Abstract
Invasive Klebsiella pneumoniae with hypermucoviscosity phenotype (HMV K. pneumoniae) is an emerging human pathogen that, over the past 20 y, has resulted in a distinct clinical syndrome characterized by pyogenic liver abscesses sometimes complicated by bacteremia, meningitis, and endophthalmitis. Infections occur predominantly in Taiwan and other Asian countries, but HMV K. pneumoniae is considered an emerging infectious disease in the United States and other Western countries. In 2005, fatal multisystemic disease was attributed to HMV K. pneumoniae in African green monkeys (AGM) at our institution. After identification of a cluster of subclinically infected macaques in March and April 2008, screening of all colony nonhuman primates by oropharyngeal and rectal culture revealed 19 subclinically infected rhesus and cynomolgus macaques. PCR testing for 2 genes associated with HMV K. pneumoniae, rmpA and magA, suggested genetic variability in the samples. Random amplified polymorphic DNA analysis on a subset of clinical isolates confirmed a high degree of genetic diversity between the samples. Environmental testing did not reveal evidence of aerosol or droplet transmission of the organism in housing areas. Further research is needed to characterize HMV K. pneumoniae, particularly with regard to genetic differences among bacterial strains and their relationship to human disease and to the apparent susceptibility of AGM to this organism.
Abbreviations: AGM, African green monkey; HMV K. pneumoniae, invasive Klebsiella pneumoniae with hypermucoviscosity phenotype; NHP, nonhuman primate; RAPD, random amplification of polymorphic DNA
Klebsiella pneumoniae is an enteric, gram-negative, lactose-fermenting bacillus with a prominent capsule. This bacterium has been associated with peritonitis, septicemia, pneumonia, and meningitis in both Old and New World primates,10,13,29 although it also is reported to constitute normal fecal and oral flora in many nonhuman primates (NHP).12 Pathogenic strains associated with the upper respiratory tract typically are heavily encapsulated.12 Over the past several decades, human medical literature indicates the emergence of an invasive K. pneumoniae disease in Taiwan and other Asian countries, in which community-acquired pyogenic liver abscesses have been attributed to strains of invasive K. pneumoniae with a unique hypermucoviscous phenotype (HMV K. pneumoniae).6,17-19,21,26,34 The hypermucoviscous phenotype has also been associated with other serious complications, including bacteremia, meningitis, and endophthalmitis. This strain of Klebsiella has become an emerging cause of pyogenic liver abscesses in some nonAsian countries, including the United States.16,20,36,39 The majority of clinical cases of HMV K. pneumoniae are in the Asian population, particularly in patients with diabetes mellitus.3,4,33 Determination of the HMV phenotype typically is based on a positive string test.8,35,39
Several virulence factors have been associated with HMV K. pneumoniae. Klebsiella spp. generally develop prominent polysaccharide capsules which increase virulence by protecting the bacteria from phagocytosis and preventing destruction by bactericidal serum factors. Capsular serotypes K1 or K2 have been reported as the major virulence determinants for human HMV K. pneumoniae liver abscesses.5,8,37,38 In addition, the mucoviscosity-associated gene magA, which encodes a structural outer membrane protein of the K1 serotype, and rmpA (regulator of the mucoid phenotype gene; located on a plasmid) have been proposed as virulence factors.9,27,31,40,41 Recently, it was suggested that 2 clones, CC23 K1 and CC82K1, are strongly associated with primary liver abscess and respiratory infection, respectively.2
Over a period of several months in 2005 to 2006, 7 African green monkeys (AGM; Chlorocebus aethiops) in the US Army Medical Research Institute of Infectious Diseases research colony developed abscesses in multiple locations and either died or were euthanized when the abscesses were determined to be nonresectable.35 HMV K. pneumoniae of the K2 serotype and carrying rmpA was determined to be the cause of the infection in 1 case, and the 6 other cases had similar clinical and pathologic features. This report35 is the only documentation, to our knowledge, of natural infection with HMV K. pneumoniae in NHP. As a result of these cases, the US Army Medical Research Institute of Infectious Diseases instituted policies to exclude HMV K. pneumoniae from the colony. The organism was included as a specific pathogen-free requirement for vendors, and K. pneumoniae culture results were reported during quarantine periods and on routine semiannual examination for all colony NHP.
