Abstract
Embryonic stem (ES) cells are under precise control of both intrinsic self-renewal gene regulatory network and extrinsic growth factor-triggered signaling cascades. How external signaling pathways connect to core self-renewal transcriptional circuits is largely unknown. To probe this, we chose BMP signaling, which is previously recognized as a master control for both self-renewal and lineage commitment of murine ES cells. Here, we mapped target gene promoter occupancy of SMAD1/5 and SMAD4 on a genome-wide scale and found that they associate with a large group of developmental regulators that are enriched for H3K27 trimethylation and H3K4 trimethylation bivalent marks and are repressed in the self-renewing state, whereas they are rapidly induced upon differentiation. Smad knockdown experiments further indicate that SMAD-mediated BMP signaling is largely required for differentiation-related processes rather than directly influencing self-renewal. Among the SMAD-associated genes, we further identified Dpysl2 (previously known as Crmp2) and the H3K27 demethylase Kdm6b (previously known as Jmjd3) as BMP4-modulated early neural differentiation regulators. Combined with computational analysis, our results suggest that SMAD-mediated BMP signaling balances self-renewal versus differentiation by modulating a set of developmental regulators.
Embryonic stem (ES) cells are pluripotent cells with the unique capacity to self-renew while maintaining the potential to differentiate into all cell types of the body, and thus hold great promise in regenerative medicine (Evans and Kaufman 1981; Martin 1981; Thomson et al. 1998). ES cells are under precise control to keep their identity or to enter determined differentiation programs upon exposure to specific differentiation cues. However, the mechanism of such coordinated control by intrinsic regulators and extrinsic stimuli is largely unknown. For intrinsic regulators, recent studies indicated that ES cells possess a core self-renewal regulatory network consisting of several critical transcription factors such as NANOG, POU5F1 (previously known as OCT4), and SOX2 (Boyer et al. 2005; Loh et al. 2006; Cole et al. 2008; Kim et al. 2008). These studies also suggested that a large set of developmental regulators whose expression is strongly associated with the cell fate switch between self-renewal and differentiation are systematically coordinated by these master transcription factors through direct promoter occupancy. This self-renewal network is further strengthened by the incorporation of the polycomb repressive complex, which co-occupies a large number of developmental regulators with those core transcription factors, thus contributing to the maintenance of ES cell pluripotency by epigenetic modification of chromatin structure (Boyer et al. 2006; Lee et al. 2006a). Extrinsic stimuli, provided by the stem cell niche, culture conditions or cell-autonomous autocrine secretion, can promote self-renewal or differentiation and usually act through various signaling transduction pathways. Among them, leukemia inhibitory factor (LIF)-STAT3, bone morphogenetic protein (BMP)-SMAD, and Wnt-beta-catenin pathways have been suggested as master signaling pathways to regulate both mouse ES cell self-renewal and differentiation (Niwa et al. 1998; Ying et al. 2003a; Sato et al. 2004).
BMP signaling plays multiple roles in ES cell biology, development, and disease (Massague et al. 2000; Mishra et al. 2005; Watabe and Miyazono 2009). As a member of the TGF-beta superfamily, BMP transduces its signal via the intracellular downstream mediators—R-SMAD proteins (SMAD1, SMAD5, SMAD8). The activated R-SMADs form a complex with Co-SMAD SMAD4 and regulate target gene expression through cooperation with other DNA-binding factors and transcription factors in the nucleus (Datto and Wang 2000; Massague and Chen 2000; ten Dijke and Hill 2004; Feng and Derynck 2005; Massague et al. 2005). Previous studies indicated that murine ES cells (mESCs) possess an autocrine BMP signaling which can promote self-renewal in collaboration with LIF-STAT3 by induction of Id proteins to suppress neural lineages differentiation (Ying et al. 2003a), or by inhibiting ERK and MAPK14 (previously known as p38) mitogen-activated protein (MAP) kinase pathways (Qi et al. 2004). BMP signaling is also widely involved in a broad spectrum of differentiation programs such as neural, hematopoietic, cardiomyogenic, and hepatic lineage formation, consistent with the notion that BMPs act as a critical signal during early embryo development and adult tissue homeostasis (Kishigami and Mishina 2005). However, whether the role of BMP signaling in maintenance of mESCs is via promoting self-renewal or via preventing differentiation is still obscure.
To better understand the role of BMP signaling in the fate determination of ES cells, in this study, we mapped the DNA-binding sites of SMAD1/5 and SMAD4 across all annotated genes’ promoter regions in mESCs by combining chromatin immunoprecipitation (ChIP) with DNA promoter array analysis (ChIP-chip) and confirmed by Illumina deep sequencing (ChIP-seq). We found that SMAD1/5 and SMAD4 occupy a group of developmental regulators which are repressed in self-renewing ES cells while rapidly induced upon differentiation. Functional analyses indicated that SMAD1/5- and SMAD4-mediated signaling is required largely to orchestrate differentiation rather than to directly promote self-renewal. Finally, we provided evidence that both Dpysl2 and Kdm6b are BMP/SMAD targets and function as early neural differentiation regulators.
