Abstract
Cancer progression in humans is difficult to infer because we do not routinely sample patients at multiple stages of their disease. However, heterogeneous breast tumors provide a unique opportunity to study human tumor progression because they still contain evidence of early and intermediate subpopulations in the form of the phylogenetic relationships. We have developed a method we call Sector-Ploidy-Profiling (SPP) to study the clonal composition of breast tumors. SPP involves macro-dissecting tumors, flow-sorting genomic subpopulations by DNA content, and profiling genomes using comparative genomic hybridization (CGH). Breast carcinomas display two classes of genomic structural variation: (1) monogenomic and (2) polygenomic. Monogenomic tumors appear to contain a single major clonal subpopulation with a highly stable chromosome structure. Polygenomic tumors contain multiple clonal tumor subpopulations, which may occupy the same sectors, or separate anatomic locations. In polygenomic tumors, we show that heterogeneity can be ascribed to a few clonal subpopulations, rather than a series of gradual intermediates. By comparing multiple subpopulations from different anatomic locations, we have inferred pathways of cancer progression and the organization of tumor growth.
As cancers progress they accumulate genomic changes, including deletions and amplifications (Hanahan and Weinberg 2000; Albertson 2006), translocations (Mitelman et al. 2007), point mutations (Sjöblom et al. 2006; Ley et al. 2008; Stratton et al. 2009), and metastable epigenetic events such as changes in DNA methylation (Widschwendter and Jones 2002; Feinberg andTycko 2004). In many cases, the discovery and classification of these changes have led to major insights into cancer. Genomic tools such as expression profiling, array-based copy number analysis, high-throughput DNA sequencing, and DNA methylation analysis have accelerated the accumulation of data about individual cancers. The resulting picture is quite complex. For example, the number of recurrent copy number changes even in specific solid cancer subtypes is very large (Loo et al. 2004; Hicks et al. 2006; Adelaide et al. 2007; Haverty et al. 2008), and it is difficult to infer the sequence of genomic alterations in any given tumor by analyzing a single sample from the tumor. Here we explore what additional information is gained, beyond studying mutations in large collections of tumors, by studying mutations in distinct subpopulations of single tumors.
As a matter of practice, histopathologists observe tumor heterogeneity (Komaki et al. 2006) when they examine tissue sections from many regions of tumors, and they classify each tumor by its highest observed malignant grade (Ignatiadis and Sotiriou 2008). Previous molecular studies have also reported heterogeneity in various forms: transcript expression (Cole et al. 1999; Bachtiary et al. 2006), protein levels (Allred et al. 2008; Johann et al. 2009), single nucleotide polymorphisms (Khalique et al. 2007), and chromosomal rearrangements (Aubele et al. 1999). Heterogeneity has also been frequently observed in the analysis of karyotypes in breast tumors from single patients (Teixeira et al. 1995, 1996; Hein et al. 1997). Several studies have also reported genetic heterogeneity in solid breast tumors using fluorescent in situ hybridization (FISH) experiments on interphase nuclei (Fiegl et al. 1995; Roka et al. 1998; Farabegoli et al. 2001). These experiments commonly report that a specific FISH probe measures different copy number signals in individual cancer cells from the same tumor. However, studies based on histopathology or just a few markers cannot have the richness of information that can be obtained by modern genomic methods. We theorized that copy number profiling of multiple sectors of a solid tumor would have the potential to greatly clarify the extent and patterns of tumor progression.
Assuming that the mutational complexity of a tumor increases with time, the history of its progression can be partially inferred by comparing the distinguishable subpopulations. To separate genomic subpopulations, we initially dissected solid breast tumors and compared the genome profiles, which revealed genomic heterogeneity and encouraged us to further separate tumor subpopulations by ploidy. Thus, we devised the Sector-Ploidy-Profiling (SPP) approach. SPP involves macro-dissecting solid tumors into multiple sectors, isolating and flow-sorting nuclei by total genomic DNA content, and analyzing the genome structure of tumor subpopulations by a form of comparative genomic hybridization (CGH) called representational oligonucleotide microarray analysis (ROMA) (Lucito et al. 2003). We then used algorithms to compare the genomes of tumor subpopulations to assess their divergence, thereby identifying genetic elements that may be involved in tumor progression. To understand the organization of tumor subpopulations at the single-cell level, we conducted further cytological studies by interphase FISH.
We applied our methods to 20 primary ductal breast carcinomas, which enable us to classify them according to whether they appear as either monogenomic (nine tumors) or polygenomic (11 tumors). We define “monogenomic” tumors to be those consisting of an apparently homogeneous population of tumor cells with highly similar genome profiles throughout the tumor mass. We define “polygenomic” tumors as those containing multiple tumor subpopulations that can be distinguished and grouped by similar genome structure. We find that polygenomic tumors may exhibit two anatomical organizations of their tumor subpopulations: segregated and intermixed. Our results show that the subpopulations in polygenomic tumors may differ by large genomic events or focal amplifications and deletions, but that in all cases the majority of chromosome breakpoints are shared. We constructed distance trees that show that tumor subpopulations share a common genetic lineage, and that each divergent subpopulation represents a branch in the evolution of a solid tumor.
Results
Copy number analysis of tumors by sector
We hypothesized that some solid tumors contain subpopulations with major variation in their genome structure, and that these might be prominent in separate sectors. To test this hypothesis, we macro-dissected four primary ductal carcinomas (T1–T4) into four sectors (S1–S4), then isolated DNA and quantified genome-wide copy number variation using ROMA (Supplemental Fig. S1; Supplemental Table S1). These tumors were randomly selected from a large collection of frozen ductal carcinomas. Two tumors analyzed by this method (T1, T2) contained minimal variation in their genomic copy number profiles in all four sectors. Our data indicated that T1 contained 39 chromosomal breakpoints that were common to all tumor sectors, and multiple amplifications and deletions present at similar copy number in every sector. Similarly, T2 contained 44 amplification and deletion breakpoints that were common in position and magnitude in all four tumor sectors. This analysis indicates that these tumors contain highly similar profiles in every sector, suggesting that T1 and T2 are each composed of a single major monogenomic tumor subpopulation or a homogeneous mixture of subpopulations that are not resolvable by dissection alone.
In contrast, when we analyzed tumors T3 and T4, we noticed a large degree of variation in the genome patterns of distinct sectors. T3 contains 21 chromosomal breakpoints common to all four sectors, but S3 of T3 also contains 16 new divergent chromosome breakpoints not present in the other tumor sectors. These chromosome breakpoints encompass three genomic amplifications (6p22.1, 6p21.1, 17q21.32) and a deletion (21q11), none of which are detectable in S1, S2, or S4. Thus at least two subpopulations are evident in this polygenomic tumor. T4 displays yet another pattern. Two sectors (S1 and S2) that contain high proportions of tumor cells as assessed by histopathology (71% and 69%, respectively) do not display prominent genomic rearrangements. Copy number variation is observed even in normal genomes (Sebat et al. 2004). Sampling from this part of the tumor (S1 and S2), and using previous genomic measures (Hicks et al. 2006), we would not judge the tumor to be highly malignant. However, had we sampled from sectors 3 and 4 (which display many prominent rearrangements, including 98 breakpoints not present in sectors S1 and S2), we would judge the tumor to be highly malignant.