Case Study
In March 2008, a 3.5-kg male rhesus macaque tested positive for HMV K. pneumoniae on an oropharyngeal sample obtained during routine semiannual physical examination. This animal was not assigned to an experimental protocol and had not been used in any previous experimental protocols. The macaque was serologically negative for cercopithecine herpesvirus 1, simian type D retrovirus, SIV, and simian T-cell leukemia virus 1 and tested negative for parasites by fecal flotation and direct smear. Although this animal showed no clinical signs of disease, concerns about transmission of HMV K. pneumoniae, which had previously caused fatal disease in AGM at the facility, led to the decision to euthanize the macaque.
Approximately 3 wk after the index case was identified and euthanized, the other 35 rhesus macaques that were housed in same room were cultured by using oropharyngeal and rectal swabs in an effort to increase sensitivity of testing. Culture results were positive for HMV K. pneumoniae in 3 male rhesus macaques in the room; 2 of the 3 animals were positive on both oropharyngeal and rectal culture, and the remaining macaque was positive on oropharyngeal culture only. At 5 wk after the index case, a 5.2-kg male cynomolgus macaque, located in a separate building and with no history of direct contact with the infected rhesus macaques, tested positive for HMV K. pneumoniae on a rectal culture during medical screening for an experimental protocol to which the animal had been assigned. This animal had cleared a 6-wk quarantine, during which he tested serologically negative for cercopithecine herpesvirus 1, simian type D retrovirus, SIV, and simian T-cell leukemia virus 1; tested culture negative by oral swab for Klebsiella spp., Salmonella spp., and Shigella spp.; and tested negative for parasites by fecal flotation and direct smear. Because of the severity of disease previously associated with this organism in AGM in our colony and because the pathogen had now been found in 2 separate buildings, we initiated screening of the entire NHP colony for HMV K. pneumoniae.
Here we describe an epidemiologic investigation of cynomolgus and rhesus macaques in our research colony that were infected with HMV K. pneumoniae. Our investigation included bacteriology, PCR, and random amplified polymorphic DNA (RAPD) typing of HMV K. pneumoniae,; we also tested the hypothesis that aerosols or droplets created during cage cleaning were responsible, in part, for transmission of the organism.
Materials and Methods
Animals.
The animals described in this report were maintained in AAALAC-accredited facility. All research was conducted as part of protocols approved by our institutional animal care and use committee and adhered to the Guide for the Care and Use of Laboratory Animals.28
The NHP colony consisted of 307 animals (51 AGM, 125 cynomolgus macaques, and 131 rhesus macaques). Rooms housing NHP contained only 1 species at a time, but multiple species were housed within a single wing or corridor of the NHP hallways and there was no defined entry order for the rooms. Semiannual evaluations of NHP included physical examination; complete blood counts and blood chemistry; serologic testing for measles, cercopithecine herpesvirus 1, simian type D retrovirus, SIV, and simian T-cell leukemia virus 1 in macaques; serologic testing for measles virus, SIV, simian T-cell leukemia virus 1, African green α herpesvirus, and rotavirus (group A) in AGM; culture of the oropharynx; and screening for parasites via fecal flotation and direct smear. Until identification of the 1st infected macaque, rectal cultures were not performed routinely unless an NHP showed clinical signs of enteric disease. All NHP were housed in 4.5-ft2 or 6.0-ft2 cages with 4 cages per rack (Allentown Caging Equipment, Allentown, NJ). The environmental conditions were maintained as recommended in the Guide for the Care and Use of Laboratory Animals28 (temperature, 16 to 29 °C; humidity, 30% to 70%; and 12:12-h light:dark cycle). Animals were fed a standard primate diet (diet 8714, Harlan Teklad, Madison, WI) supplemented with fruit and other food treats. Fresh water, provided ad libitum, was chlorinated at the municipal level and filtered (Edstrom Industries, Waterford, WI). Environmental enrichment (Challenge Ball, Kong, and Hercules Dental Device, Bio-Serv, Frenchtown, NJ) was provided, and cages were arranged so that the animals were facing each other across the room.