Results
Promoter occupancy of SMAD1/5 and SMAD4 in murine ES cells
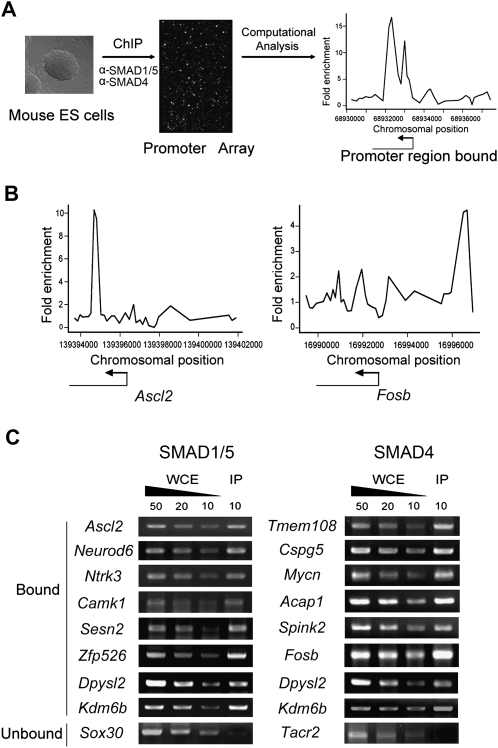
To investigate the role of BMP in cell fate determination of mESCs, we tried to identify the direct targets of BMP signal mediators, SMAD1/5 and SMAD4, by ChIP with anti-SMAD1/5 and anti-SMAD4 antibodies (Supplemental Fig. S1) in undifferentiated R1 ES cells. Although SMAD8 is also a BMP-regulated R-SMAD, it is poorly recognized by anti-SMAD1/5 antibody (Supplemental Fig. S1), and its mRNA level is low in R1 cells (data not shown). Genomic DNA fragments enriched by ChIP were amplified and subjected to hybridization to Agilent mouse promoter array, which contains 60-mer oligonucleotides probes ∼200 base pairs (bp) apart covering the region from –5.5 kilobases (kb) to +2.5 kb relative to the transcriptional start sites (TSS) for ∼17,000 annotated mouse genes (Fig. 1A; Supplemental Methods). Potential binding sites were defined as continuous peaks of signal intensity (Fig. 1B; Supplemental Tables S1, S2). We then mapped these binding sites to the mouse genome and finally identified 562 SMAD1/5-associated genes and 2518 SMAD4-associated genes, respectively (Supplemental Tables S3, S4). We then validated the SMAD–DNA binding from randomly selected target genes using a modified ChIP-PCR method as described previously (Lee et al. 2006b) and confirmed the SMAD association in 72 out of the 91 examined genomic regions (Fig. 1C; Supplemental Fig. S2), suggesting an estimated false positive rate of ∼20%, which falls into a normal level compared with many other such types of works (Martone et al. 2003; Odom et al. 2004; Hartman et al. 2005; Zheng et al. 2007; Mathur et al. 2008). We also subjected ChIP DNA of SMAD1/5 and SMAD4 to Illumina sequencing and found that the majority (62.5%) of SMAD1/5 ChIP-chip target sites and 40.5% of the SMAD4 ChIP-chip target sites can be validated by either SMAD1/5 or SMAD4 ChIP-seq (Supplemental Tables S3, S4).
Figure 1.
Genome-wide analysis of SMAD1/5- and SMAD4-binding sites in R1 ES cells. (A) Strategy of mapping SMAD1/5- and SMAD4-binding sites in the genome of R1 cells. (B) A representative view of SMAD-bound regions. Plots display ChIP-enriched ratios or the signal intensity ratio of the immunoprecipitated DNA (IP)/whole cell extract (WCE) DNA within the chromosomal region indicated by the x-axis. The corresponding gene is depicted below the plot, and the transcriptional start sites (TSS) and transcriptional directions are denoted by arrows. (C) Representative results of confirmation of SMAD binding to DNA by ChIP-site specific PCR. Immuno-enriched (anti-SMAD IP) DNA (10 ng) from R1 cells and a range of unenriched WCE DNA amounts (10, 20, 50 ng of DNA) are used for each primer pair which is designed according to the predicted bound regions.
We then examined the SMAD-binding site distribution along TSS (−5.5 kb to +2.5 kb) and found that a great amount of SMAD1/5-binding sites were located at −1.5 kb to +1.5 kb, whereas SMAD4 sites were relatively enriched within +0.5 kb to +2.0 kb, with the rest evenly distributed along the examined promoter regions (Supplemental Fig. S3). Such distributions are consistent with the general occurrence of transcriptional regulation near the TSS (Guenther et al. 2007) and with the biological relevance of SMAD–DNA interaction.
SMAD co-occupancy, de novo prediction, and validation of cis SMAD-binding elements
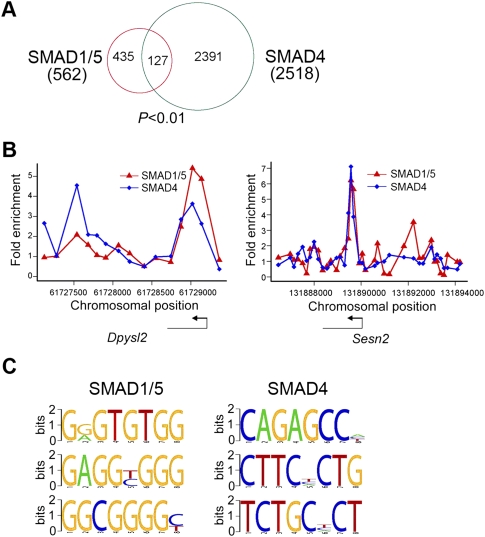
In the canonical SMAD-dependent BMP signaling pathway, SMAD1/5 and SMAD4 form a heterocomplex to regulate target gene transcription (Massague et al. 2005). We found that, of the 562 SMAD1/5-associated genes, 127 (∼23%) were co-occupied by SMAD4, which is significantly more than random expectation (empirical P < 0.01; Fisher's exact test P = 6.76 × 10−24; Fig. 2A,B).
Figure 2.
Co-occupancy of SMAD1/5 and SMAD4 in a subset of genes and de novo prediction of SMAD DNA-binding motifs. (A) A Venn diagram showing the overlap among genes bound by SMAD1/5 and SMAD4. Numbers in parentheses are total numbers of genes associated by respective SMAD. (B) A representative view of co-occupancy by two SMADs. Plots display unprocessed ChIP-enriched ratios within the chromosomal region indicated in the x-axis. The corresponding gene is depicted below the plot, and the TSS and transcriptional direction are denoted by arrows. (C) De novo prediction of SMAD-interacting DNA motifs within SMAD-binding sites obtained by DME with default parameters. Three representative motifs are shown here.