Copy number analysis of tumors by sector and ploidy
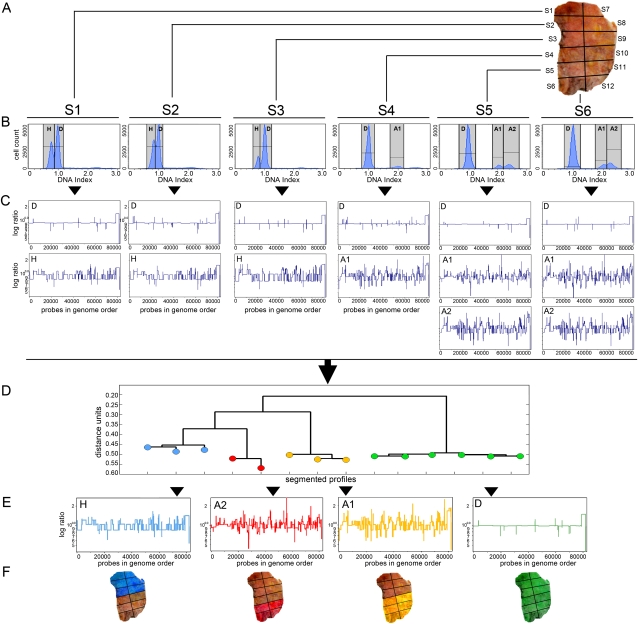
To gain a clearer picture of the number of subpopulations and their clonal relationship, we added a further tool for separating subpopulations, fluorescence-activated cell sorting (FACS). Previous studies have shown that FACS can be used to separate tumor cells by ploidy for genomic analysis (Corver et al. 2008). FACS separates subpopulations of tumor cells, and tumor cells from normal cells, by differences in their total genomic DNA content, or ploidy. We combined sectoring and FACS to isolated tumor subpopulations, prepared DNA from all separable fractions, and applied ROMA to 16 tumors (Supplemental Table S1). We illustrate the SPP method with a single example, tumor T10 (Fig. 1A–F).
Figure 1.
Sector-Ploidy-Profiling (SPP) approach. The SPP approach separates tumor subpopulations by macro-dissection and cell sorting by ploidy. (A) Macro-dissection of tumor sectors. (B) Sorting of DAPI-stained nuclei using FACS by differences in total genomic DNA content. (C) Profiling of chromosome breakpoints across the genome by ROMA CGH. (D) Calculation of neighbor-joining trees using copy number profiles. (E) Coalescence of highly similar copy number profiles. (F) Topography of subpopulations in the tumor. Tumor sectors S7–S12 are colored according to the adjacent subpopulations in S1–S6.
T10 was cut in half along one axis, and six cuts were made along an orthogonal axis, resulting in 12 pieces (Fig. 1A). Nuclei were prepared from six of these pieces and then separated by FACS into subpopulations distinguishable by total DNA content (Fig. 1B; Supplemental Fig. S2). DNA from each peak was prepared and analyzed using ROMA, and then the raw ratio profiles were segmented using a circular binary segmentation (CBS) algorithm (Venkatraman and Olshen 2007). The segmented profiles were always clearly related but sometimes distinguishable by their chromosome breakpoint pattern (Fig. 1C). We also used Pearson correlations and neighbor-joining algorithms to form distance trees that clustered the profiles into similar and distinguishable subgroups (Fig. 1D). In each case where we claim that a genomic breakpoint distinguished two subgroups, we examined the raw data to rule out the possibility of segmentation artifacts, namely, that the differences were not merely of degree. To facilitate further comparisons between subgroups, we coalesced profiles within subgroups by calculating the means of the segmented values from subgroups of individual CGH profiles (Fig. 1E). To reveal the topography of the subpopulations, we colored the sectors of the tumor in Figure 1F.
Classification of tumors
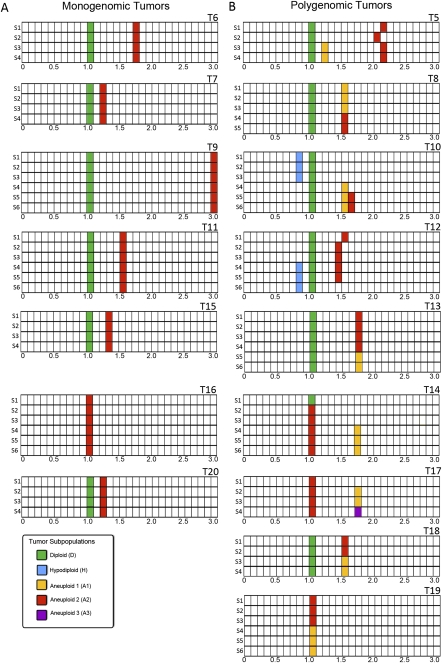
We classified 16 tumors into monogenomic and polygenomic by SPP (Fig. 2). Seven tumors were considered monogenomic. Six of the monogenomic tumors (T6, T7, T9, T11, T15, and T20) contained in all sectors a single distribution of aneuploid nuclei with DNA indices of 1.2 to 3.0 along with the expected diploid fraction of index 1.0, presumably composed of stroma and immune cells. The aneuploid fractions all showed abnormal CGH profiles, but within each tumor this profile was highly similar in every sector. One tumor (T16) had a single FACS peak (with a DNA index of 1.0), but this peak contained a highly rearranged pseudo-diploid tumor population in every sector, as revealed by CGH.
Figure 2.
Summary of Sector-Ploidy-Profiling (SPP) results for tumors T5–T20. (A) Monogenomic tumors. (B) Polygenomic tumors. Tumors were cut into four to six sectors. Nuclei were isolated from each sector and sorted by FACS according to differences in total genomic DNA content. DNA content is plotted on the x-axis (calibrated with a normal diploid control with a DNA index 1.0). Tumor sectors are plotted on the y-axis (S1–S6). Filled blocks indicate FACS peaks. Colors represent different subpopulations as distinguished by their CGH profiles: (blue) hypodiploid; (green) normal diploid; (orange, red, purple) distinguishable aneuploid tumor subpopulations. The total number of colors used in the schematic of a given tumor is the same as the total number of subpopulations distinguished in that tumor. For example, tumor T12 contains four subpopulations: one diploid subpopulations present in all sectors, one hypodiploid subpopulation present only in sectors 1–3, one aneuploid subpopulation present only in sectors 4–6, and a second aneuploid subpopulation present only in sectors 5–6.
Nine tumors were classified as polygenomic and displayed considerable complexity. Eight had multiple peaks of ploidy. In every case, subpopulations distinguishable by total DNA content were also clearly distinguishable by variation in their CGH profiles. Three tumors had more than one aneuploid subpopulation distinguishable by FACS (T5, T10, T12). Three tumors had subpopulations of pseudo-diploid cells exhibiting aberrant CGH profiles (T14, T17, and T19). Five tumors had subpopulations with genomic transitions that were not evident from ploidy, but were distinguishable by sector when analyzed by CGH (T8, T13, T17, T18, T19). Two tumors had hypodiploid subpopulations (T10 and T12).