Bacteriology.
Oropharyngeal and rectal swabs were taken from 307 cynomolgus macaques, rhesus macaques, and AGM. Any animal that tested positive was removed from the room and housed in a room that contained only NHP that cultured positive for HMV K. pneumoniae. The rooms from which these NHP originated then underwent 6 wk of quarantine, during which weekly oropharyngeal and rectal swabs were cultured in an effort to eliminate all infected NHP from general housing. A total of 2297 oropharyngeal and rectal samples were collected and cultured from midMarch to early September 2008.
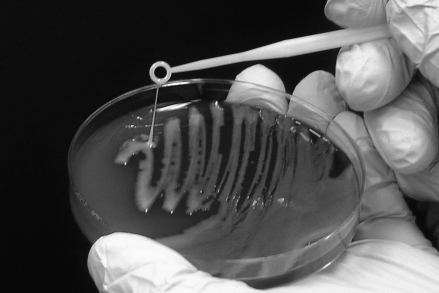
All swabs were collected by using culture collection and transport systems (BBL Culture Swab, Becton Dickinson, Franklin Lakes, NJ). The swabs were delivered to the microbiology laboratory, used to inoculate MacConkey agar plates, and incubated at 35 °C without CO2. The plates were examined the day after inoculation for the presence of hypermucoviscous, lactose-fermenting, gram-negative rods. All suspect colonies were analyzed by using an automated bacterial identification system (Vitek 2, bioMerieux, Hazelwood, MI) and subcultured on a 5% sheep blood agar plate to ensure purity. Any isolate yielding a probability of 90% or greater for K. pneumoniae was further analyzed through the string test, in which a bacterial loop was touched to a suspect colony on a 5% sheep blood agar plate and withdrawn slowly. Bacteria forming a ‘string’ of at least 5 mm were determined to be positive (Figure 1). Isolates determined to be HMV K. pneumoniae were boiled and the resulting DNA was frozen at −70 °C for molecular analysis.
Figure 1.
Positive string test. The HMV phenotype of K. pneumoniae is defined by a positive-string test. The test is performed by touching a colony with a bacterial loop and gently lifting. If a mucoid ‘string’ of 5 mm or more forms, the string test is considered positive.
Real-time PCR.
Rapid real-time PCR assays (TaqMan and TaqMan Minor Groove Binder assays, Applied Biosystems, Foster City, CA) were used to detect rmpA and magA, genes that are highly associated with the HMV phenotype; all HMV K. pneumoniae cultures from midMarch 2008 to September 2008 were tested as described previously.15 A total of 78 HMV K. pneumoniae samples were tested via PCR for the magA and rmpA genes as part of the current epidemiologic study.
RAPD analysis.
Bacterial genomic DNA was extracted (Illustra Genomic Prep Mini Spin Kit, GE Healthcare, Buckinghamshire, UK) according to the manufacturer's instructions. RAPD analysis (Ready-To-Go RAPD Beads, GE Healthcare) was performed by using a primer provided in the kit (primer 2, 5′ GTT TCG CTC C 3′); we tested six different random 10mer primers provided and selected primer 2 because it yielded easily readable patterns and the most bands that were well separated. AmpliTaq polymerase, dNTPs (0.4 mM each), bovine serum albumin (2.5 µg), buffer (3 mM MgCl2, 30 mM KCl, 10 mM Tris [pH 8.3]), and Stoffel fragment were provide as dried beads that were stable at room temperature. Primer 2 (10 µL; 5 pmol/µL), template DNA (10 µL; 5 ng/µL), and 5 µL sterile distilled water were added to the reaction beads for a total reaction volume of 25 µL. PCR amplification was performed in a thermocycler (Mastercycler, Eppendorf, Hamburg, Germany) by using 5 min at 95 °C followed by 45 cycles of 95 °C for 1 min, 36 °C for 1 min, and 72 °C for 2 min. PCR products were separated on a 1.5% agarose gel, which was stained with ethidium bromide and visualized by UV transillumination. DNA fragment patterns were compared by visual inspection.
Environmental testing.
A total of 326 samples were collected from various NHP housing rooms in the facility and divided among 3 test methods. For the first test, 258 environmental swabs were collected from predetermined sites over 6 wk. Swabs were taken at midday, after rooms had been cleaned and sanitized by the animal caretaker. Each week, 42 swabs were taken from the door knob, room corner, drain cover, handle of hose spray nozzle, lixit, air handling vent, and rubber surface of floor squeegee in 6 NHP housing rooms (total, 252 swabs). These rooms contained either infected, quarantined (HMV K. pneumoniae status unknown), or culture-negative animals. During week 5, additional swabs were taken from cage fronts of 3 different NHP cages in the room housing infected animals, and during week 6, swabs were taken from feces in pans of 3 infected macaques (total, 6 swabs).
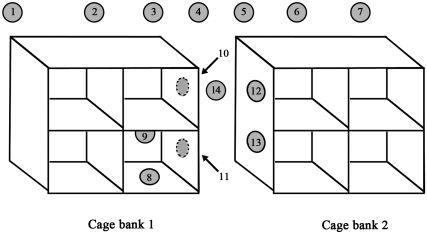
The second test involved affixing 14 MacConkey plates to the wall and on the surfaces of 2 banks of cages in a grid pattern (Figures 2 and 3), removing lids, allowing the caretaker to spray solid and liquid organic material out of 1 bank of cages, and then promptly removing the culture plates. This test was performed twice: once while the first bank of cages was sprayed out, and then again when the second bank of cages was sprayed. A total of 28 MacConkey plates were submitted for culture by using this test method.
Figure 2.
Culture plates affixed in a grid pattern around and on NHP cages. Once the plates were in place, the animal caretaker performed routine spray-down of organic material from the cages. The plates then were removed and cultured to determine the extent of environmental spread of HMV K. pneumoniae during routine cage-cleaning procedures.
Figure 3.
Grid pattern configuration of culture plates. MacConkey cultures plates were taped in positions 1 through 13, and the lids were removed. Plates 8 and 9 were placed in an empty cage, directly below and beside infected NHP. The animal caretaker then followed routine procedures to spray waste material out of cage bank 1, avoiding wetting of NHP in cages. Once the gross contamination was removed from cage bank 1, all culture plates were covered and removed. A second set of MacConkey plates were taped in positions 1 through 13 and the experiment repeated for cage bank 2.
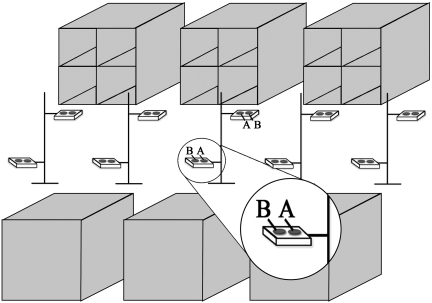
The third test involved exposing MAC culture plates, placed on trays throughout the room immediately after the caretaker cleaned the cages by using a high-pressure spray. Culture plates remained in place for various periods of time. Tray stands were constructed by using polyvinyl chloride material and contained 2 trays each: 1 at the height of upper cages, and the other at the height of the lower cages. The tray stands were placed in the center of the room, at 5-ft intervals along the length of the room (Figure 4). Each tray held 2 culture plates. One set of 10 plates (position A in Figure 4) was left in place from the completion of spraying to 60 min afterward. The second set of 10 plates (position B in Figure 4) was in place from the completion of spraying until 5 min after spraying; these plates then were replaced with 10 additional plates that remained in place from 5 min to 10 min after spraying. The plates were exchanged again; the final set of plates was placed in position B from 10 min to 60 min after spraying. A total of 40 culture plates were submitted for culture by using this third test method.
Figure 4.
Room configuration for culturing for HMV K. pneumoniae after cage spray. In an effort to determine the environmental spread of HMV K. pneumoniae, covered MacConkey culture plates were placed on trays while an animal caretaker sprayed waste material from cages. As soon as the caretaker had completed the spray-down, plate covers were removed. Plates in position A (inset) remained in place from 0 to 60 min after spraying. In position B, 3 sets of plates were rotated at predetermined time points: set 1 remained in place from 0 to 5 min, set 2 from 5 to 10 min, and set 3 from 10 to 60 min after spraying.
Results
Bacteriology.
From midMarch 2008 to early September 2008, a total of 2297 swabs from 307 rhesus macaques, cynomolgus macaques, and AGM were tested by oropharyngeal and rectal culture for HMV K. pneumoniae. Although 195 samples were positive for K. pneumoniae, only 81 of those samples were of the HMV phenotype. The 81 HMV K. pneumoniae samples represented 19 infected animals, of which 14 had multiple positive cultures during the monthly physical examinations. The single index animal (a macaque) was euthanized promptly after yielding a single positive sample, and another NHP had a single positive culture and then tested negative on multiple subsequent cultures. Another 3 animals were swabbed and then moved into biocontainment as part of an experimental protocol; positive results were identified after the animals were in biocontainment, which made further evaluation inadvisable. As a result, 78 positive HMV K. pneumoniae isolates were evaluated as part of this study.
PCR gene analysis.
Real-time PCR testing of the 78 HMV K. pneumoniae samples resulted in 49 rmpA+/magA− isolates, 17 rmpA−/magA+ isolates, and 12 rmpA−/magA− isolates. PCR results from 16 NHP (the index case and 15 NHP that were available for repeated cultures) are presented in Table 1. In some cases, an animal had PCR results that were variable between culture sites or over time.
Table 1.
HMV K. pneumoniae and rmpA/magA status in macaques
| HMV K. pneumoniae statusa by date |
|||||||||||
| Animal ID | Culture type | 3/6/08 | 3/13/08 | 4/7/08 | 4/28/08 | 5/9/08 | 5/20/08 | 5/28/08 | 6/17/08 | 7/21/08 | 8/18/08 |
| Rh1b | Oral | +/– | |||||||||
| Rectal | |||||||||||
| Rh2 | Oral | Neg | +/– | Neg | +/– | Neg | Neg | +/– | Neg | ||
| Rectal | Neg | Neg | +/– | +/– | +/– | +/– | Neg | ||||
| Rh3 | Oral | Neg | +/– | Neg | Neg | Neg | Neg | Neg | Neg | ||
| Rectal | +/– | +/– | +/– | Neg | +/– | +/– | +/– | ||||
| Rh4c | Oral | Neg | +/– | Neg | Neg | Neg | Neg | Neg | Neg | ||
| Rectal | +/– | +/– | +/– | +/– | Neg | –/+ | Neg | ||||
| Rh5 | Oral | Neg | +/– | +/– | +/– | Neg | +/– | +/– | |||
| Rectal | +/– | +/– | +/– | +/– | Neg | +/– | |||||
| Rh6 | Oral | Neg | Neg | Neg | Neg | Neg | Neg | Neg | |||
| Rectal | +/– | Neg | +/– | Neg | Neg | Neg | |||||
| Rh7 | Oral | Neg | +/– | +/– | Neg | Neg | Neg | ||||
| Rectal | +/– | +/– | +/– | +/– | Neg | ||||||
| Rh8 | Oral | Neg | +/– | Neg | Neg | Neg | |||||
| Rectal | +/– | +/– | –/+ | ||||||||
| Rh9 | Oral | Neg | Neg | −/− | Neg | