Abstract
Endogenous retroviruses (ERVs) contribute to a range of germline, as well as somatic mutations in mammals. However, autonomous retrotransposition of potentially active elements has not been demonstrated in the rat genome. We cloned an insertion that disrupted the normal splicing of the Cntrob gene that was subsequently identified as a nonautonomous, novel endogenous retrovirus of the RnERV-K8e family. The RnERV-K8e family is closely related to the recently reported MmERV-K10c elements, but differs from the autonomous mouse MusD or IAP families. In addition, we identified a novel, unexpectedly close relative of RnERV-K8e in the mouse, suggesting ERV-K cross-species transmission between mice and rats. We cloned a potentially autonomous RnERV-K8e element identified by in silico analysis and, using an in vitro retrotransposition assay, demonstrated that it is capable of retrotransposition. This particular element (named Rat-ρ, pronounced “retro”) encodes a retroviral envelope gene (env); however, env is not required for de novo retrotransposition events. Significant levels of RnERV-K8e-associated genetic polymorphisms were detected among inbred rat strains, suggesting ongoing retrotransposition in the rat genome. This study identifies an ERV-K-type family in rats that shows obvious signs of recent activity. Ongoing retrotranspositional activity may significantly add to genomic variability among inbred rat strains.
Endogenous retroviruses (ERVs) are the result of ancestral infections by retroviruses that became incorporated into germline DNA, and have been transmitted in a Mendelian manner. ERVs and ERV-derived sequences have been detected in all vertebrates studied, and they comprise around 9% of the rat genome (Gibbs et al. 2004). In contrast to lentiviruses that primarily amplify by infection and rarely function as endogenous retroviruses (Katzourakis et al. 2007), ERVs are prone to have two distinct ways of amplifying their genomes. First, they can function as conventional retroviruses and infect the host. Second, they acquire an intracellular lifestyle, and amplify by a “copy-and-paste” mechanism of retrotransposition, in a way similar to other retrotransposons. During the process of endogenization, the ERVs tend to lose their retroviral envelope gene along with their ability to leave their host cells, even though the retrotransposition reaction may remain dependent on env function (Dewannieux et al. 2006). Recent activities of endogenized retroviruses being responsible for a range of germline, as well as somatic mutations, have been reported in mammals (Maksakova et al. 2006; Moyes et al. 2007). Somatic integration events influencing gene expression have been reported in the rat genome since 1975 (Mori et al. 1975), indicating ERV activity in rat populations. However, all of the reported transcripts or novel insertions of rat ERV elements are fragmented, and contain inactivating mutations. In silico searches identified several families that belong to β-ERVs in the rat genome, including the young, potentially autonomous Rattus norvegicus RnERV-β5 elements (Baillie et al. 2004). Furthermore, in silico sequence analyses predicted two ERV1, as well as four ERV2 families, that may still be active in rats (Gibbs et al. 2004). Among these, RnNICER2 and RnNICER3 (ERV1) are closely related to the mouse-specific MuLV retrovirus, whereas RnERVK3 (ERV2) is distantly related to the simian retroviruses (Gibbs et al. 2004). Nevertheless, autonomous retrotransposition of these potentially active elements has not been demonstrated. In contrast, the “master” copies of several endogenized retrovirus families responsible for most of the insertional germline mutations in mice (Zhang et al. 2008) have been described, including the intracisternal A particle (IAP), the MusD/early transposon (ETn), and the GLN families (Dewannieux et al. 2004; Ribet et al. 2004, 2008b).
During our positional cloning efforts to identify the rat hd mutation (Krenova et al. 1999), we performed extensive sequencing of the critical region on chromosome 10 on the genomic and cDNA level (data not shown). Compared to wild-type reference rats, we noticed a chimeric transcript of Cntrob (previously known as Lip8) in hd/hd mutant rats. Here we establish that the hd locus in rats contains a recent integration of a copy of the novel ERV-K8e family, resulting in the inactivation of the Cntrob gene through aberrant splicing. We demonstrate that one particular copy of the highly reiterated RnERV-K8e family, closely related to the element that leads to the aberrant splicing in Cntrob, is capable of retrotransposition, and encodes an intact env ORF with the potential to infect rat cells. Thus, we identified an autonomous element (that we named Rat-ρ) of a yet uncharacterized ERV-K family in rats. This family, classified as RnERV-K8e, shows obvious signs of recent activity in rats and indications of occasional transmission and recombination events with related ERV-K elements in mice. We demonstrate that, due to its ongoing activity, the RnERV-K8e family is polymorphic among inbred rat strains. We propose that RnERV-K8e activity is an underestimated source of genome variability in rats, and speculate that ongoing activity of RnERV-K8e may contribute to phenotypic variability among rat strains.
Results
A retrotransposon insertion introduces cryptic splice signals that override normal Cntrob splicing
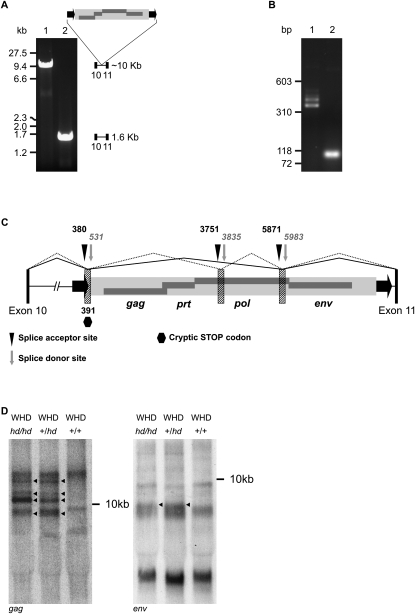
We discovered a large insertion in intron 10 of Cntrob in hd/hd mutant animals (Fig. 1A). Using long-range genomic PCR spanning exon 10, intron 10, and exon 11 of Cntrob, the hd/hd homozygous rats showed a ∼10 kilobases (kb) product, whereas wild-type DNA shows the expected size of intron 10 of about 1600 bp (Fig. 1A). The 8-kb insertion maps 40 bp upstream of exon 11, and shows all the hallmarks of a recent integration of a retrovirus (6-bp target site duplication flanking an element consisting of identical long terminal repeats (LTRs) and internal sequence containing gag, prt, pol, and env genes).
Figure 1.
Insertion of a retrovirus into Cntrob. (A) Long-range PCR spanning exons 10 and 11 shows a ∼10-kb insertion in hd/hd homozygotes (lane 1), whereas the SHR control PCR product (lane 2) has the expected size (1578 bp). The Cntrob gene contains insertion of an endogenous retrovirus (ERV) in the sense orientation, containing all typical retroviral genes (gag, pol, prt, and env, dark gray rectangles) flanked by long terminal repeats (LTRs, black arrows). Exons 10 and 11 are represented by black rectangles. (B) Aberrant splicing of Cntrob due to the retroviral insertion. RT-PCR of total RNA from testes spanning exons 10 and 11 of Cntrob. Lane 1 shows multiple PCR products in a size range of 400–500 bps in mutant (hd/hd) rats. Lane 2 shows the expected 98-bp PCR product in SHR controls (+/+). (C) Cntrob-ERV chimeric transcripts identified by RT-PCR. Splice signals are designated as vertical, black triangles (splice acceptor) or as vertical, gray arrows (splice donor). Up to three short segments of the retrovirus (striped rectangles [not to scale]) incorporate into the mature Cntrob mRNA. Exons 10 and 11 of Cntrob are depicted as black bars (not to scale). Two out of five different splicing variants are shown (solid and dotted lines). STOP codon is marked as a hexagon. (D) Northern blotting of total RNA from testes of WHDhd/hd, WHD+/hd, and WHD+/+. The probe corresponded to either a gag (left panel) or an env (right panel) fragment of the Cntrob-ERV. A <9-kb transcript is detectable in all the samples. The multiple hd-specific bands are marked by arrows.
Because the retroviral integration occurred in an intron of the Cntrob gene, the mutagenic effect of the retroviral insertion on gene expression required further investigation. In order to analyze hd-specific transcripts, RT-PCR was performed on hd/hd mutant and control samples by using a primer combination spanning exons 10 and 11 (Fig. 1B). The multiple mRNA products were detected that were all longer than the expected size in wild-type control (98 bp), but shorter than the full-length retroviral mRNA. Sequence analysis revealed that the longer products were chimeric transcripts formed by Cntrob- and retrovirus-derived sequences, generated by various splicing events imposed by cryptic splice signals donated by the retrovirus (Fig. 1C; Supplemental Table S1). All identified aberrant splice donor/acceptor sites positioned a premature STOP codon in frame of Cntrob (position 391 bp in the ERV, Fig. 1C), giving rise to 480–496-aa long fusion protein products (GenBank accession nos. EF532346–EF532350). The truncated CNTROB protein (centrobin) products contain only the first 10 exons of Cntrob, while the wild-type protein is encoded in 19 exons and has 897 aa (GenBank accession nos. EF532342–EF532345). The truncated centrobin was detected by Western blot analysis in hd/hd homozygous rats using testis tissue. Heterozygous +/hd rats showed two bands: the expected full-length protein and the truncated protein; whereas homozygous wild-type rats expressed only the full-length protein (data not shown). Less abundant, but similar splicing products were detected from heart and brain, suggesting that alternative splicing is not specific to the testis (Supplemental Fig. S1). A band corresponding approximately to a full-length retrovirus transcript (∼9 kb) was detected on a Northern blot using either a gag or an env-specific probe (Fig. 1D). Additional, env-specific transcripts appeared in the hd/hd and +/hd samples; these bands were not detectable in the wild-type Wistar Hypodactylous (WHD) sample (Fig. 1D). The multiple bands are proposed to be various splicing variants of the Cntrob-ERV fusion. The majority of the splice products were detectable by the gag probe only, suggesting that the putative cryptic splice signals are not utilized with equal frequency. In sum, the data presented in Figure 1 indicate that some ERV-associated sequences are actively transcribed specifically in hd rats, which may reflect activity of the retroviral insertion in Cntrob.
The element inserted in Cntrob represents a novel ERV family
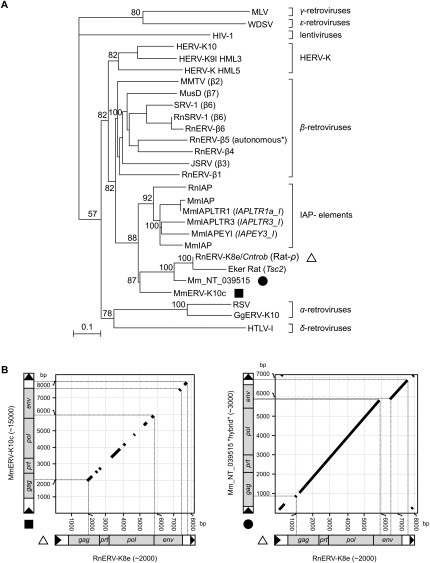
The finding of a de novo mutagenic insertion in the Cntrob gene suggests a recent retroviral integration event in the WHD inbred strain. The inserted element had characteristics of a full-length retrovirus, coding for the gag, prt, pol, and env genes (Fig. 1C). In addition to the ERV inserted into Cntrob, other ERVs, including IAP elements (Furter et al. 1989), as well as numerous ERV subfamilies of β-retroviruses have been reported from both the rat and mouse genomes (Baillie et al. 2004). Phylogenetic analysis based on nucleotide sequence alignments of retroviral pol genes of both exogenous retroviruses and ERVs (Fig. 2A) revealed that the Cntrob-element clusters together with the poorly characterized MnERV-K10c family in mouse, and does not group together with the IAP elements. Both the Cntrob- and the IAP elements form a cluster separate from β-ERVs, where the potentially autonomous RnERV-β5 (Baillie et al. 2004) is grouped (Fig. 2A). However, the LTRs of the Cntrob-element have no significant similarity to the LTRs of the rat IAP elements (Repbase, http://www.girinst.org), and show only partial similarity (95% identity over a 70-bp stretch of the 429-bp-long LTRs) to MmERV-K10c (Fig. 2B). Accordingly, the new element discovered in Cntrob was classified as RnERV-K8e (Repbase). Based on our analyses, the insertion in the Tsc2 gene that was previously classified as an IAP element (Xiao et al. 1995) can now be identified as a truncated copy of RnERV-K8e (Fig. 2A). Importantly, all but one of the IAP copies (Ribet et al. 2008a), and the MusD family lack the env ORF and form strictly intracellular virus-like particles. In contrast, several copies of both the rat and the mouse ERV-K family members contain intact env ORFs. The significant dissimilarity of LTR and env sequences between the RnERV-K8e and MmERVK-K10c families (Fig. 2B) supports a distinct phylogenetic history of the two subfamilies. However, occasional transmission events between the murine hosts might explain the emergence of novel ERV families (e.g., NT_039515.6) (Fig. 2A,B), represented by ∼3000 copies in the mouse genome (not yet annotated by Repbase). This new family is significantly more closely related to RnERV-K8e, consistent with recent transmission between species (Fig. 2). Most of the mouse and rat ERV-K elements with intact ORFs also possess identical or near-identical 5′ and 3′ LTRs (data not shown), a signature of recent and autonomous retrotransposition or infection of ERV-K families in murine species.
Figure 2.
Phylogenetic characterization of the RnERV-K8e family. (A) Phylogeny of retroviruses based on the neighbor-joining method using highly conserved nucleotide sequence regions of the pol proteins (Baillie et al. 2004) rooted using a non-LTR retrotransposon. Percent bootstrap values, obtained from 1000 replicates, are indicated. The ERV families are indicated on the right. MLV, Moloney murine leukemia virus; WDSV, walleye dermal sarcoma virus; HIV-1, human immunodeficiency virus type 1; HERV-K, human endogenous retrovirus type K; MMTV, mouse mammary tumor virus; MusD, Mus musculus type D element; SRV-1, Simian retrovirus type D; RnSRV-1, SRV-1 from Rattus norvegicus (Rn); RnERVβ(1-6), β(1-6)-type ERVs from Rattus norvegicus; JSRV, jaagsiekte sheep retrovirus; IAP elements, Intracisternal-A particle elements from Mus musculus (Mm), from Rattus norvegicus (Rn), or Mesocricetus auratus (Ma); RnERV-K8e (Rat-ρ), endogenous retrovirus identified in this study (indicated by a triangle); Eker rat, a nonautonomous element inserted into the Tsc2 gene; Mm_NT_039515 (indicated by a filled circle); MmERV-K10c (indicated by a filled square), ERV-K-type element from Mus musculus; RSV, Rous sarcoma virus; GgERV-K10, ERV-K-type element from Gallus gallus; HTLV-1, human T-cell leukemia virus type-1. All sequences are accessible from GenBank or Rebpase (see Supplemental material). Scale bar indicates nucleotide divergence. (B) Dot plots of RnERV-K8e and its two closest relatives were created using BLAST2 algorithm (parameters are given in Supplemental material). The RnERV-K8e element (triangle) is plotted on the x-axis; the y-axis represents MmERV-K10c (filled square, left) and Mm_NT_039515.6 (filled circle, right). For each subfamily, the estimated copy number is indicated in brackets. The thicknesses of the diagonal lines are out of scale. Accession numbers are given in the Supplemental material.
An RnERV-K8 copy on chromosome 20 is an autonomous element
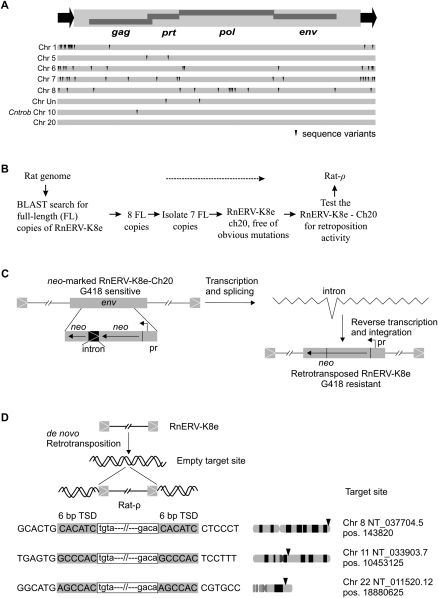
Although the particular copy of RnERV-K8e that disrupted Cntrob possessed all components of a full-length retrovirus, it is unlikely to be functional due to a single inactivating mutation in the gag gene (Fig. 3A). In silico analysis predicted ∼2000 copies of RnERV-K8e in the rat genome. While eight copies, including the Cntrob-element, were full-length elements (8.3 kb) (Fig. 3A), the rest of the copies were found incomplete due to truncations at the 5′-end (typical of retroelements), or at the env gene (data not shown)—a clear sign of an ongoing endogenization process (Ribet et al. 2007). Eight full-length copies of RnERV-K8e were analyzed in detail (Fig. 3A). Seven copies harbored sequence variants, but had 99% identity at the nucleotide level, consistent with recent activity. The spectrum of variants found in the different copies of RnERV-K8e includes also several potentially inactivating mutations. Only a single copy, a potentially autonomous element localized on chromosome 20 within the MHC complex was free of obvious mutations (Fig. 3A).
Figure 3.
Identification of an autonomous RnERV-K8e element. (A) Genomic organization of a full-length, autonomous RnERV-K8e element, with the 429-bp-long LTRs (black arrows) flanking a 8332-bp sequence containing four complete open reading frames (ORFs) encoding the retroviral gag, prt, pol, and env genes. Eight full-length copies of the RnERV-K8e were found in the rat genome (assembly RGSC v3.1, see Supplemental material). The dark triangles indicate sequence variants that likely render the RnERV-K8e nonfunctional (see also Supplemental Table S2). The copy inserted into Cntrob contains a single point mutation disrupting the gag gene. The copy found on chromosome 20 (of the BN strain) contains no obvious mutations and was tested for functionality in a retrotransposition assay. (B) Flowchart of the identification of an autonomous copy of the RnERV-K8e family. (C) A reporter neo-cassette was incorporated into the env gene of the RnERV-K8e-Ch20 element with the neo gene and a promoter (pr) in reverse orientation. The marker cassette is inactive due to the insertion of an intron. Retrotransposition is assayed by the activation of the neo gene. The estimated retrotransposition frequency is 5 × 105–2 × 106, one and two orders-of-magnitude lower than those of IAP (Dewannieux et al. 2004) and MusD elements (Ribet et al. 2004), respectively. (D) Examples of de novo retrotransposition events of the RnERV-K8e-Ch20 element isolated from genomic DNA of G418 resistant HeLa colonies. Insertion junctions are depicted with endogenous sequences in uppercase and proviral sequences in lowercase. The retrovirus is flanked by canonical 6-bp target site duplication sequences (TSD, shaded). Arrows mark the approximate chromosomal locations of the de novo retrotransposition events. Exact positions of the target sites are shown on the right.
Next we investigated whether this particular copy of RnERV-K8e is capable of retrotransposition (Fig. 3B). We constructed a genetically marked RnERV-K8e-Ch20 element that contained a neomycin cassette disrupted by an intron in reverse orientation in the env gene. An ex vivo retrotransposition assay based on transfection of the marked element into human HeLa cells followed by G418-selection (Fig. 3C; Dewannieux et al. 2004) was used to reveal retrotransposition activity. The resulting retrotransposition frequency was 5 × 10−5–2 × 10−6. Molecular analysis of three G418-resistant clones confirmed de novo retrotransposition events of the marked element into chromosomes 8, 11, and 22 flanked by 6-bp target site duplications (Fig. 3D). Thus, the ERV copy located on chromosome 20 is an autonomous element that we named Rat-ρ. Rat-ρ is the first, naturally occurring, autonomous copy of a young ERV-K endogenous retrovirus family demonstrated to be active in rats.
Transcription levels of RnERV-K8e are tissue- and strain-specific
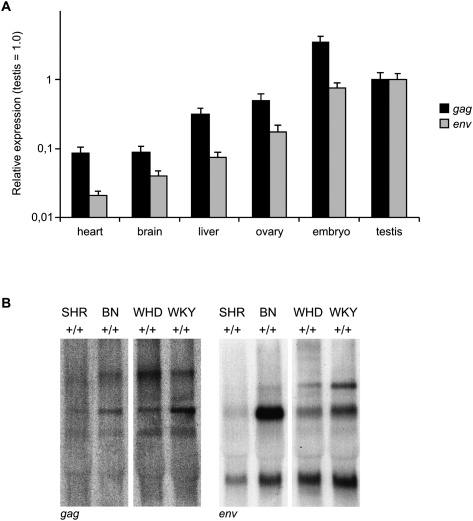
Using a quantitative PCR assay, a gag- or env-specific primer set detected different levels of ERV transcription in different tissues of Brown Norway (BN) animals (Fig. 4A). Notably, the level of env expression is about an order-of-magnitude higher than that of gag. In comparison to somatic tissues, both probes detected elevated levels of ERV transcription in embryonic- and testis-derived germinal tissues (Fig. 4A). Interestingly, varying transcription levels of RnERV-K8e were observed in testes of different wild-type strains by Northern blotting. In comparison to WHD, the abundance of the potential full-length transcript was relatively low in spontaneously hypertensive rat (SHR). In contrast, robust transcription was revealed in Wistar Kyoto rat (WKY) (probes gag and env) and in the BN sample (probe env) (Fig. 4B). The variation in ERV expression levels in the testes among different wild-type strains would suggest that ERV activity could potentially generate genetic polymorphism between inbred strains, as well as between different colonies of the same inbred strain.
Figure 4.
Characterization of RnERV-K8e-specific transcripts. (A) Quantitative TaqMan PCR analysis of RnERV-K8e transcription in different tissues of BN rats, with primers specific for the gag or env genes. Heart, brain (frontal lobe), and testis samples were collected from three males; ovaries were obtained from two females. The three midgestation (14.5 dpc) embryos were littermates. Each sample was run in six replicates, including the GAPDH control. Columns represent mean values, error bars show the standard error of the mean (SEM) (for statistical comparison see Supplemental material). Expression in testis is set to 1. (B) Polymorphic status of RnERV-K8 by Northern blotting of total RNA from testes of SHR+/+, BN+/+, WHD+/+, and WKY strains. Probe corresponds to gag (left panel) and env (right panel) fragment of the Cntrob-ERV, respectively. A <9-kb transcript (corresponding to the full-length of RnERV-K8e) is detectable in all the samples. The specific bands have various intensities in the different strains. Notably, the predicted full-length transcript detected by the env probe is highly abundant in BN testis.
The polymorphic status of RnERV-K8e indicates recent activity in inbred rat strains
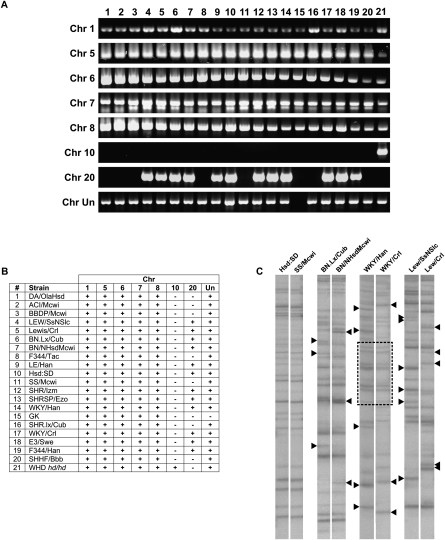
In order to investigate the polymorphic status of RnERV-K8e in different inbred strains, genomic PCR was employed to screen eight genomic loci for the presence or absence of the identified full-length elements on a panel of 21 inbred strains. RnERV-K8e copies were detected at genomic loci of chr 1, chr 5, chr 6, chr 7, and chr 8 in all of the tested inbred lines, indicating that RnERV-K8e insertions at these particular loci arose prior to divergence of inbred strains (Fig. 5A,B). In contrast, the elements at loci on chr 10, chr 20, and chr Un were polymorphic among the inbred strains (Fig. 5A,B). As expected, the de novo insertion at the Cntrob locus on chr 10 locus was detectable in the mutant strain (hd/hd) only (Fig. 5A,B). Notably, the autonomous Rat-ρ element located at the chr 20 locus, demonstrated to be active in retrotransposition, displays the highest degree of polymorphism. The polymorphic band at chr 20 was not detected in nine out of 21 strains, including the Cntrob mutant rat (hd/hd) (Fig. 5). Accordingly, the insertion into the Cntrob gene cannot be directly associated with the retrotranspositional activity of the chr 20 element (see Discussion). Importantly, the polymorphic elements are estimated to be fairly young on the basis of absence of sequence variants (chr 20) or the low number of inactivating mutations (1–3) on chr 10 and chr Un (Supplemental Table S2). Thus, the period when these loci became polymorphic and the inbreeding process were obviously overlapping. Based on the number of inbred strains involved, the emergence of the polymorphic status at the genomic loci occurred in the order of chr Un, chr 20, and chr 10 (Cntrob). The relatively high polymorphic status of the tested genomic loci (three out of eight) indicates a frequently occurring, recent retroviral activity in the inbred strains.
Figure 5.
Insertional polymorphism of RnERV-K8e. (A) Polymorphic status of eight genomic loci for RnERV-K8e in 21 inbred rat strains. One PCR primer is specific to the particular genomic locus, while the second is specific to RnERV-K8e (see Supplemental material). (B) Tabular representation of A. RnERV-K8e copies are present (+) or absent (−) at the particular genomic loci. (C) Transposon display performed on closely related inbred strains. Polymorphic bands are indicated by black triangles. Patterns identical between WKY/Han and WKY/Crl are framed.
In order to visualize genome-wide polymorphism of RnERV-K8e, a PCR-based transposon display assay (Waugh et al. 1997; Behura 2006) was performed that amplified genomic junction sequences flanking RnERV-K8e integrations. Because potential mobility is usually associated with young ERV copies, we chose to amplify the junctions of young elements. By designing the primers to highly conserved regions of RnERV-K8e, we visualized the junctions of around 25 copies (Fig. 5C). No detectable retroviral activity was observed in the related strains of Hsd:SD and SS/Mcwi (Fig. 5C, lanes 1, 2). In contrast, significant polymorphism was observed between either of the Rat-ρ-harboring BN.Lx/Cub and BN/NHsdMcwi or the LEW/SsNSlc and the Lewis/Crl strains. Similarly, a high degree of polymorphism was observed between the WKY/Han and WKY/Crl strains. Notably, due to the complicated breeding history of WKY, the substrains carry large chromosomal segments of different origins (Kurtz et al. 1989; Saar et al. 2008). Therefore, the complex pattern visualized by transposon display might not be explained exclusively by retrotransposition. In order to estimate the genomic diversity that is unrelated to retrotransposition, AFLP analysis was performed on the polymorphic strains shown in Figure 5C. The relatively low divergence revealed by the AFLP analysis (Supplemental Fig. S2) supports our assumption that the RnERV-K8e element actively contributes to genomic polymorphism in inbred rat strains. A large number of polymorphic loci (SNPs, microsatellites) were already characterized (Steen et al. 1999; Saar et al. 2008), indicating extensive genomic polymorphism among the 21 rat strains. However, our transposon display approach could be especially valuable for comparison of either closely related strains, or animals belonging to the same strain from different sources. In such cases, the genomic differences are expected to be low; thus, these polymorphisms might be attributed to retroviral activity with high probability.
Discussion
Earlier reports of recent insertions of ERV sequences suggested that active copies of ERVs might exist in rat populations (Xiao et al. 1995; Baillie et al. 2004). Here, we identified a novel, active family of ERVs in the rat genome, RnERV-K8e, that was discovered as a recent mutagenic insertion in the Cntrob gene associated with the hd phenotype (Liška et al. 2009). The mutagenic insertion in the Cntrob gene in an inbred strain argues for a relatively recent integration event, suggesting a currently ongoing, active phase of RnERV-K8e expansion in rats. However, the Cntrob copy of RnERV-K8e-carries an inactivating mutation (Fig. 3A). Thus, we also aimed at the identification and characterization of autonomous RnERV-K8e copies. By definition, autonomous elements are capable of producing all of the protein factors required for their amplification, and can mediate retrotransposition events. Finding an autonomous copy of an ERV is a challenging task. For example, despite ERV-associated polymorphisms in certain populations suggesting recent activity, no naturally occurring, autonomous HERV-K element was identified in humans (Steinhuber et al. 1995; Medstrand and Mager 1998; Barbulescu et al. 1999; Turner et al. 2001; Hughes and Coffin 2004; Moyes et al. 2007). The failure to find a HERV-K master copy inspired Dewannieux and colleagues to predict and reconstruct the sequence of an active, ancestral element (Dewannieux et al. 2006; Zhang et al. 2008), in a manner similar to the resurrection of the Sleeping Beauty transposon (Ivics et al. 1997). In a systematic search for an autonomous RnERV-K8e element, we have cloned and analyzed eight full-length copies, from which only one, residing on chromosome 20, seemed free of inactivating mutations (Fig. 3A). We demonstrated that this particular copy was capable of de novo transposition in a cell culture-based retrotransposition assay. Thus, we have successfully identified an autonomous copy of the rat RnERV-K8e family that we named Rat-ρ.
In contrast to the MusD family (Ribet et al. 2007), but similar to the human HERV-K elements (Dewannieux et al. 2005), Rat-ρ is encoding an intact env gene. The occasional lack of the env gene and the relatively large copy number (>2000 per haploid genome) suggest the RnERV-K8e family is undergoing endogenization (Fig. 6). An important step in this process is the loss of the env gene that might be followed by further adaptations to promote the intercellular life cycle of the ERV. Depending on the status of endogenization, ERV amplification might occur by either infection or retrotransposition only or by both of the processes. Strikingly, defective elements carrying different inactivating mutations or even large deletions can be often mobilized in trans by active copies (Mager and Freeman 2000; Horie et al. 2007; Ribet et al. 2008a). The transferred sequences can even be packaged into virions released from the newly infected cells (Evans et al. 2009).
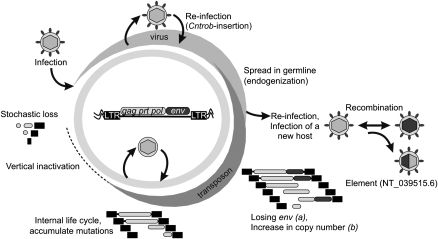
Figure 6.
Model of RnERV-K8e life cycle. An infectious retrovirus integrates into the germline of a host organism and is transmitted vertically. Such ERV may retain the capability to produce infectious particles with occasional re-infection or horizontal transmission. We propose that the Cntrob-RnERV-K8e insertion is a potential reinfection event. In addition to reinfection, the retrovirus has the potential to infect a new host and recombine with related ERVs. Emergence of a new ERV family, represented by NT_039515.6, might be the result of a transmission event between the rat and mouse hosts (Fig. 2A,B). Upon endogenization the infectivity is usually decreased, with eventual loss of the env gene and/or the plasma membrane targeting function (a). In addition, retrotransposition efficiency and copy number might dramatically increase (b). Considering the env–independent, but low retrotranspositional activity of Rat-ρ, the endogenization status of RnERV-K8e is proposed to be between steps a and b. The endogenized virus could finish its life cycle as a real retrotransposon. Strikingly, the exogenous and endogenous life cycles of the same retrovirus might coexist, giving rise to subfamilies (Ribet et al. 2008a).
The results of the in vitro retrotransposition assay raised two points regarding the endogenization status of RnERV-K8e. The assay demonstrated that the Rat-ρ autonomous element was capable of “copy-and-paste” retrotransposition without the requirement of a functional env. This feature highlights a significant difference between RnERV-K8e and HERV-K families, as HERV-K requires an active env gene for its retrotransposition (Dewannieux et al. 2006), possibly explained by their different stage of the endogenization process. Alternatively, the requirement for an active env gene in the retrotransposition reaction could be a strategy to prevent HERV-K-type elements from losing their env, and consequently prevent extinction. The functional independence of retrotransposition on env function, its robust transcription, as well as the significant polymorphism of RnERV-K8e observed in the BN and WKY strains, are signs of successful endogenization. On the other side, since the autonomous Rat-ρ copy could not be detected in WHD rats (Fig. 5A,B), it cannot be excluded that the Cntrob insertion itself was an infection event. The relatively low retrotransposition frequency of Rat-ρ (5 × 10−5–2 × 10−6) and the presence of an intact env gene in several copies suggest that RnERV-K8e elements may need env for efficient retrotransposition. Thus, similarly to the HERV-K family (Belshaw et al. 2004), it is possible that RnERV-K8e elements might amplify by both retrotransposition and infection, and infectious elements enter the rat genome by occasional re-infections (Fig. 6). Indeed, multiple autonomous copies might exist in the same genome, as it was reported for the IAP families (Ribet et al. 2008a). The presence of highly related ERV-K families in murine species (RnERV-K8e and a novel family represented by NT_039515.6) suggests that occasional transmissions between the rat and mouse hosts are an ongoing process (Fig. 6). Considering their high reiteration, both of these ERV-K families appear to have successfully established themselves in their host genome (Fig. 2B).
Although most polymorphic ERVs might have no significant effect on host genes, new retroviral insertions could affect gene expression in several ways (Zhang et al. 2008). The mutagenic effect is most obvious when the ERV directly hits an exon of a gene. Yet, in the majority of the cases, the strong transcriptional regulatory elements of ERVs affect host gene expression when the insertion occurs in an intron or nearby a coding sequence (Maksakova et al. 2006). For example, an intronic insertion can generate a truncated protein product by overriding the normal splicing process (Maksakova et al. 2006). Indeed, our analysis of the transcription of the hd allele revealed that the intronic RnERV-K8e insertion interferes with the normal splicing process of the Cntrob transcript, giving rise to multiple chimeric mRNA species. The identified aberrant splicing events (Fig. 1C; Supplemental Table S1) positioned a premature STOP codon in frame with the Cntrob ORF. Thus, the chimeric mRNAs are translated to truncated proteins containing only the first 10 of 19 exons of Cntrob, followed by a few amino acids derived from the retrovirus. The orientation of inserting transposable elements might have an impact on its mutagenic potential: For the L1 retrotransposons, sense intronic insertions are usually more mutagenic, while antisense insertions frequently escape purifying selection (Chen et al. 2006). Depending on the position of the insertion, it might be deleterious to protein function or can leave some function of the protein intact, potentially leading to dominant negative effects (Zhou et al. 2007). The lack of any abnormalities in hd heterozygotes argues against an interference of the truncated centrobin product with normal protein function (Liška et al. 2009). Furthermore, it is well documented that the strong promoter/enhancer activity of integrated retroviruses can influence transcription of nearby genes (Druker et al. 2004; Maksakova et al. 2006; Horie et al. 2007). This can appear as a germline mutation (Boeke and Stoye 1997; Maksakova et al. 2006) or manifest as somatic oncogene activations (Mori et al. 1975; Kung et al. 1991; Callahan and Smith 2000; Druker et al. 2004). This feature of retroviral promoters is utilized in specific genetic screens to identify oncogenes (Collier et al. 2005). It has been reported that ERVs can exhibit substantial transcriptional transactivation on genomic regions flanking the integration sites, occasionally generating chimeric transcripts between IAP sequence and endogenous genes (Horie et al. 2007).
Our finding of a naturally occurring, endogenous retrovirus that is active in the rat germline raises interesting questions as to what extent these novel elements contribute to genetic variation among rats or across mammalian genomes. In humans, de novo ERV insertions are rare and ERV-associated insertional polymorphism is very low (Moyes et al. 2007). In contrast, ERV-associated insertional polymorphism is extremely high in mice (Zhang et al. 2008). The polymorphic nature of a set of RnERV-K8e elements, confirmed by genomic PCR and transposon display (Fig. 5A–C), suggests that ongoing RnERV-K8e activity could result in a significant insertional polymorphism among inbred rat strains. In addition, RnERV-K8e might not be the only transposing ERV responsible for genetic variability in rats. Indeed, a functional L1 retrotransposon has been identified in rat chloroleukemia cells (Servomaa and Rytomaa 1990; Kirilyuk et al. 2008). Furthermore, elements identified in silico might add to the list of potentially active ERVs (Baillie et al. 2004; Gibbs et al. 2004). In addition to autonomous elements, SINE mobilization might also contribute to insertional polymorphism (Gibbs et al. 2004). Therefore, ERV/transposable element-generated polymorphisms probably represent an underestimated, but important source of genetic variability among inbred rat strains. Accordingly, it would be interesting to test the impact of ERV activation on global gene expression. We propose that a comprehensive effort to document ERV/transposable element polymorphisms among inbred strains would complement other established sources of genetic variability detection, e.g., SNP data.
Methods
Animals
All animal experiments were approved by The Charles University Animal Care Committee. As hd/hd males are sterile, the strain carrying the hd mutation (WHD) was propagated by backcrossing heterozygous (+/hd) males to homozygous (hd/hd) females.
Molecular analysis of a retrovirus insertion in the Cntrob gene
The retrovirus inserted into Cntrob was amplified by long-range PCR encompassing intron 10 of Cntrob using Takara LA Taq polymerase, cloned into pCR-XL-TOPO Cloning Kit (Invitrogen) and sequenced by primer walking. The GenBank accession number is EF532341. Genotyping of the hd allele was performed by PCR using a reverse primer in exon 11 of Cntrob and two forward primers in intron 10 and within the retroviral LTR, respectively, resulting in differently sized amplicons for the wild-type (WT) and the hd allele.
Analysis of chimeric splice products generated by retroviral insertion into Cntrob
Full-length and partial (between exons 10 and 11) Cntrob cDNA was amplified from testicular RNA of a hd/hd rat, cloned into pDrive (PCR cloning kit, Qiagen) and pCR-XL-TOPO (TOPO XL PCR Cloning Kit, Invitrogen), respectively, and multiple clones preselected to comprise all different insert sizes were sequenced. The results were deposited in GenBank under accession numbers EF532346–EF532350, and control samples as EF532342–EF532345.
Northern blotting
RNA from whole testes from SHR+/+, BN+/+, WHD+/+, WHD+/hd, and WHDhd/hd strains was extracted using TRIzol reagent (Invitrogen) and purified using RNeasy Mini kit (Qiagen) according to manufacturer's recommendations. For each lane, 10 μg of RNA was denatured, electrophoresed in a 1.2% FA gel in 1× MOPS buffer, transferred overnight to a BrightStar membrane (Ambion) according to manufacturer's recommendations and UV cross-linked. A probe specific for the env gene and the gag gene of the ERV was synthesized by PCR and labeled with 32P using random priming (Stratagene). Membranes were prehybridized with Ultrahyb buffer (Ambion) for 30 min at 42°C, hybridized overnight at the same temperature in fresh Ultrahyb buffer, washed in 6× SSC followed by 0.2 SET/0.2% SDS wash, and exposed overnight.
Analysis of the full-length RnERV-K8e copies
Eight full-length RnERV-K8e elements were amplified from the rat genome (see Supplemental material). Rat-ρ was amplified from the genomic DNA of Brown Norway (BN) and cloned into pCR-XL-TOPO. PCR was performed using Phusion DNA polymerase (New England BioLabs) according to the manufacturer's protocol, with 9 min for primer extension.
TaqMan real-time quantitative PCR probing gag or env genes of RnERV-K8
Total RNA was extracted from the testes of wild-type and Cntrob mutant rats by RNeasy Mini Kit (Qiagen). TaqMan assay targeted to detect gag or env was used together with GAPDH as control. TaqMan assays were purchased from ABI (Applied Biosystems). A cDNA equivalent of 12.5 ng of the initial RNA template was used in each PCR reaction. Each sample was performed in six replicates using the 7700 sequence detection system from ABI (Applied Biosystems). Default baselines and threshold levels were selected as recommended by the manufacturer to obtain Ct values. For each run, a ΔCt (Gag Ct − GADPH Ct) value was calculated for each sample. For normalization of the expression, ΔΔCt values were obtained by subtracting the average testis ΔCt value from each individual ΔCt value. The formula, 2−ΔΔCt was used to calculate the expression relative to testis.
Retrotransposition assay
Retrotransposition of genetically marked elements was done as previously described (Dewannieux et al. 2004). A test construct was generated from RnERV-K8e-Ch20/pCR-XL-TOPO construct: A NdeI–AclI (1.1 kb) fragment of the env gene was exchanged with the neo marker cassette of Pkh15 (Horie et al. 2007). HeLa cells were used for retrotransposition assay. One day before transfection, 5 × 105 cells were plated out per 50-mm dish. Cells were transfected using jetPEI (RGD) with 1 μg of plasmid DNA. Six days after transfection, cells were transferred to a 100-mm dish and were allowed to settle for a day before starting the selection by adding G418 (600 μg/mL). After 10–15 d of selection, cells were fixed, stained, and counted or individually plated on new dishes for further studies. For analysis of de novo retrotransposition events, genomic DNA was isolated from G418r colonies and subjected to inverse PCR.
Integration-site analysis by inverse PCR
Genomic DNA was digested with Sau3AI or Tsp509I followed by ligation with T4 DNA ligase under diluted conditions. Nested PCRs amplifying the left and right flanks of Rat-ρ were performed by using primers IAP21R and InversF1 followed by IAPR2 and LTR-3-F. PCR products were purified with QIAquick gel extraction kit (Qiagen) and directly sequenced.
Transposon display protocol
Transposon display was performed according to Waugh et al. (1997), except that BfaI was used to digest the genomic DNA (see also Supplemental material). Primer sequences are given in the Supplemental material.
Acknowledgments
This work was partially supported by Czech Science Foundation grant 301/07/P178 (F.L.), Czech Science Foundation grant 304/06/0116 (D.K.); EU grants LSGH-2004-005235 and LSHG-CT-2005-019015 (N.H.); and the German Ministry for Science and Education (NGFN-Plus) (N.H.).
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. EF532341–EF532350.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100073.109.
References
- Baillie GJ, van de Lagemaat LN, Baust C, Mager DL. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J Virol. 2004;78:5784–5798. doi: 10.1128/JVI.78.11.5784-5798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- Behura SK. Molecular marker systems in insects: Current trends and future avenues. Mol Ecol. 2006;15:3087–3113. doi: 10.1111/j.1365-294X.2006.03014.x. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retrolements. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Callahan R, Smith GH. MMTV-induced mammary tumorigenesis: Gene discovery, progression to malignancy and cellular pathways. Oncogene. 2000;19:992–1001. doi: 10.1038/sj.onc.1203276. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: Lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Blaise S, Heidmann T. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol. 2005;79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LH, Alamgir AS, Owens N, Weber N, Virtaneva K, Barbian K, Babar A, Malik F, Rosenke K. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J Virol. 2009;83:2429–2435. doi: 10.1128/JVI.01926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter CS, Heizmann CW, Berchtold MW. Isolation and analysis of a rat genomic clone containing a long terminal repeat with high similarity to the oncomodulin mRNA leader sequence. J Biol Chem. 1989;264:18276–18279. [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, Takeda J. Retrotransposons influence the mouse transcriptome: Implication for the divergence of genetic traits. Genetics. 2007;176:815–827. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc Natl Acad Sci. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Lower R, Buschmann C, Horn AV, Traub P, Schumann GG. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenova D, Pravenec M, Housa D, Liška F, Bila V, Kasparek R, Kren V. Linkage mapping of rat hypodactyly locus to chromosome 10. Transplant Proc. 1999;31:1620. doi: 10.1016/S0041-1345(99)00060-3. [DOI] [PubMed] [Google Scholar]
- Kung HJ, Boerkoel C, Carter TH. Retroviral mutagenesis of cellular oncogenes: A review with insights into the mechanisms of insertional activation. Curr Top Microbiol Immunol. 1991;171:1–25. doi: 10.1007/978-3-642-76524-7_1. [DOI] [PubMed] [Google Scholar]
- Kurtz TW, Montano M, Chan L, Kabra P. Molecular evidence of genetic heterogeneity in Wistar-Kyoto rats: Implications for research with the spontaneously hypertensive rat. Hypertension. 1989;13:188–192. doi: 10.1161/01.hyp.13.2.188. [DOI] [PubMed] [Google Scholar]
- Liška F, Gosele C, Rivkin E, Tres L, Cardoso MC, Domaing P, Krejči E, Šnajdr P, Lee-Kirsch MA, de Rooij DG, et al. Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol Reprod. 2009 doi: 10.1095/ biolreprod.109.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Freeman JD. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J Virol. 2000;74:7221–7229. doi: 10.1128/jvi.74.16.7221-7229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Kato K, Ushimaru Y, Hirono I. Intracisternal A particles in transplantable reticulum cell sarcoma of rats. Gann. 1975;66:693–695. [PubMed] [Google Scholar]
- Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: A new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326–333. doi: 10.1016/j.tig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: Retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Harper F, Dewannieux M, Pierron G, Heidmann T. Murine MusD retrotransposon: Structure and molecular evolution of an “intracellularized” retrovirus. J Virol. 2007;81:1888–1898. doi: 10.1128/JVI.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 2008a;18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Harper F, Esnault C, Pierron G, Heidmann T. The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J Virol. 2008b;82:4413–4419. doi: 10.1128/JVI.02141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servomaa K, Rytomaa T. UV light and ionizing radiations cause programmed death of rat chloroleukaemia cells by inducing retropositions of a mobile DNA element (L1Rn) Int J Radiat Biol. 1990;57:331–343. doi: 10.1080/09553009014552441. [DOI] [PubMed] [Google Scholar]
- Steen RG, Kwitek-Black AE, Glenn C, Gullings-Handley J, Van Etten W, Atkinson OS, Appel D, Twigger S, Muir M, Mull T, et al. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 1999;9:AP1–AP8. [PubMed] [Google Scholar]
- Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich MP, Vogetseder W. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet. 1995;96:188–192. doi: 10.1007/BF00207377. [DOI] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BB, Powell W. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Mol Gen Genet. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Jin F, Yeung RS. Germ-line Tsc2 mutation in a dominantly inherited cancer model defines a novel family of rat intracisternal-A particle elements. Oncogene. 1995;11:81–87. [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Bouhassira EE, Tsai HM. An IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Blood. 2007;110:886–893. doi: 10.1182/blood-2007-01-070953. [DOI] [PMC free article] [PubMed] [Google Scholar]