MnCl2, 120 nM Na2B4O7, 640 nM ZnSO4, 0.5 nM CuCl2, 0.1 nM NaMoO4, 0.1 nM VOSO4, 0.03 nM CoSO4, and 0.09 nM NiSO4. The final pH of the solution was adjusted to pH 3.5 with H2SO4. Where mentioned, glucose, cellobiose, and avicel (microcrystalline cellulose) were used as the sole carbon source at a final concentration of 0.4% (w/v). The cells were grown to mid-log phase at 80°C at 170 rpm, and growth was monitored using direct cell counts with a light microscope.
Total mRNA isolation of S. solfataricus
Total RNA was isolated from S. solfataricus using the TRIzol method. Fifty milliliters of cells pelleted from mid-log phase culture was washed in 1 mL of medium and resuspended in 100 μL of buffer containing 25 mM Tris/HCl (pH 8.0), 1 mM EDTA, and 100 mM NaCl (Buffer A). After 15 min of incubation on ice, 100 μL of Buffer A supplemented with 1.6% N-lauryl sarcosine (w/v) (sodium salt; Sigma), 0.12% (v/v) Triton X-100 (Buffer B; Sigma), and incubated for 30 min at room temperature. One milliliter of TRIzol reagent (Invitrogen) was added to the solution. The resulting mixture was mixed by inverting the tube and incubated for 5 min at room temperature. Two hundred microliters of chloroform (Sigma) was added the tube, and the resulting mixture was shaken vigorously and centrifuged at 14,000 rpm for 15 min. Following phase separation, the upper phase was removed and pipetted into a separate centrifuge tube. Five hundred microliters of isopropanol was added to the separated supernatant, and the tube was centrifuged at 12,000 rpm for 15 min. The resulting pellet was washed with 70% ethanol, air-dried, and resuspended in 40 μL of dH2O.
cDNA preparation
5′-end RNA-seq
For each sample, 10 μg of total RNA was incubated with 2 U of TAP (Epicentre) for 1.5 h at 37°C to generate 5′ monophosphate RNAs. 3′ Ends were blocked by incubating the TAP-treated RNA with 2.5 μL of 100 mM NaIO4 for 1.5 h at 4°C in the dark. Excess NaIO4 was saturated by adding 1/10 volume of 1 M lysine and incubating for 10 min at room temperature. Each reaction was then passed through NucAway spin columns (Ambion) to remove salts as instructed by the manufacturer. The 56-bp Illumina 5′ adapter (in a single-strand RNA form) was ligated to the resulting treated RNA by using T4 RNA ligase (Invitrogen) in the presence of RNase inhibitor (Invitrogen) in a final volume of 50 μL as instructed. The reaction was incubated overnight at 16°C. Five microliters of 5 M NH4OAc and 125 μL of ethanol were then added, and the reaction was chilled for 20 min at −20°C. The sample was spun at maximum speed for 15 min, and ethanol was removed. After a second wash with 70% ethanol, RNA was dissolved in 10 μL of RNase-free water. For the first strand synthesis of cDNA, the SuperScript First Strand Synthesis system (Invitrogen) was used according to the manufacturer's instructions. The primers used were random hexamers attached to the Illumina 3′ adapter. RT products were cleaned using Montage Clear columns (Millipore) to remove primer dimers. The cleaned RT product was then amplified using primers matching the 5′ and 3′ Illumina adapters for 20 cycles of PCR. The resulting cDNA was run on a 3% low melting agarose gel, a gel slice containing cDNA sized 150–500 bp was taken, and cDNA was extracted. The retrieved cDNA was sent for sequencing using the Illumina Genome Analyzer.
Whole-transcript RNA-seq
Ten micrograms of total RNA was treated with RQ1 RNase-free DNase in a final volume of 50 μL. The reaction was incubated for 30 min at 37°C and stopped by addition of RQ1 DNase stop solution and incubation for 10 min at 65°C. Double-strand cDNA (ds-cDNA) was generated using the Double Stranded cDNA Synthesis kit (Invitrogen) as instructed, except that the oligo(dT) primers were replaced by random hexamers for first-strand synthesis. The resulting ds-cDNA was cleaned as described above and sent for sequencing using the Illumina Genome Analyzer per the manufacturer's instructions (including the nebulization and adapter ligation shown in Fig. 1).
Read mapping and analysis
Sequencing reads were mapped to the S. solfataricus P2 genome (GenBank: NC_002754) using BLASTN with an E-value of 0.0001 and the “–F F” flag. Only reads mapped by at least 33 bp to the genome with up to two mismatches were taken into account. Under these parameters, BLASTN produces alignments similar to those of Maq (Pearson correlation ∼ 1, P = 0). The alignment with the best bit-score was accepted as the correct mapping for each read. Reads having more than one best-scoring position on the genome, as well as reads mapped to rRNA, were discarded. Since rRNA molecules are larger, on average, than mRNAs, there was more rRNA representation in the whole-transcript data (Table 1). A transcript coverage map was calculated based on the alignment of whole-transcript reads. The background expression level was determined as the expression in the tenth percentile of the lowest expressed genes. The calculated background was 0.8 reads/bp.
For each read, the position where the alignment began (associated with the alignment strand relative to the genome) was recorded. “Reproducible 5′-end sites” were defined as positions supported by at least five reads in the cellobiose sample, four reads in the cellulose sample, and two reads in the glucose sample. These parameters normalize the differences between sample sizes, that is, the different number of reads obtained for each of the growth conditions.
Gene annotation for S. solfataricus P2 was downloaded from GenBank. For each gene, reproducible sites that overlapped its protein-coding region, as well as those residing in the intergenic region upstream of its beginning, were associated with the gene. Gene-associated sites were defined as such only if their strand matched the direction of transcription of their respective genes. Sites were divided to “upstream sites” (those that occur upstream of the gene start or overlap the first nucleotide of the ORF) and “middle sites” (those occurring within the ORF). The dominant upstream site, that is, the one supported by most sequences, was further considered as the TSS of the gene. All the associated sites for each gene are summarized in Supplemental Table S1.
Operon definition
Genes that start an operon were defined as genes with an upstream reproducible 5′-end site, or genes whose upstream genes were located on the other strand. A new operon was defined when a significant change (greater than twofold) of read coverage occurred between genes for >36 bp (read size). In cases in which expression data were absent, genes that were <40 bp apart and located on the same strand were defined as belonging to the same operon (Tolstrup et al. 2000). Operon definitions obtained through this approach were manually refined to account for large fluctuations in sequencing coverage.
ncRNA discovery
Reproducible sites that were not found to overlap (or to be upstream of) any annotated gene in the correct strand were analyzed as possibly encoding the 5′ ends of noncoding RNAs. These sites were scanned for downstream ORFs. Detected ORFs were tested using BLASTP for homology with known proteins (E-value = 1 × 10−4). ORFs having similarity to known proteins were classified as misannotated genes, and ORFs sized greater than 50 amino acids with no homologs were classified as possible novel proteins.
The transcript length of ncRNAs was estimated by scanning the read coverage, produced by the whole-transcript sequencing, downstream to the reproducible site. A sharp decline in the read coverage was considered the end of the transcript based on a manual assessment. The predicted sequences were tested for homology with known ncRNA genes using BLASTN (E-value = 1 × 10−6, word = 4).
Conservation of novel ncRNAs detected in this study was determined by comparing them with sequence homologs in six S. islandicus genomes (downloaded from NCBI: NC_012588, NC_012589, NC_012622, NC_012623, NC_012632, NC_012726) using BLASTN (E-value = 1 × 10−6, word = 7). As a control, random intergenic regions of the same sizes that were not predicted as ncRNAs were similarly compared to the six S. islandicus genomes. The negative control process was repeated 2000 times to generate a statistical P-value for ncRNA conservation.
Identification of previously known ncRNAs
Sequences of known ncRNAs were downloaded from NCBI. These sequences were aligned to the genome using BLASTN (E-value = 1 × 10−6, word = 4) and were searched for expression. In cases in which an ncRNA was found in at least one of the growth conditions, it was determined as identified. ncRNAs aligned to more than one genomic position with the same score, for example, ncRNAs from repetitive elements, were determined as identified if reads that aligned to these genomic positions were found. Coverage of whole-transcript sequencing data in isolated intergenic regions that lacked a sequenced 5′ end was also considered as ncRNA.
COG analysis
Genes were sorted to the different COG functional groups according to the COG data available on the NCBI site (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=Retrieve&dopt=Protein+Table&list_uids=180). Only genes that showed expression were analyzed. Genes that didn't show any expression, as well as those for which no COG was assigned, were excluded from further analysis. COG functional groups with less than 20 genes that showed expression were discarded from subsequent analysis because of lack of statistical power. Enrichment of antisense or genes with 5′ UTRs within a specific COG group was tested using hypergeometric probability followed by Bonferroni correction for multiple testing.
RNA cleavage/degradation
Internal sites were defined as reproducible sites that were found in the middle of a gene sequence, on the same strand. Only sites found more than 50 bp downstream from the annotated ORF beginning (ATG) were analyzed as internal sites. In cases where several sites were closely spaced on the transcript, only the dominant site was selected, that is, the site supported by most reads in windows of 50 bp. Dominant sites (n = 1240) were used for the preparation of the plot in Figure 4D.