Lineage of subpopulations
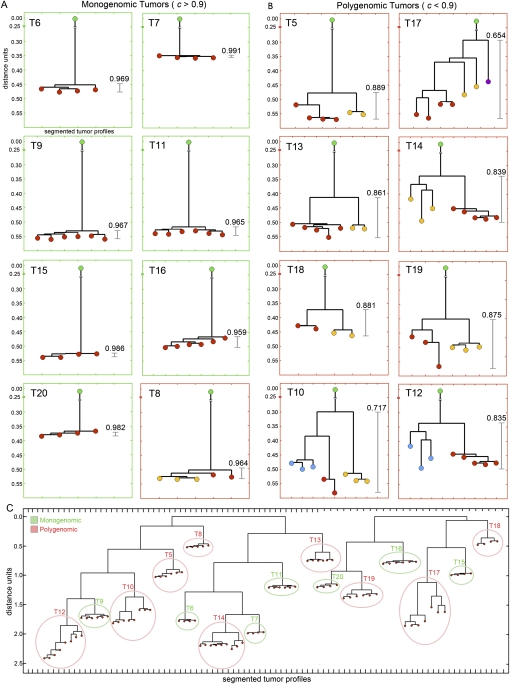
Similarities and differences between the profiles of subpopulations within a tumor were often obvious by plotting segmented profiles, but to discern variation with more rigor, we used computational methods that scale with large numbers of profiles. In order to equalize the dynamic range of amplifications and deletions, we used the log of the intensity ratios in the segmented profile. We computed the matrix of Pearson correlations between each individual profile and used a neighbor-joining algorithm (Saitou and Nei 1987) computed from one minus the correlation to construct distance trees between the profiles. We omitted the sex chromosomes to diminish extraneous correlation, and computed the distance using the segmented profiles to avoid the noise inherent in raw copy number data. The trees were rooted using flow-sorted diploid copy number profiles. The resultant trees for each profile are shown in Figure 3. The trees divide into two groups: those with a high correlation, >0.9 between all subpopulations (Fig. 3A), and others that were less correlated (Fig. 3B). The former group corresponds to the monogenomic tumor class and the latter to polygenomic tumors, with one exception (T8). In this case, the number of events that distinguishes subpopulations is very small: three focal amplifications on chromosome 12q21.1 (Fig. 4A). These differences are readily apparent by examining graphs of the segmented profiles, but less so by the mathematical measures. The color coding of the leaves of the tree match the color coding of Figure 2 and represent profile subgroups.
Figure 3.
Distance trees of copy number profiles. Neighbor-joining trees were constructed from distance trees by calculating 1-correlation matrices of all copy number profiles in a single tumor. (Green) The trees were rooted with a single coalesced diploid profile. (Green) Monogenomic tumors; (red) polygenomic tumors. (Red, yellow, blue) The leaves are colored to show different subpopulations as determined by comparing ROMA copy number profiles. (A) Tumor trees with a minimum correlation coefficient > 0.9. (B) Tumor trees with a minimum correlation coefficient < 0.9. (C) Distance trees of all tumor profiles without a diploid root node. Two trees were calculated separately: one from 85K experiments (T4–T14) and one from the 390K experiments (T15–T20).
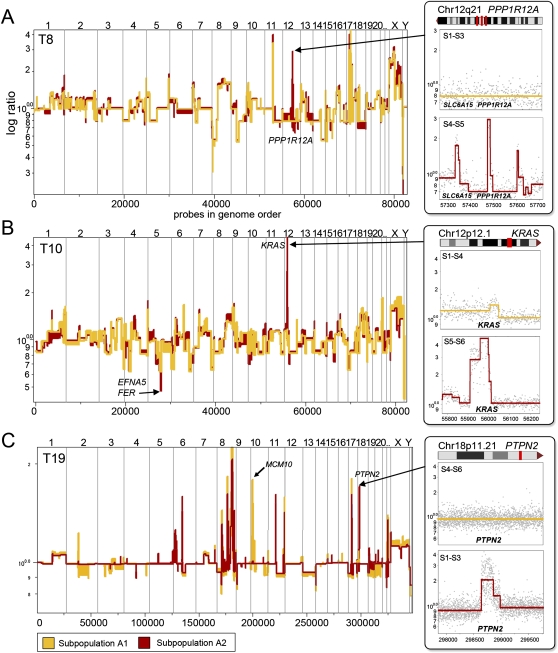
Figure 4.
Focal lesions that differ between subpopulations in single tumors. Segmented log ratio CGH data from coalesced tumor profiles are plotted in genome order. (A) Tumor T8 contains three focal amplifications, including the amplification of the PPP1R12A locus on Chr12q21, which is present in the A2 tumor subpopulation (red), but absent in A1 (yellow). (B) Tumor T10 contains a focal amplification of the KRAS locus on Chr12p12.1, which is present in the A2 tumor subpopulation (red), but absent in A1 (yellow). T8 also contains a homozygous deletion of the EFNA5 and FER locus on Chr5q21.3 in the (red) A2 subpopulations that is hemizygously deleted in A1 (yellow). (C) Tumor T19 contains a focal amplification of the PTPN2 locus on Chr18p11.21, which is present in the A2 subpopulation (red), but absent in A1 (yellow). T19 also contains a focal amplification of the MCM10 locus on Chr10p13 in the A1 tumor subpopulation that is absent in A2.
Overall, subpopulations within a tumor are very similar and share many or most chromosome breakpoints. On the other hand, we see very few common breakpoints between different tumors. This strongly implies that all subpopulations within a tumor have a common clonal origin. Given the potential importance of this conclusion, we felt it useful to validate it by purely computational analysis. The result of distance clustering of all tumor subpopulations clearly confirms that the subpopulations within a tumor are vastly more related to each other than the subpopulations between tumors (Fig. 3C). We cannot rule out that some tumors are mixtures of totally distinct clones, but we have never seen evidence for this alternate hypothesis (e.g., by observing two completely unrelated subpopulations within the same tumor).
Tumor progression
The order of progression can be inferred from subpopulation data if we make two assumptions. The first assumption is that the tumor subpopulations have arisen from a common progenitor tumor cell. The second assumption is that there is no “reversion to normal” in a lineage once a change occurs. In other words, observable mutations only accumulate. There can be violations of this assumption, for example, if a chromosome with changes is subsequently lost. Also, violations of this assumption can arise due to observing mixtures of subpopulations.
In almost all cases, the subpopulations within a tumor have many similar copy number changes (Fig. 4), but have few in common with other tumors, justifying the assumption of a common origin for subpopulations in each individual tumor. However, tumor T4 had sectors with essentially no discernible copy number changes (“flat” profiles), and other sectors with many chromosomal breakpoints (Supplemental Fig. S1). The sectors with flat profiles nevertheless were full of malignant cells as judged by histopathology. Thus a common origin for tumor cells with flat profiles and for those with copy number changes cannot strictly be inferred.