Using the DME software (Smith et al. 2005), we extracted highly enriched DNA sequences from the SMAD-binding regions and found the canonical SMAD1-bound GC-rich elements and SMAD4-bound CAGA elements among the top 10 scored motifs (Fig. 2C; Supplemental Fig. S4A; Supplemental Table S5; Massague et al. 2005). To examine whether these de novo predicted motifs have real SMAD-binding activity, we selected the first four motifs and performed oligonucleotide pull-down analysis with biotinylated DNA oligonucleotides consisting of four repeats of the predicted 8-mer motifs. All of them could apparently pull down their corresponding SMAD proteins although the fourth motif of SMAD4 (M4) showed a relatively weak SMAD-binding activity (Supplemental Fig. S4B). Taken together, the binding profiles obtained here represent authentic SMAD-binding events and are valid for further transcriptional regulation analysis.
Correlation of SMAD–DNA interaction with BMP-modulated expression
To confirm that the SMAD1/5- and SMAD4-associated genes are direct transcriptional regulators in mESCs in response to BMP, we treated undifferentiated R1 ES cells with BMP4 or with the BMP antagonist noggin that can inhibit BMP signaling effectively (Supplemental Fig. S5) for 4 h. We then extracted mRNA from these cells and analyzed gene expression profiles by Affymetrix cDNA microarray. Differentially expressed genes were found through the RankProd algorithm with a cutoff of P-value < 0.05 (Supplemental Table S6). We found that SMAD1/5- and SMAD4-associated genes were more enriched among noggin up-regulated or BMP4 down-regulated genes than noggin down-regulated or BMP4 up-regulated genes (Supplemental Fig. S6). This suggests that both SMAD1/5 and SMAD4 predominately mediate transcriptional repression by the BMP signaling pathway in mESCs.
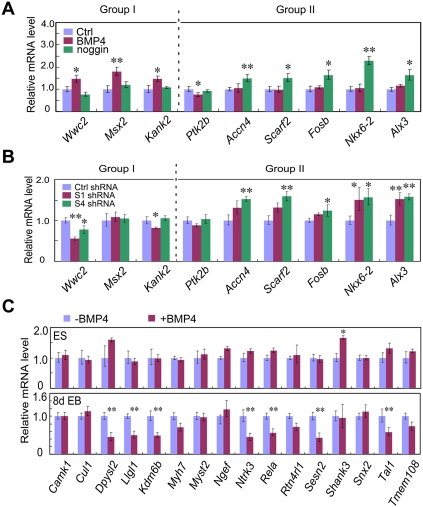
We then separated the differentially expressed genes into two subgroups (Supplemental Table S6). The Group I includes the genes up-regulated by BMP4 and/or down-regulated by noggin whereas the Group II includes the genes down-regulated by BMP4 and/or up-regulated by noggin. To confirm the microarray data, some of the genes with SMAD binding in the two groups were examined for their responses to BMP4 and noggin treatment by quantitative RT-PCR (qRT-PCR). As shown in Figure 3A, these genes were indeed differentially regulated by BMP4 and noggin treatment. To further examine the role of SMAD proteins in mediating the BMP-regulated transcription, we specifically knocked down Smad1 and Smad4 by stable expression of shRNA constructs in R1 cells (Supplemental Fig. S7). The expression profiles for most of the tested genes in these Smad knockdown cells were in agreement with those upon BMP4/noggin treatment (Fig. 3B). For example, the Group I genes Wwc2 and Kank2 that were up-regulated by BMP4 exhibited reduced expression in Smad1 knockdown cells, whereas the Group II genes Accn4, Scarf2, Nkx6-2, and Alx3 that were up-regulated by noggin showed enhanced expression in Smad1 and Smad4 knockdown cells. Some of the genes like Msx2 exhibited no changes in Smad knockdown cells. It could be because the transcriptional effect is only detectable in the presence of additional cooperative transcription factors upon BMP stimulation.
Figure 3.
Expression analysis of SMAD-associated genes. (A) Validation of genes expression change upon ligand stimulation. Group I includes genes that are up-regulated in BMP4 and/or down-regulated in noggin according to microarray analysis and genes in Group II are oppositely regulated. (B) Gene expression in Smad1 and Smad4 knockdown ES cells. (C) SMAD-associated genes are regulated in ES cells and EB upon BMP4 treatment. ES cells and 8-d EB were treated with 20 ng/mL BMP4 for 4 h. Total RNA was extracted and gene expression was analyzed by qRT-PCR. The significance of expression was analyzed by Student's t-test and data are presented as mean ± SEM (n = 3; **P < 0.01; *P < 0.05).
Although SMAD-binding activity was identified in the self-renewing state, it is possible that SMAD pre-occupancy does not initiate transcriptional change until the cells are subjected to differentiation cues. Therefore, we also examined gene expression in ES-derived day 8 embryoid body (EB) as a representative differentiation stage. ES cells and day 8 EB were treated with BMP4 for 4 h and total RNA was extracted for qRT-PCR analysis. More SMAD-binding targets were randomly selected from both SMAD1/5 and SMAD4-binding targets (Supplemental Tables S3, S4) such as Kdm6b, Ntrk3, Sesn2, and Tal1. As shown in Figure 3C, Shank3 was significantly up-regulated in ES cells, and several other genes (Dpysl2, Llgl1, Kdm6b, Ntrk3, Rela, Sesn2, and Tal1) were significantly repressed by BMP4 in day 8 EB. Taken together, these results indicate that the DNA binding of SMAD proteins does mediate BMP-induced transcriptional control both in the self-renewal state or in certain differentiation context and that endogenous BMP signaling mainly plays a repressive role in self-renewing ES cells.
Enrichment of developmental regulators in SMAD-associated genes
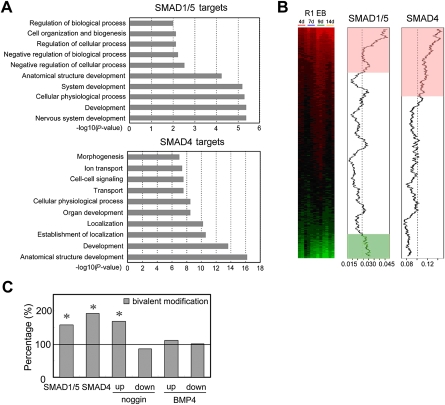
To assess the functionality of SMAD-associated genes in self-renewal and differentiation, we analyzed the association of these genes in Gene Ontology (GO) biological processes. Both SMAD1/5 and SMAD4 targets were significantly enriched in development-related processes (Fig. 4A). Interestingly, the nervous system development category was the most significantly represented in SMAD1/5 targets functional groups, supporting the notion that BMP promotes ES cell self-renewal by inhibiting neural differentiation (Ying et al. 2003b). We also observed a significant enrichment of development functions, especially neurodevelopment functions enrichment among the subset of Illumina-sequencing confirmed target genes (Supplemental Tables S7, S8).
Figure 4.
Developmental regulators are enriched in the SMAD-associated genes. (A) GO analysis of SMAD-associated genes. The y-axis shows the GO terms, and the x-axis shows the enrichment significance P-values for the top 10 enriched GO terms. (B) Genome-wide analysis of SMAD1/5 and SMAD4 targets during EB differentiation. The expression profile data are from Hailesellasse Sene et al. (2007). Genes are rank-ordered by the degree of induction (red) and repression (green) relative to undifferentiated ES cells (left). The two plots (middle and right) show moving average of the frequency of probes for genes that have SMAD-binding sites in a 2000-probe sliding window. The pink and light-green shaded areas indicate the genes whose SMAD-binding frequency is higher than the background level. The dashed line indicates the expected average (background level or the ratio of the number of probes for SMAD targets over the total number of interrogated gene probes). (C) Percentage of SMAD-bound genes, BMP4- and noggin-regulated genes that contain both H3K4me3 and H3K27me3 bivalent modifications were compared to that over all promoters. Asterisk indicates Fisher's exact test P < 0.001. Proportion test also showed similar results (data not shown).
To explore the possible role of SMAD1/5- and SMAD4-assoicated genes in differentiation process, we analyzed the gene expression profile during EB formation of ES cells reported by Hailesellasse Sene et al. (2007). We ranked genes according to their expression levels during differentiation so that the up-regulated and down-regulated genes are grouped to the top and bottom of the panel, respectively (Fig. 4B). The SMAD-binding frequency in a sliding window from the up- to down-regulated genes revealed that both SMAD1/5 and SMAD4 targets were enriched among the up-regulated genes upon differentiation (see pink area in Fig. 4B). SMAD1/5 targets also showed modest enrichment in the down-regulated group (green area), whereas there was no enrichment of SMAD4 targets in the down-regulated group. The observation that a significant amount of SMAD1/5- and SMAD4-associated genes are silenced in self-renewing ES cells while tending to be rapidly induced upon differentiation resembles the pattern of the bivalently modified genes in ES cells that have both H3K4 trimethylation and H3K27 trimethylation and largely encode developmental regulators (Bernstein et al. 2006).
We then compared SMAD-bound target genes to the bivalently modified genes identified by Mikkelsen et al. (2007). Both SMAD1/5 and SMAD4 targets are indeed enriched for bivalently modified genes (Fig. 4C; Fisher's exact test P = 2.44 × 10−4 and 4.38 × 10−22. The subset of target genes confirmed by ChIP-seq have a similar level of enrichment), consistent with the direct binding of SMADs to many developmental regulators suggested by GO annotations (Fig. 4A). It is also consistent with the gene expression profiles during ES cell to EB transition, where SMAD1/5 and SMAD4 targets were enriched among genes repressed in ES cells (Fig. 4B). Intriguingly, bivalent histone modifications are highly over-represented only among the noggin up-regulated genes (Fig. 4C, Fisher's exact test P = 2.47 × 10−18), but not in noggin down-regulated or those changed in response to exogenously added BMP4, suggesting that bivalent modification may be associated with endogenous BMP-mediated gene silencing in self-renewing ES cells and rapid activation during early development. Indeed, we observed a correlation between a decreased H3K27me3 modification and an increased expression of the Nkx6-2 and Alx3 genes upon noggin treatment (Supplemental Fig. S8).
SMAD-mediated BMP signaling is largely required for early differentiation rather than direct self-renewal maintenance
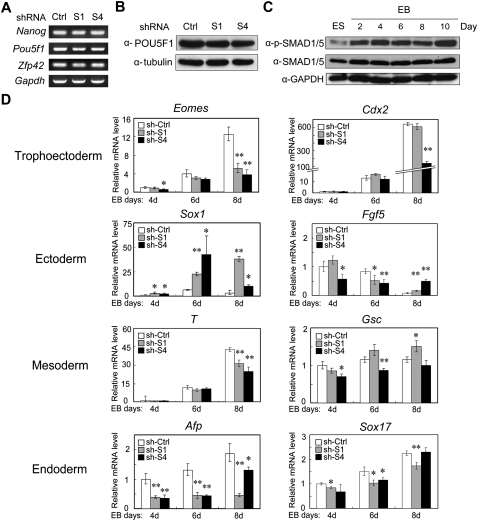
It was reported that BMP signaling can cooperate with LIF signaling to maintain mouse ES cell self-renewal by inhibiting Id expression and then neural lineage entry (Ying et al. 2003a). How SMAD-mediated BMP signaling orchestrates self-renewal and early differentiation has not been systematically analyzed. To test whether SMAD-mediated BMP signaling is necessary for ES cell self-renewal, we examined the mRNA expression of several widely used self-renewal markers—Nanog, Pou5f1, and Zfp42 (previously known as Rex1)—in Smad1 and Smad4 knockdown ES cells. Consistent with the lack of morphological changes of the ES cells, we found no significant difference of those self-renewal markers at the mRNA level in Smad1 or Smad4 knockdown ES cells compared to control cells (Fig. 5A). The protein level of POU5F1 also remained unchanged in the Smad knockdown ES cells (Fig. 5B). Pluripotent ES cells undergo rapid proliferation, but Smad knockdown did not influence the proliferation rate of ES cells (Supplemental Fig. S9), implying that SMAD1 and SMAD4 may not be crucial for self-renewal. This result is in accordance with the observation that Smad4 knockout ES cells show no defect of self-renewal and proliferation (Sirard et al. 1998). When examining endogenous BMP signaling activity during ES cell to EB differentiation by monitoring SMAD1/5 phosphorylation, the phospho-SMAD1/5 level was lower in ES cells as compared with EB (Fig. 5C), suggesting that BMP signaling activity is low and then elevated during differentiation.
Figure 5.
SMAD1 and SMAD4 are largely involved in differentiation regulation rather than direct self-renewal maintenance. (A) Knockdown of Smad1 or Smad4 has no effect on the expression of self-renewal markers. Total RNA extracted from R1 cells expressing Smad shRNA or control shRNA constructs were subjected to RT-PCR. Gapdh served as a loading control. (B) No difference of the POU5F1 protein levels in R1 cells expressing Smad shRNA or control shRNA. Protein expression was examined by immunoblotting. Tubulin served as a loading control. (C) Phosphorylation of SMAD1/5 is enhanced during EB formation. EB was formed in serum-free KO-SR culture conditions, and total proteins were collected at indicated times and subjected for anti-phopho-SMAD1/5 and anti-SMAD1/5 immunoblotting. GAPDH served as a loading control. (D) Smad knockdown alters the expression profile of germ layer markers. Quantitative RT-PCR analysis was performed to examine marker expression in Smad shRNA and control shRNA cells during the course of EB differentiation. The significance of expression was analyzed by Student's t-test, and data are presented as mean ± SEM (n = 3; **P < 0.01; *P < 0.05). This experiment was repeated three times and similar results were obtained.
Next, we examined whether Smad knockdown influences early differentiation. ES cells were induced to form EB, and various germ layer markers were examined by qRT-PCR. As shown in Figure 5D, the expression of trophoblast lineage markers Eomes in both Smad1 and Smad4 knockdown cells and Cdx2 in Smad4 knockdown cells was down-regulated. Sox1, an ectodermal marker, was elevated in both Smad1 and Smad4 knockdown cells, consistent with BMP inhibition of early neural fate (Liu and Niswander 2005). The expression pattern of another ectodermal marker Fgf5 was also altered. Both Smad1 and Smad4 knockdown led to significantly reduced expression of endodermal marker—alpha fetoprotein (Afp). Two mesodermal markers, brachyury (T) and goosecoid (Gsc), were modestly decreased in Smad4 knockdown cells in certain time points whereas no significant change was observed in Smad1 knockdown cells. Considering that SMAD1 and SMAD5 may have overlapping functions in ES cells, the observed effects of Smad1 knockdown here should be an underestimation of BMP-regulated R-SMAD functions. Nonetheless, these changes together reflect the requirement of BMP signaling for early embryogenesis (Winnier et al. 1995). Therefore, SMAD1- and SMAD4-mediated signaling is mainly required for proper differentiation programming rather than for direct self-renewal maintenance, consistent with the above observation that a significant portion of SMAD1/5 and SMAD4 target genes are development- or differentiation-related.
Functional validation of SMAD target genes controlled by BMP signaling
BMP can potently inhibit early neural differentiation in various in vitro ES cells to neural lineage differentiation systems (Finley et al. 1999; Ying et al. 2003b). To validate the functional importance of the SMAD target genes, we employed a serum-free monolayer differentiation system to monitor the process from ES cells to SOX1- and nestin-positive neural precursors (Ying et al. 2003b). In agreement with a previous report (Ying et al. 2003b), BMP4 strongly inhibited early neural differentiation as shown by reduced expression of Sox1 and nestin (Supplemental Fig. S10).
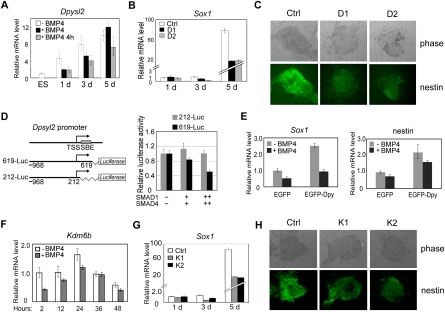
Dpysl2, also known as Crmp2, has been implicated to function in axon guidance and in neural diseases such as Alzheimer's disease, neuroinflammation, and schizophrenia (Zhao et al. 2006; Cole et al. 2007; Vuaillat et al. 2008). As the Dpysl2 promoter is co-occupied by both SMAD1/5 and SMAD4 (Figs. 1C, 2B) and its expression is attenuated by BMP4 in day 8 EB (Fig. 3C), we explored its function in BMP-regulated early neural differentiation. Consistent with the above data, BMP4 inhibited Dpysl2 expression at the initial stage of neural differentiation but had less or no effect at day 5 (Fig. 6A). Using two independent specific Dpysl2 shRNA constructs (D1 and D2) (Supplemental Fig. S11), we found that Dpysl2 knockdown dramatically reduced the expression of Sox1 especially at day 5 of neural differentiation (Fig. 6B), suggesting a requirement of DPYSL2 for proper neural precursor commitment. This was further confirmed by examining nestin expression, which was also down-regulated in Dpysl2 knockdown cells (Fig. 6C; Supplemental Fig. S12). To further demonstrate that Dpysl2 is a direct target of SMAD-mediated BMP signaling, we cloned the Dpysl2 promoter region (−968 to +619) into pGL3-Basic luciferase plasmid to examine the SMAD effect on the Dpysl2 promoter transcriptional activity (Fig. 6D). As expected, the construct (619-Luc) covering a predicted SMAD-binding element (SBE) exhibited SMAD1/4 concentration-dependent transcriptional repression in HEK293T cells whereas the construct (212-Luc) without the SBE had no response. This result was further confirmed by constitutively active BMP receptor ALK6 (BMPR1B) (Supplemental Fig. S13). In addition, using oligonucleotides derived from the putative SMAD binding DNA sequences of the Dpysl2 promoter as probes, oligonucleotide pull-down assay also revealed that both SMAD1 and SMAD4 could directly bind to the Dpysl2 promoter (Supplemental Fig. S14). To confirm the function of Dpysl2 in neural differentiation, we transiently overexpressed DPYSL2 as a GFP fusion protein in R1 cells (Supplemental Fig. S15). As shown in Figure 6E, DPYSL2 overexpression elevated the expression of Sox1 and nestin at day 3 of neural differentiation. These data together strongly suggest that Dpysl2 is a bona fide BMP/SMAD target and plays a role in early neural differentiation.
Figure 6.
BMP-regulated Dpysl2 and Kdm6b play an important role in early neural differentiation. (A) Dpysl2 expression is increased during neural differentiation, and BMP4 impairs this increase at the early stage (day 1, 3). R1 cells were cultured in neural differentiation medium in the presence (+) or absence (−) of 10 ng/mL BMP4 for 5 d or treated with BMP4 for 4 h before harvest (4 h). Dpysl2 expression was determined by qRT-PCR. (B) Dpysl2 knockdown reduces Sox1 expression. Sox1 expression was determined in the Dpysl2 shRNA D1- or D2-expressing cells that were subjected to neural differentiation. (C) Dpysl2 knockdown reduces nestin expression. Nestin expression was determined by immunofluorescence in the shRNA D1- or D2-expressing cells at day 5 of neural differentiation. (D) SMAD represses the Dpysl2 promoter activity. (Left) Schematic map of the Dpysl2 promoter-driven luciferase reporters. TSS, transcriptional start sites; SBE, SMAD-binding element (for both SMAD1/5 and SMAD4) that is predicted from ChIP-chip analysis (Supplemental Tables S1, S2). Reporter plasmid (0.1 μg), Renilla plasmid (10 ng), SMAD1 and SMAD4 (0, 0.05 μg, or 0.1 μg, each) were cotransfected into HEK293T cells. At 36 h post-transfection, cells were harvested for luciferase assay. The experiment was performed in triplicate, and the data are presented as mean ± SEM of three independent experiments after normalization to Renilla activity. (E) DPYSL2 increases Sox1 and nestin expression during early neural differentiation. R1 ES cells were transfected with pEGFP-Dpysl2 or pEGFP-C3 empty vector respectively and then replated for neural differentiation. RNA was extracted at day 3 for qRT-PCR analysis. (F) Kdm6b expression is reduced by BMP4 in the early stages of neural differentiation. (G) Kdm6b knockdown reduces Sox1 expression. Sox1 expression was determined in the shRNA K1- or K2-expressing cells that were subjected to neural differentiation similarly as in B. (H) Kdm6b knockdown decreases nestin expression. Nestin expression was determined by immunofluorescence in shRNA K1- or K2-expressing cells at day 5 of neural differentiation.
KDM6B, a histone H3K27 demethylase, has been implicated in early embryo development, inflammation, and other biological processes (Agger et al. 2007; De Santa et al. 2007). Like Dpysl2, the Kdm6b promoter was co-occupied by both SMAD1/5 and SMAD4 (Fig. 1C) and its expression was significantly reduced by transient BMP4 treatment in day 8 EB (Fig. 3C), implying its possible function in a certain differentiation process. We then tested this possibility in neural differentiation and found that Kdm6b expression increased at the early stages of neural differentiation and then declined gradually (Fig. 6F). Interestingly, BMP4 inhibited Kdm6b expression just at the early stage (Fig. 6F), and enhanced SMAD protein association with the Kdm6b promoter was also observed upon BMP4 treatment during this early stage (Supplemental Fig. S16A). Further, the correlation of decreased SMAD binding with Kdm6b up-regulation during neural differentiation also indicates a repressive role of BMP/SMAD signaling on Kdm6b in self-renewing ES cells (Fig. 6F; Supplemental Fig. S16B). When Kdm6b was knocked down by two specific shRNA constructs (K1 and K2) (Supplemental Fig. S11), neural differentiation is significantly impaired as shown by the decreased expression of Sox1 and nestin (Fig. 6G,H). This is consistent with the recently reported role of KDM6B in early neural differentiation (Burgold et al. 2008). Therefore, our data revealed that Kdm6b is involved in early neural differentiation under the control of SMAD-mediated BMP signaling.
Discussion
By employing ChIP-chip technology combined with computational analysis and experimental validation, we mapped target gene promoter occupancy of SMAD1/5 and SMAD4 in mESCs on a genome-wide scale. We found that the SMAD-bound genes are significantly associated with differentiation and development.
Genome-wide identification of SMAD-associated targets
We verified 91 genes from our ChIP-chip data by ChIP-site specific PCR and obtained a confirmation rate of ∼80%. Our data also revealed several novel SMAD-binding motifs in addition to the previously identified ones (for review, see Massague et al. 2005). We found that SMAD4 occupies a larger number of genes than SMAD1/5 does, and a significant portion of SMAD1/5-bound genes is not co-occupied by SMAD4. The following reasons may account for the low co-occupancy. First, SMAD4, in addition to transducing BMP signaling, also mediates TGF-beta/Activin/Nodal signaling. Indeed, recent studies also suggested that Activin/Nodal signaling is required for mouse ES cells propagation (Ogawa et al. 2007). Second, SMAD1/5 may function in a SMAD4-independent manner (He et al. 2006; Zhao et al. 2008; Retting et al. 2009).
Through cDNA microarray and qRT-PCR, we then established the link between SMAD binding and BMP-regulated transcription. We not only observed a correlation between SMAD binding with BMP4-mediated expression change in the self-renewing ES cells, but also found that there is a portion of SMAD-binding genes which exhibit a context-dependent regulation of transcription under BMP4 treatment. It was previously suggested that Id genes (Id1 and Id3) are primary targets of BMP/SMAD signaling in inhibiting neural differentiation (Ying et al. 2003a), and our cDNA array also identified Id1, Id2, Id3 as significantly up-regulated genes by BMP4 in self-renewing ES cells (Supplemental Table S6). However, these genes were not in our SMAD-bound genes list although ChIP-site specific PCR revealed SMAD1/5-immunoprecipetated Id1 promoter regions from ES cells (data not shown). Missing Id genes in this study could be due to relatively limited binding of SMAD under endogenous BMP signaling and/or stringent peak identification algorithm cutoff.
Association of the SMAD-bound genes with developmental control
Computational analysis of the SMAD-associated targets revealed that SMAD1/5 and SMAD4 occupied a large set of important developmental regulators, and most of the SMAD1/5-bound genes are categorized into the neural development-related functional group. This result supports the notion that SMAD-mediated BMP signaling, in collaboration with LIF, can promote self-renewal by inhibiting neural differentiation (Ying et al. 2003a). By analyzing gene expression profile during EB differentiation, we found SMAD-bound genes are predominantly repressed in ES cells and rapidly up-regulated upon differentiation, which is further confirmed by computational analysis of histone H3K4 and H3K27 trimethylation. Both SMAD1/5 and SMAD4 targets are indeed enriched in bivalently modified genes. H3K4 and H3K27 trimethylation are generally indicative of genes activation and repression, respectively (Mikkelsen et al. 2007), while genes with bivalent modification are found to largely encode important developmental regulators in ES cells and are silent but “poised” for rapid activation (Bernstein et al. 2006). However, it remains unclear whether such histone modifications are directly regulated by SMAD binding. Examination of the responsiveness of SMAD-bound genes to BMP4 or noggin also indicated that most of those genes are repressed by BMP in self-renewing ES cells, further confirming that BMP–SMAD signaling controls the expression of developmental regulators.
During early embryo development, BMP functions as a morphogen to pattern germ layers and regulates multiple cell differentiation programs. However, little is known about direct targets of BMP to interpret BMP signaling in specific mammalian developmental process. Our data have provided some potential target genes. For instance, we found that Dpysl2, a gene highly expressed in the neural system, had an important function in early neural differentiation. Our results have also revealed that another BMP/SMAD target, the histone H3K27 demethylase Kdm6b, is required for early neural differentiation, implying that BMP signaling may regulate certain differentiation processes through transcriptional control of some key epigenetic regulators.
Role of BMP/SMAD signaling in fate determination of ES cells
Our data suggest that BMP/SMAD signaling coordinates self-renewal and differentiation by modulating the expression of developmental regulators. Knockdown of Smad1 or Smad4 expression in ES cells did not impair their self-renewal ability but led to alterations in the expression pattern of germ layer markers during EB differentiation. Consistent with this, studies on in vitro differentiation of ES cells showed that SMAD-mediated BMP signals at least take part in neural, cardiomyogenic, hematopoietic, and hepatic lineages commitment (Loebel et al. 2003). Although BMP has been found to promote ES cell self-renewal in the presence of LIF, it is not needed for self-renewal under NANOG overexpression (Ying et al. 2003a). Furthermore, Smad4−/− ES cells show no defect in self-renewal ability (Sirard et al. 1998), and the embryos deficient of several BMP signaling components can all survive past the blastocyst stage when ES cells are derived and exhibited progressively embryonic lethality between embyonic day 6.5 and 11.5 (for reviews, see Datto and Wang 2000; Chang et al. 2002), indicating a nonessential role of BMP signaling for ES cell self-renewal but rather necessary for proper lineage commitment. The fact that BMP combined with LIF can support mouse ES cell self-renewal is basically through inhibition of neural differentiation but not as a direct self-renewal enhancer (Ying et al. 2003a; data not shown). As we focus our studies on SMAD-mediated BMP signaling, we cannot rule out the possibility that BMP may somewhat support self-renewal via other SMAD-independent pathways such as MAP kinases.
Although our results suggest that SMAD-mediated BMP signaling does not directly participate in maintenance of ES cell pluripotency, BMP/SMAD signaling may exert its function to coordinate with the core self-renewal network and modulate the balance between ES cell self-renewal and differentiation by influencing those shared targets. Examination of the overlap between the genes bound by SMAD and those bound by nine critical self-renewal related transcription factors (Kim et al. 2008) revealed that the SMAD1/5 targets intersect significantly with those of all nine factors, and the SMAD4 targets also have a substantial overlap with those of SOX2, NR0B1 (previously known as DAX1), and KLF4 (Supplemental Fig. S17). When the SMAD targets were compared with another data set that mapped NANOG-, POU5F1-, and TCF3-bound genes (Cole et al. 2008), similar results were obtained (data not shown).
A recent study has identified a connection of SMAD1 targets with the core self-renewal network (Chen et al. 2008). Although both the SMAD1-binding enhancer sites in their study and the promoter sites in ours share significant overlap to the core self-renewal transcription factor target sites, our study revealed that SMAD target genes are mostly associated with development and differentiation, with similar pattern to bivalently modified genes in mouse ES cells, which has not been observed by the study of Chen and colleagues. This discrepancy could be due to the different binding profiles of SMAD1/5: Chen et al. (2008) found that a large portion of the SMAD1-binding regions were in the intergenic region without the canonical SMAD1-binding motif, whereas we focused our analysis on SMAD binding in the promoter regions and found that many of the SMAD1/5-binding sites were enriched with GC elements as identified in the canonical SMAD1-binding motifs (Massague et al. 2005).
Consistent with our conclusion that BMP/SMAD signaling is not required for ES cell self-renewal per se, a recent study showed that extrinsic stimuli are dispensable for self-renewal when differentiation-inducing signaling from MAP kinase is eliminated (Ying et al. 2008). Interestingly, NANOG can also bind to SMAD1 to block BMP-induced mesoderm differentiation (Suzuki et al. 2006), and POU5F1 and NANOG were suggested to maintain ES cell self-renewal by association with transcriptional repression complexes (Liang et al. 2008). Thus it is possible that BMP signaling coordinates with core self-renewal factors to balance self-renewal versus differentiation via modulating those development-associated target genes when ES cells switch from the self-renewal state into a differentiation state.
Methods
Growth conditions for murine embryonic stem cells
R1 ES cells were cultured under typical ES cell conditions (Boyer et al. 2006). When prepared for ChIP assay, cells were cultured on gelatinized tissue culture plates for two passages to eliminate MEF cells. KO-DMEM and Knockout Serum Replacement (KO-SR) (Invitrogen GIBCO) were used to substitute DMEM and fetal calf serum for feeder-free culture.
Antibodies and chromatin immunoprecipitation assay
We use affinity-purified SMAD1/5 and SMAD4 antibodies for ChIP assay. A detailed description of ChIP assay is provided in Supplemental materials.
Array design and data normalization and identification of SMAD-bound regions
The chip assay was performed with Agilent Mouse Promoter two-color Array (NCBI 35, UCSC mm7). The probe set provides coverage for ∼17,000 of the best defined mouse transcripts as defined by RefSeq, which cover −5.5 kb upstream to +2.5 kb downstream from the transcriptional start sites. The raw data were processed by R package limma. The log-ratio of IP over WCE was normalized within arrays using “loess” method and the normalized log-ratios were then used for SMAD targets selection using a previously reported method (Kim et al. 2005). For identification of bound regions and more detailed information, see Supplemental materials.
Gene Ontology (GO) analysis
The GO biological functions enriched among the SMAD1/5 and SMAD4 target genes were analyzed using Cytoscape v2.3.2 with plug-in BiNGO 2.0.
Gene expression and histone modification analysis
The cDNA microarray was carried out with Affymetrix's mouse genome 430 2.0 array. Data processing and normalization are described in Supplemental materials. EB differentiation microarray data of R1 cells (Hailesellasse Sene et al. 2007) were used in the expression analysis for the SMAD1/5 and SMAD4 target genes during ES cell to EB transition. The histone modification analysis was done by overlapping the H3K4me3 and H3K27me3 modified genes in mES cells (Mikkelsen et al. 2007) with our SMAD targets.
RNA isolation and quantitative real-time RT-PCR (qRT-PCR)
To determine mRNA expression by RT-PCR, RNA was isolated from ES cells, EB, or neural precursor cells using TRIzol reagent (Invitrogen). Reverse transcriptase (Toyobo) was employed for oligo(dT) primed first-strand cDNA synthesis. Quantitative real-time PCR was carried out on Mx3000P (Stratagene) using EvaGreen dye (Biotium). The ΔΔCt method was used to comparatively quantify the amount of mRNA level. The oligonucleotides used for PCR are listed in Supplemental Table S9.
Derivation of neural precursor cells from mouse ES cells
Serum-free monolayer neural differentiation was modified from the protocol described previously (Ying et al. 2003b). Detailed information can be found in Supplemental materials.
Statistic analysis
Enrichment significance P-values were determined by either Monte Carlo simulation or by Fisher's exact test. Details are described in Supplemental materials.
Acknowledgments
We thank Duanqing Pei for R1 cells; Qi Zhou and Lingsong Li for help with ES cell culture; and Jun Li, Jie Hao, Wei Zhang, and Junsong Han for technical assistance. This work was supported by grants from the National Natural Science Foundation of China (30671033, 30890033, 30588001, 30620120433), 973 Program (2006CB943401, 2006CB910102, 2006CB910700), and 863 Program (2006AA02Z172, 2006AA02A114).
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE18629.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092114.109.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-β superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- Chen X, Han Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole AR, Noble W, van Aalten L, Plattner F, Meimaridou R, Hogan D, Taylor M, LaFrancois J, Gunn-Moore F, Verkhratsky A, et al. Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer's disease progression. J Neurochem. 2007;103:1132–1144. doi: 10.1111/j.1471-4159.2007.04829.x. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto M, Wang XF. The Smads: Transcriptional regulation and mouse models. Cytokine Growth Factor Rev. 2000;11:37–48. doi: 10.1016/s1359-6101(99)00027-1. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Finley MF, Devata S, Huettner JE. BMP-4 inhibits neural differentiation of murine embryonic stem cells. J Neurobiol. 1999;40:271–287. [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, Campbell PA, Rudnicki MA, Andrade-Navarro MA. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes & Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16:265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006a;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006b;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat Rev Neurosci. 2005;6:945–954. doi: 10.1038/nrn1805. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc Natl Acad Sci. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes & Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes & Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L, Derynck R, Mishra B. Transforming growth factor-β signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D, Morishita Y, Watabe T, Niwa H, Miyazono K. Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci. 2007;120:55–65. doi: 10.1242/jcs.03296. [DOI] [PubMed] [Google Scholar]
- Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136:1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern SE, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes & Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodriguez-Esteban C, Izpisua Belmonte JC. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGF-beta–Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vuaillat C, Varrin-Doyer M, Bernard A, Sagardoy I, Cavagna S, Chounlamountri I, Lafon M, Giraudon P. High CRMP2 expression in peripheral T lymphocytes is associated with recruitment to the brain during virus-induced neuroinflammation. J Neuroimmunol. 2008;193:38–51. doi: 10.1016/j.jneuroim.2007.09.033. [DOI] [PubMed] [Google Scholar]
- Watabe T, Miyazono K. Roles of TGF-β family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003b;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Tang R, Xiao Z, Shi Y, Feng G, Gu N, Shi J, Xing Y, Yan L, Sang H, et al. An investigation of the dihydropyrimidinase-like 2 (DPYSL2) gene in schizophrenia: Genetic association study and expression analysis. Int J Neuropsychopharmacol. 2006;9:705–712. doi: 10.1017/S1461145705006267. [DOI] [PubMed] [Google Scholar]
- Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-β signaling. J Biol Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]