Acknowledgments
We thank Naama Barkai, Dvir Dahary, Zohar Biron-Sorek, Debbie Lindell, Oded Beja, Bareket Dassa, Gil Amitai, Yonit Halperin, Shula Michaeli, Igor Ulitsky, and Gadi Schuster for scientific discussion and comments on earlier versions of the manuscript. We also thank David Bernick for assistance in the RNA preparation protocols, and Shirley Horn-Saban and Daniella Amann-Zalcenstein for assistance in Illumina sequencing. R.S. was supported by the Alon Fellowship, the Y. Leon Benoziyo Institute for Molecular Medicine, The Crown Human Genome Center, and the M.D. Moross Institute for Cancer Research. O.W. was supported by the Kahn Center for Systems Biology of the Human Cell, and is grateful to the Azrieli Foundation for the award of an Azrieli Fellowship.
Footnotes
[Supplemental material is available online at http://www.genome.org. Raw sequence data, reads alignment files, and a coverage map for all samples have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE18630. Transcriptome viewer is available at http://www.weizmann.ac.il/molgen/Sorek/Sulfolobus_solfataricus_transcriptome/index.html.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100396.109.
References
- Andersson AF, Lundgren M, Eriksson S, Rosenlund M, Bernander R, Nilsson P. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr Opin Microbiol. 2007;10:189–198. doi: 10.1016/j.mib.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Bell SD, Jackson SP. Transcription and translation in Archaea: A mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- Benelli D, Maone E, Londei P. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol Microbiol. 2003;50:635–643. doi: 10.1046/j.1365-2958.2003.03721.x. [DOI] [PubMed] [Google Scholar]
- Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Brenneis M, Hering O, Lange C, Soppa J. Experimental characterization of cis-acting elements important for translation and transcription in halophilic Archaea. PLoS Genet. 2007;3:e229. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Chen Q, Crosa JH. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J Biol Chem. 1996;271:18885–18891. doi: 10.1074/jbc.271.31.18885. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Klug G. RNA degradation in Archaea and Gram-negative bacteria different from Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:275–317. doi: 10.1016/S0079-6603(08)00807-6. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide T, Reiss DJ, Bare JC, Pang WL, Facciotti MT, Schmid AK, Pan M, Marzolf B, Van PT, Lo FY, et al. Prevalence of transcription promoters within archaeal operons and coding sequences. Mol Syst Biol. 2009;5:285. doi: 10.1038/msb.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- McHardy AC, Goesmann A, Puhler A, Meyer F. Development of joint application strategies for two microbial gene finders. Bioinformatics. 2004;20:1622–1631. doi: 10.1093/bioinformatics/bth137. [DOI] [PubMed] [Google Scholar]
- Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- Numata K, Okada Y, Saito R, Kiyosawa H, Kanai A, Tomita M. Comparative analysis of cis-encoded antisense RNAs in eukaryotes. Gene. 2007;392:134–141. doi: 10.1016/j.gene.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH. The structure and complexity of a bacterial transcriptome. J Bacteriol. 2009;191:3203–3211. doi: 10.1128/JB.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, Yu L, Assefa SA, He M, Croucher NJ, Pickard DJ, et al. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 2009;5:e1000569. doi: 10.1371/journal.pgen.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Hudepohl U, Zillig W. Mutational analysis of an archaebacterial promoter: Essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci. 1990;87:9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci. 2009;106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. Normalized nucleotide frequencies allow the definition of archaeal promoter elements for different archaeal groups and reveal base-specific TFB contacts upstream of the TATA box. Mol Microbiol. 1999;31:1589–1592. doi: 10.1046/j.1365-2958.1999.01274.x. [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Straub J, Brenneis M, Jellen-Ritter A, Heyer R, Soppa J, Marchfelder A. Small RNAs in haloarchaea: Identification, differential expression and biological function. RNA Biol. 2009;6:281–292. doi: 10.4161/rna.6.3.8357. [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- Tolstrup N, Sensen CW, Garrett RA, Clausen IG. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles. 2000;4:175–179. doi: 10.1007/s007920070032. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM, Sorek R. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci. 2009;106:3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago MA, Dennis PP, Omer AD. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:1812–1828. doi: 10.1111/j.1365-2958.2005.04505.x. [DOI] [PubMed] [Google Scholar]
- Zillig W, Palm P, Reiter WD, Gropp F, Puhler G, Klenk HP. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988;173:473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]