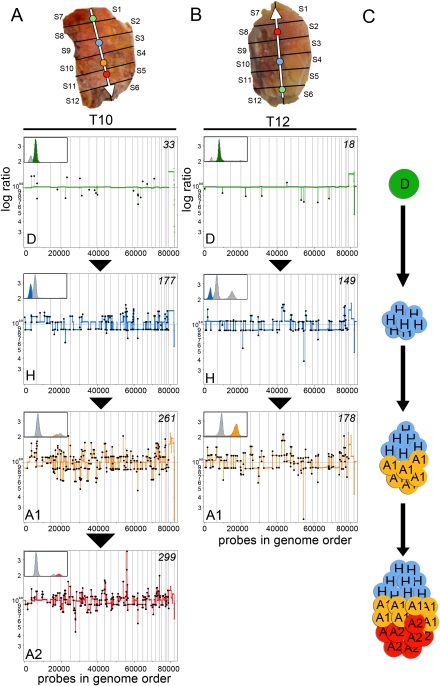
In the general case, we assume a common clonal origin and make inferences about the order of progression. Two of the most extreme examples of progression are seen in tumors T10 and T12, which have a hypodiploid state and an aneuploid state (Figs. 2B, 5). Using the assumption of irreversibility, we can assert that the aneuploid state derives from the hypodiploid state, as the aneuploid tumor cells display many more chromosomal breaks (Fig. 5). The T10 and T12 hypodiploid cancer genomes have what we previously called a “saw-toothed” profile (Hicks et al. 2006). This pattern is associated with basal-like expression breast cancer subtypes (Bergamaschi et al. 2006; Chin et al. 2006). In these two examples, the hypodiploid subpopulation progress to aneuploid and acquire focal amplifications and deletions.
Figure 5.
Genomic progression from hypodiploid to hyperaneuploid. Coalesced, segmented copy number profiles are ordered in increasing numbers of chromosome breakpoints. The topography of the subpopulations in the tumor sectors is shown with a white vector to indicate the direction of progression. FACS histograms are shown with the gated subpopulation highlighted in color. (A) Tumor T10 progresses from diploid (D) (green) to hypodiploid (H) (blue), to hyperaneuploid (A1) (yellow), to hyperaneuploid (A2) (red), as the number of chromosome breakpoints increases. (B) Tumor T12 progresses from diploid (D) (green) to hypodiploid (H) (blue) to hyperaneuploid (A1) (yellow). (C) Illustration of the clonal expansion of subpopulations that occur as the tumor grows.
The most prominent differences between populations were changes in the copy number of broad chromosomal regions. However, many polygenomic tumor subpopulations diverged by a small number of focal (narrow) genetic events, and we may infer that these focal changes occurred “late,” after tumor initiation and considerable expansion. Overall, we identified 24 focal lesions that differed between tumor subpopulations: 12 amplifications and 12 deletions (Supplemental Table S2). As we expected, many focal amplifications encompassed known oncogenes, including KRAS, PPP1R12A, HRASLS, MYC, RAD52, and RARA; while the deletions eliminated known tumor suppressors: CDKN2A, CASK, EFNA5, FER, PAX8, and ERCC3 (Futreal et al. 2004). Furthermore, we identified many focal deletions and amplifications containing single genes not previously implicated in cancer, including CACNA1C, HYDIN, SLC6A15, DCLK2, DNER, and C11ORF87.
We illustrate focal differences with three polygenomic tumors (T8, T10, and T19). The T8 tumor subpopulations diverged by only three tandem genomic amplifications on chromosome 12q21.1 present in the A1 tumor subpopulations in sectors 4 and 5, but not sectors 1 to 3 (Fig. 4A). These focal regional amplifications encompassed three single genes—BC061638, SLC6A15, and PPP1R12A—the former of which have not previously been implicated in cancer. The T10 tumor subpopulations diverged by only a single genomic amplification and a single deletion (Fig. 4B). The region of chromosome 12p12.1 contains the KRAS oncogene and was present at greater than 10 copies in the A2 subpopulation in sectors 5 and 6, but was only present in three copies in the A1 subpopulation. The T19 tumor subpopulations diverged by two amplifications on chromosome 10p14-p12.33 and 18p11.21 containing the MCM10 and PTPN2 oncogenes, respectively. In the next section, we use these focal changes to analyze the spatial relationship of subpopulations by FISH.
Spatial organization of subpopulations
It is evident even from our crude dissections that some tumor subpopulations are regionally segregated, while in other cases, two or more subpopulations co-occupy the same sector. To explore this further, we used interphase FISH to visualize single tumor cells using the subpopulation-specific chromosome markers in tumor T10 (Fig. 7B, see below). Tumor T10 is made up of one hypodiploid subpopulation (H) occupying sectors 1–3 and two distinct aneuploid tumor subpopulations (A1 and A2) that co-occupy sectors 5 and 6 (Fig. 1). The A2 tumor subpopulation diverges from the A1 subpopulation by only two genetic lesions: a homozygous deletion on chromosome 5q21.1-22.1 and the amplification of more than 10 copies of the KRAS locus at 12p12.1 (Fig. 4C). Both of the other tumor subpopulations (A1 and H) carry three copies of KRAS according to their CGH profiles. Thus, a FISH probe to the amplified KRAS locus serves to distinguish A2 from both A1 and H subpopulations.
Figure 7.
Intermixing of tumor subpopulations in tissue sections. A FISH probe strategy was used to mark chromosomes that are differentially amplified in two tumor subpopulations (A1 and A2) in tissue sections from sector 5 and sector 6 of T10. (A) Tumor T10 contains four sectors (S11, S12, S5, S6) with similar FACS histograms. The FACS histogram from sector 5 is shown and contains one diploid peak (green) and two aneuploid peaks that were gated and analyzed by CGH (yellow and red). (B) Segmented copy number data are plotted with FISH probes annotated to show the strategy for distinguishing the diploid cells from the A1 and A2 tumor subpopulations. The MYC probe on chromosome 8q24.21 (orange) detects two copies in the diploid cells and three copies in both of the tumor subpopulations (A2 and A3). LCON (purple) and RCON (blue) are control FISH probes on Chr12p12.1 that report two copies in all of the subpopulations. The KRAS (red) and ETNK (green) probes report six to 10 copies in the A2 subpopulation, but not in A1. (C,D) Tissue sections from T10 sector 5 show three types of cells: D diploid, A1 tumor cells, and A2 tumor cells. Diploid cells contain two copies of all of the probes. A1 tumor cells contain three copies of MYC and two copies of the other probes. The A2 tumor cells display a bright yellow signal resulting from the colocalization of the KRAS and ETNK probes, which are present in high copy number. (E,F) DAPI channels are false-colored to show the location of the three cell types: D (green), A1 (yellow), and A2 (red) in the tissue sections from panels C and D. The three cell types are stochastically intermixed in the tissues.
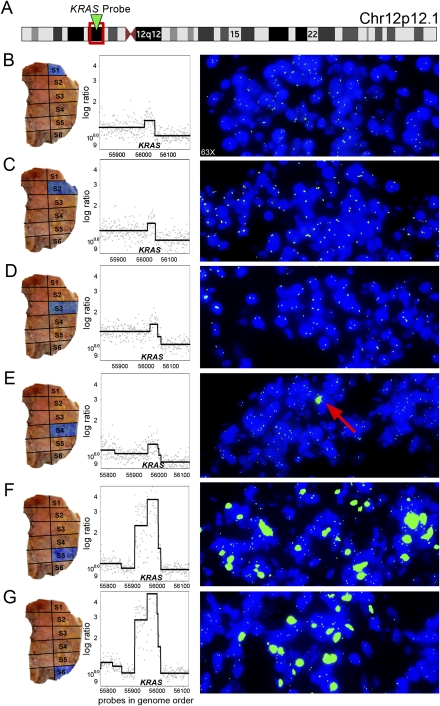
The regional segregation of tumor subpopulations predicted by ROMA is confirmed in T10 through interphase FISH by hybridizing a KRAS probe to the six tissue sections corresponding to the sectors analyzed by ROMA (Fig. 6). Many of the tumor cells from sectors 5 and 6 contained a highly amplified KRAS locus. Within the other sectors (1–4), the stroma and tumor cells exhibited just two or three copies of the KRAS locus expected from the CGH profiles. However, in two microscopic fields of about 500 tumor cells in sector 4, we observe one isolated cell that was highly amplified for KRAS (Fig. 6E).
Figure 6.
Regional amplification of the KRAS locus. Tissue sections from sectors 1–6 from tumor T10 are hybridized with a single FISH probe specific to the KRAS locus. (B–G, left) The topography of each tumor sector from which the tissues sections are cut. The log ratio and segmented copy number data of the KRAS amplification are also shown for each tumor sector. (A) Ideogram showing the cytobands and location of the KRAS FISH probe on chromosome 12p12.1. (B–D) Tissue sections from sectors 1–3 show two or three copies of the KRAS locus in the stromal and tumor cells. (E) Sector 4 contains a majority of tumor and stromal cells with two or three copies of the KRAS locus; however, one tumor cell shows a massive amplification of the KRAS locus. (F–G) Sectors 5 and 6 show numerous tumor cells with a high copy number of KRAS as a homologous staining region intermixed with other stromal and tumor cells that contain two or three copies of the KRAS locus.
The presence of multiple tumor subpopulations in sectors is obvious in tumors where the FACS histograms contain multiple aneuploid peaks. It is not clear from FACS, however, whether these co-occupied sectors result from our gross dissection crossing a boundary between segregated neighborhoods, or, alternatively, from an organization in which the subpopulations physically intermix. To further explore this level of organization in tumor T10, we used a complex of FISH probes capable of distinguishing subpopulations A1 and A2 from normal stroma and from each other. To distinguish A1 and A2 from normal stroma, we used a MYC probe present in both the A1 and A2 at a copy number of three. To distinguish A2 from A1, we used two probes (ETNK and KRAS) that colocalize to the region with a highly amplified KRAS locus in A2. We visualized all cells, tumor and diploid, using two probes, LCON and RCON, that map just outside the amplified region on A2. The probe scheme and location of the mixed sector 5 of T10 are shown in Figure 7B. The results of multicolor FISH performed on tissue sections from sector 5 are shown in Figure 7, C and D. These FISH experiments allowed us to clearly identify the diploid cells, the A1 subpopulation and the A2 subpopulation (D, A1, and A2 in Fig. 7C,D) and reveal that single A1 and A2 tumor cells are intermixed, rather than occupying separate domains (Fig. 7E,F).
Discussion
Dissecting the clonal composition of tumors at the genetic level is key to understanding the nature and progression of cancer and assessing prognosis and treatment. Genomic heterogeneity has long been reported in breast tumors, but with conflicting results, some suggesting that breast tumors are homogeneous (Noguchi et al. 1992, 1994; Endoh et al. 2001) and some heterogeneous (Teixeira et al. 1995, 1996; Farabegoli et al. 2001; Shipitsin et al. 2007). These reports were based on analysis of single samples from whole tumors, in which the subpopulations were not separated by differences in topography or ploidy. Only one study examined genomic variation in regionally separated tumor quadrants using CGH and concluded that some breast tumors had genetically distinct quadrants (Torres et al. 2007). Our preliminary analysis of T1–T4 in which we used sectoring and CGH is consistent with this earlier study. In our full study, we analyze a larger number of samples, and more sectors per tumor, and use separation of subpopulations by ploidy as well as FISH to study the clonal composition of tumors. As a result, we describe heterogeneity in both greater breadth and detail, enabling us to infer the progression of subpopulations.
In summary, we find that clonal genomic heterogeneity in breast cancers is very common. We identified 11 polygenomic tumors in our sample of 20. In heterogeneous tumors, we observed that the subpopulations may be anatomically separate or intermixed. We also find that these tumors consist of only a few major subpopulations. As we showed for one case, differences in the genome of subpopulations can be exploited to visualize the population substructure of a solid tumor by FISH, enabling us to unravel the developmental organization of tumor growth and the migratory pattern of cells within the tumor. From the shared chromosomal breakpoints, we infer that tumor subpopulations have a common genetic lineage. By comparing subpopulations, we can infer the order of certain genomic events.
In some tumors (T4, T5, T10, T12, and T14) the subpopulations differ by many genomic events. In the case of T4, we observe one subpopulation without discernible genomic copy number changes and another subpopulation with many events. In a previous study (Hicks et al. 2006), we reported that ∼10% of breast cancers had profiles with no discernible events. Perhaps those profiles arose from analysis of breast cancers in very early stages or from sampling only one subpopulation in the tumor. In all the other cases reported here, the subpopulations share many chromosomal events, but the total number of events is substantially greater in certain subpopulations. In T10 and T12 the subpopulations with lower numbers of events are hypodiploid, and the subpopulations with higher numbers are clearly aneuploid, strongly suggesting that a hypodiploid state preceded the aneuploid state. These two were the only tumors displaying the “sawtooth” pattern of genomic breaks (Hicks et al. 2006). Recent experiments have shown evidence that the basal-like expression subtype of breast cancer and BRCA1 tumors display the sawtoothed genome profile, with extensive low-level chromosomal loss and gains (Bergamaschi et al. 2006; Chin et al. 2006). Our results suggest that the extensive chromosomal loss may represent a common early stage in the evolution of basal-like subtypes, which is then followed by increased ploidy.
In contrast, in some tumors the subpopulations differ by only a few focal events. Events common to two profiles are “early” (prior to their divergence), while events unique to the profiles are “late” (after their divergence). In Supplemental Table 2 we list those focal changes that we classify as “late” and are therefore implicated in progression as opposed to initiation. These loci contain many well-known cancer genes, such as KRAS, which were first discovered on the basis of being able to initiate malignancy; however, many loci contain single genes that have not previously been implicated in cancer (Supplemental Table S2) and are worthy of more study. Many of the focal amplifications and deletions that we identified are regionally segregated in the tumor (Supplemental Table S2). Regional amplifications have previously been reported in glioblastomas, where the amplification of EGFR was shown to occur only in specific anatomical locations (Nafe et al. 2004). Our data show that regional amplifications and deletions occur frequently in the polygenomic breast tumors.
Several, but not all, polygenomic tumors showed evidence of two tumor subpopulations co-occupying a tumor sector. SPP is insufficient to determine if the co-occupying subpopulations are intermixed at the cellular level. However, once subpopulations are identified, molecular markers can be used to examine the spatial organization of the subpopulations at the cellular level. For example, tumor T10 had three tumor subpopulations: H, A1, and A2, with the latter two intermixed. A1 and A2 were very similar, differing by a massive amplification of the KRAS locus. This amplification, and the amplification of nearby genes, provided us with FISH markers to distinguish A2 from A1 in tissue sections. Based on the discrete breakpoints of the amplicon in ROMA profiles of both S5 and S6, we believe that this amplification occurred in a single cell similar to the A1 subpopulation that subsequently underwent clonal expansion and finally diverged to become the A2 subpopulation present throughout these sectors. We observed a pattern of extensive intermixing of A2 and A1 in sectors 5 and 6, and very limited penetration of A2 in sector 4. We can think of three reasonable and nonexclusive explanations for intermixing subpopulations. First, the subpopulations A1 and A2 cooperate, and their mutual presence has a selective advantage. Second, A1 provides a hospitable environment into which A2 can invade, whereas normal stroma mixed with H does not. Last, A2 originated in sector 6 and has only begun invading its way back into the remainder of the tumor. The last explanation is consistent with recent experiments suggesting that the overexpression of KRAS leads to increased cell migration (Fotiadou et al. 2007).
In our study, we analyzed only histological grade III (18/20) and grade II (2/20) ductal carcinomas (see Supplemental Table S1). Thus we could not correlate different tumor grades with the monogenomic or polygenomic classes. However, the fact that we observe both classes in grade III tumors suggests that they do not represent exclusive stages of progression. We also tested for correlation of clinical parameters including ER, PR, and Her2 status (when available) for each tumor with the monogenomic and polygenomic classes using the Fischer's exact test, but did not find any significant correlations (data not shown). Some triple negative tumors, for example, were classified as monogenomic and some as polygenomic tumors. While our samples were limited to only 20 tumors, our current data suggest that the ER, PR, and Her2 clinical parameters show no specific correlation with either class of genomic heterogeneity. Furthermore, we scored the tumor grade in H&E-stained tissue sections from the four to six sectors of T1–T10 to see if a change in tumor grade correlated with the polygenomic tumors. We found no significant correlations: Polygenomic tumors often contained the same high grade (III) in all four to six tumor sectors. We do not have expression data for the tumors we studied, so we cannot say if the expression subtype correlates with genomic heterogeneity, or if heterogeneity accounts for the failure of some breast cancer expression profiles to classify neatly into subtypes.
Much can be learned by discerning the subpopulations in a tumor and their spatial organization. Such analysis can be used to explore theories of cancer progression, patterns of growth (Norton and Massague 2006), migration, and metastasis (Liu et al. 2009) and may be of use in clinical settings. For example, clinical pathologists have long been aware of tumor heterogeneity and report the highest tumor grade observed after a fairly exhaustive survey of the tumor mass. However, as we have shown here, histological heterogeneity does not by itself imply genomic heterogeneity or vice versa. Genome-wide measures derived by sampling a single region may not be representative of the entire tumor when subpopulations are anatomically segregated. The degree of genomic heterogeneity itself might be a useful clinical parameter and could be missed entirely if not deliberately sought.
The clonal evolution models for tumor progression are consistent with our results in the polygenomic tumor subpopulations. The primary assumption of the clonal evolution models (monoclonal and polyclonal) is that the majority of cancer cells are capable of unlimited proliferation. This assumption contrasts with the fundamental assumption of the cancer stem cell hypothesis, which states that only a rare subpopulation of tumor cells is capable of unlimited proliferation, while the vast majority are only capable of normal cell division potential. In the polygenomic tumors, we observed that the majority of chromosome breakpoints are persistent throughout the tumor in all subpopulations, suggesting that the majority of cells are capable of unlimited proliferation.
Clearly, cancers must evolve by a series of discrete events, so finding heterogeneity is not unexpected. What is perhaps surprising is that our data show that the genomic heterogeneity of tumors can be ascribed to relatively few homogeneous subpopulations. While we do see evidence of gradualism in some subpopulations, there are often large gaps in some of the distance trees constructed from profiles of subpopulations from the polygenomic tumors. Similar observations of gaps in the fossil records plague models of biological evolution (Eldredge and Gould 1972). Moreover, in all cases, the “inferred” common progenitor of subpopulations is already at a great distance from “normal” (Fig. 3). Apparent gaps in the distance tree can be explained several ways. Perhaps only after the slow accumulation of multiple changes does a cancer subpopulation suddenly emerge with an enhanced capacity for clonal expansion. Alternatively, sudden changes in genomic profile occur by catastrophic mitotic events or by cell fusion, with the subsequent destabilization of the chromosomes. In some cases, something even more radical might be occurring: The cancer gradually evolves off-site at a distant metastasis, acquiring a dramatically altered profile, and then returns to the primary and greatly expands its mass.
We observe a significant proportion of tumors that are apparently monogenomic, and even in the polygenomic tumors we never distinguish more than three major tumor subpopulations. However, our assessment of tumor heterogeneity is likely to be an underestimate. Minor and very heterogeneous subpopulations will be averaged into main subpopulations if they share DNA index. Moreover, the tumor dissection will not in general follow the natural boundaries of subpopulations, further blurring our assessments. We are limited in our method of separating subpopulations by sector and ploidy. However, we are currently exploring a method that does not share these limitations, namely, the analysis of copy number in tumors by single-cell DNA sequencing. Although not without its own limitations, single-cell analysis has the potential to further clarify the extent and origins of tumor heterogeneity, and more importantly, the genetic pathways of tumor progression.
Methods
Patient samples
Twenty frozen primary ductal carcinomas were obtained from the Cooperative Human Tissue Network (T1–T7), Peggy Kemeny at North Shore University Hospital (T7–T8), Asterand Corporation (T16–T17), Larry Norton at Memorial Sloan-Kettering Cancer Center (T12–T14), and from Hanina Hibshoosh at Columbia University (T19–T20).
Sector macro-dissection
The 1–2-cm2 frozen tumors were macro-dissected into eight to 16 sectors of equal size using surgical scalpels. Half of the sectors from each tumor were used to prepare tissue sections at 6 μm in size using a cryomicrotome. The other half of the adjacent tumor sectors were used to isolate nuclei for SPP.
FACS
Nuclei were isolated from tumor samples by finely mincing a tumor sector in a Petri dish in 1.0–2.0 mL of NST-DAPI buffer (800 mL of NST [146 mM NaCl, 10 mM Tris base at pH 7.8, 1 mM CaCl2, 21 mM MgCl2, 0.05% BSA, 0.2% Nonidet P-40]), 200 mL of 106 mM MgCl2, 10 mg of DAPI, and 0.1% DNase-free RNase A using two no. 11 scalpels in a cross-hatching motion. Minced tissue was stored on wet ice for 15 min. Before flow cytometric analysis, samples were filtered through 37-μm plastic mesh. In all LSRII and FACS Vantage analysis, a small amount of prepared nuclei from each tumor sample was mixed with a diploid control sample (derived from a lymphoblastoid cell line of an apparently normal person) to accurately determine the diploid peak position within the tumor DNA content distribution and establish FACS collection gates. Nuclei were sorted with a Becton Dickinson FACS Vantage DiVa Flow Cytometer and Cell Sorter by gating cellular distributions with differences in their total genomic DNA content according to DAPI intensity. Additionally, a small sample of cells (n < 5000) from the adjacent sectors (that were used for histology) had nuclei isolated and stained with DAPI for analysis by a Becton Dickinson LSRII flow cytometer to generate a histogram of the DNA distributions in order to determine if they were consistent with the flow-sorted tumor sectors.
Comparative genomic hybridization using ROMA
DNA was isolated from the flow-sorted nuclei using the QIAGEN Genomic DNA Isolation Kit. A total of 200 ng of DNA was used to make complexity-reducing representations of genomic DNA for whole-genome copy number analysis by ROMA as described by Grubor et al. (2009). ROMA greatly increases signal-to-noise ratios and diminishes the amount of sample required for analysis; therefore, no additional whole-genome amplification step was required from the tumor sectors. Samples were hybridized on two array platforms: 85K arrays based on BglII representations (samples T1–T14), and 390K arrays based on DpnII representations, depleted of DpnII fragments containing AluI sites (T15–T20). The microarrays were custom designed with probes complementary to the complexity-reducing representations and manufactured by NimbleGen. Hybridizations of the 85K experiments were performed in color reversal to prevent color bias and ensure data quality, while 390K experiments were performed without a dye swap. All tumor samples were cohybridized with a reference genome from fibroblast DNA.
Informatics
The ROMA experiments were scanned, gridded, and normalized with a Lowess curve-fitting algorithm followed by a local normalization as described by Hicks et al. (2006). The data were imported and analyzed using Splus (Insightful) and Matlab (Mathworks), and the geometric mean ratio was computed from each color channel. In color-reversal experiments, the geometric mean of two log ratios was calculated. The data were then segmented to define nonoverlapping genomic regions that vary in copy number across the human genome using both the Kolmogorov-Smirnov algorithm (Grubor et al. 2009) and the circular binary segmenter (Venkatraman and Olshen 2007). The segmented genomic copy number profiles from each sector were then used for the statistical analysis.
Fluorescence in situ hybridization
FISH probes were constructed by one of two methods. The KRAS probe used in Figure 6 was designed using the PROBER algorithm and pooled from PCR products 500–1400 bp in length (Navin et al. 2006). The LCTR, RCTR, ETNK, and KRAS probes were designed using bacterial artificial chromosomes from the UCSC Genome Browser. FISH analysis was conducted on interphase cells in 10-μm frozen tissue sections. These probes were hybridized to frozen tissue sections that were fixed in methanol overnight and moved to 70% ethanol. The FISH experiments were performed as reported by Hicks et al. (2006) with DAPI staining to visualize the nucleus. Selected cells were photographed in a Zeiss Axioplan 2 microscope equipped with an Axio Cam MRM CCD camera and Axio Vision software.
In order to mitigate the analysis of shaved nuclei, we employed three precautionary steps. First, we cut relatively large (7 μm) tissue sections using a cryomicrotome in order to encompass whole nuclei. Second, we captured Z-planes that contained 40–50 images from each 63× objective microscope using a mechanical stage. Using Axiovision Software, we generated Z-plane images of the DAPI-stained nuclei, which we used to exclude any partially shaved nuclei in the quantification of FISH probe signals. Third, we hybridized two diploid control probes to all nuclei (RCON and LCON) that surround the KRAS amplification on chromosome 12p12.1 and a MYC control probe on chromosome 8. These control probes served as indicators that the nucleus was not shaved on chromosome 12p12.1. When we did not observe two copies of each control probe in the nucleus, it was not scored for copy number. Using these three criteria, we observed that the majority of cells that we scored (89.69%) showed copy number signals consistent with one of three subpopulations: D, A1, or A2. However, some nuclei (10.31%) did report patterns of copy number that were inconsistent with the predicted subpopulations. We cannot distinguish if these nuclei represented a minor subpopulation or if they were shaved nuclei. Finally, in order to avoid probe artifacts, we did not score any nuclei where the probes did not overlap the DAPI channel.
Statistics
In order to identify highly similar copy number profiles in single tumors for profile coalescing, we calculated a matrix of Pearson correlations between profiles and used a neighbor-joining algorithm (Saitou and Nei 1987). The neighbor-joining algorithm was used in place of an ultrametric method because we did not assume an equal distance from each copy number profile to the root node. In our calculations of correlation matrices, we used segmented data from the autosomes in order to exclude extraneous correlations from the sex chromosomes, and since our reference sample was male. The correlation matrix was converted to a distance matrix using (1-correlation). Clusters of highly similar copy number profiles were then “coalesced” into mean segmented profiles to represent each subpopulation in a single tumor. The pairwise difference between coalesced profiles was then calculated to identify subpopulation-specific amplifications and deletions. Each genomic lesion was annotated to identify UCSC genes (Hsu et al. 2006) and cancer genes. Cancer genes were identified using a compiled database from the cancer gene consensus (Futreal et al. 2004) and the NCI cancer gene index (Sophic Systems Alliance Inc., Biomax Informatics A.G). Distance trees were calculated using the same methods for coalescing profiles (1-Pearson correlations and neighbor-joining). A single distance tree was calculated for each tumor (Fig. 3A,B). Additionally, the minimum correlation between all tumor profiles is reported as the clonal correlation (cc), a measure of intratumor heterogeneity in Supplemental Table S1. In a separate analysis, we used the same methods to construct a distance tree using all tumor copy number profiles. In this analysis, we clustered the 85K (T4–T14) and 390K (T15–T20) tumor profiles separately and did not use any diploid profiles as a root node (Fig. 3C).
Acknowledgments
We thank Pamela Moody and Tara Spencer at the CSHL FACS facility; Stephen Hearn at the CSHL microscope facility; Michael Ronemus and Diane Esposito for useful discussions; and Deepa Pai, Yamrom Boris, and Anthony Leotta for informatics support. We also thank Patrick Blake and Nancy Navin at Sophic Systems Alliance Inc. for support with the NCI Cancer Gene Index annotations. This work was supported by the NCI T32 Fellowship to N.N., and grants to A.Z. from the Swedish Cancer Society (0046-B04-38XAC), the Stockholm Cancer Society (03:171 and 02:144), and the Stockholm Cancer Society (03:17). This work was also supported by grants to M.W. from the Department of the Army (W81XWH04-1-0477) and the Breast Cancer Research Foundation. M.W. is an American Cancer Society Research Professor.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE16672.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099622.109.
References
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;8:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A, Medina D. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- Aubele M, Mattis A, Zitzelsberger H, Walch A, Kremer M, Hutzler P, Hofler H, Werner M. Intratumoral heterogeneity in breast carcinoma revealed by laser-microdissection and comparative genomic hybridization. Cancer Genet Cytogenet. 1999;110:94–102. doi: 10.1016/s0165-4608(98)00205-2. [DOI] [PubMed] [Google Scholar]
- Bachtiary B, Boutros PC, Pintilie M, Shi W, Bastianutto C, Li JH, Schwock J, Zhang W, Penn LZ, Jurisica I. Gene expression profiling in cervical cancer: An exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, Kim H, Wang P, Sørlie T, Hernandez-Boussard T, Lonning E, Tibshirani R, Børresen-Dale A, Pollack R. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman T, Roydasgupta R, Kuo WL, Lapuk A, Neve R, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Cole K, Krizman B, Emmert-Buck M. The genetics of cancer—a 3D model. Nat Genet. 1999;21:38–41. doi: 10.1038/4466. [DOI] [PubMed] [Google Scholar]
- Corver W, Middeldorp A, ter Haar N, Jordanova S, van Puijenbroek M, van Eijk R, Cornelisse J, Fleuren J, Morreau H, Oosting J, et al. Genome-wide allelic state analysis on flow-sorted tumor fractions provides an accurate measure of chromosomal aberrations. Cancer Res. 2008;24:10333–10340. doi: 10.1158/0008-5472.CAN-08-2665. [DOI] [PubMed] [Google Scholar]
- Eldredge N, Gould J. Punctuated equilibria: An alternative to phyletic gradualism. In: Schopf TJM, editor. Models in paleobiology. Freeman; San Francisco: 1972. pp. 82–115. [Google Scholar]
- Endoh Y, Tamura G, Kato N, Motoyama T. Apocrine adenosis of the breast: Clonal evidence of neoplasia. Histopathology. 2001;38:221–224. doi: 10.1046/j.1365-2559.2001.01095.x. [DOI] [PubMed] [Google Scholar]
- Farabegoli F, Santini D, Ceccarelli C, Taffurelli M, Marrano D, Baldini N. Clone heterogeneity in diploid and aneuploid breast carcinomas as detected by FISH. Cytometry. 2001;46:50–56. doi: 10.1002/1097-0320(20010215)46:1<50::aid-cyto1037>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;2:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Fiegl M, Tueni C, Schenk T, Jakesz R, Gnant M, Reiner A, Rudas M, Pirc-Danoewinata H, Marosi C, Huber H, et al. Interphase cytogenetics reveals a high incidence of aneuploidy and intra-tumour heterogeneity in breast cancer. Br J Cancer. 1995;72:51–55. doi: 10.1038/bjc.1995.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadou PP, Takahashi C, Rajabi HN, Ewen ME. Wild-type NRas and KRas perform distinct functions during transformation. Mol Cell Biol. 2007;27:6742–6755. doi: 10.1128/MCB.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, Yamrom B, Alex G, Pai D, Navin N, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis ROMA. Blood. 2009;113:1294–1303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, Lohr S, Wu TD, Cavet G, Zhang Z, Chant J. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- Heim S, Teixeira MR, Dietrich CU, Pandis N. Cytogenetic polyclonality in tumors of the breast. Cancer Genet Cytogenet. 1997;95:16–19. doi: 10.1016/s0165-4608(96)00322-6. [DOI] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Ignatiadis M, Sotiriou C. Understanding the molecular basis of histologic grade. Pathobiology. 2008;75:104–111. doi: 10.1159/000123848. [DOI] [PubMed] [Google Scholar]
- Johann J, Rodriguez-Canales J, Mukherjee S, Prieto A, Hanson C, Emmert-Buck M, Blonder J. Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J Proteome Res. 2009;5:2310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalique L, Ayhan A, Weale E, Jacobs J, Ramus J, Gayther A. Genetic intra-tumour heterogeneity in epithelial ovarian cancer and its implications for molecular diagnosis of tumours. J Pathol. 2007;3:286–295. doi: 10.1002/path.2112. [DOI] [PubMed] [Google Scholar]
- Komaki K, Sano N, Tangoku A. Problems in histological grading of malignancy and its clinical significance in patients with operable breast cancer. Breast Cancer. 2006;13:249–253. doi: 10.2325/jbcs.13.249. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions in cancer causations. Nat Rev Cancer. 2007;4:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Nafe R, Glienke W, Burgemeister R, Gangnus R, Haar B, Pries A, Schlote W. Regional heterogeneity of EGFR gene amplification and nuclear morphology in glioblastomas. An investigation using laser microdissection and pressure catapulting. Anal Quant Cytol Histol. 2004;26:65–76. [PubMed] [Google Scholar]
- Navin N, Grubor V, Hicks J, Leibu E, Thomas E, Troge J, Riggs M, Lundin P, Maner S, Sebat J, et al. PROBER: Oligonucleotide FISH probe design software. Bioinformatics. 2006;22:2437–2438. doi: 10.1093/bioinformatics/btl273. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Motomura K, Inaji H, Imaoka S, Koyama H. Clonal analysis of human breast cancer by means of the polymerase chain reaction. Cancer Res. 1992;52:6594–6597. [PubMed] [Google Scholar]
- Noguchi S, Motomura K, Inaji H, Imaoka S, Koyama H. Clonal analysis of predominantly intraductal carcinoma and precancerous lesions of the breast by means of polymerase chain reaction. Cancer Res. 1994;54:1849–1853. [PubMed] [Google Scholar]
- Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- Roka S, Fiegl M, Zojer N, Filipits M, Schuster R, Steiner B, Jakesz R, Huber H, Drach J. Aneuploidy of chromosome 8 as detected by interphase fluorescence in situ hybridization is a recurrent finding in primary and metastatic breast cancer. Breast Cancer Res Treat. 1998;48:125–133. doi: 10.1023/a:1005937305102. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;3:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MR, Pandis N, Bardi G, Andersen JA, Mitelman F, Heim S. Clonal heterogeneity in breast cancer: Karyotypic comparisons of multiple intra- and extra-tumorous samples from 3 patients. Int J Cancer. 1995;63:63–68. doi: 10.1002/ijc.2910630113. [DOI] [PubMed] [Google Scholar]
- Teixeira MR, Pandis N, Bardi G, Andersen JA, Heim S. Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 1996;56:855–859. [PubMed] [Google Scholar]
- Torres L, Ribeiro FR, Pandis N, Andersen JA, Heim S, Teixeira MR. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat. 2007;102:143–155. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]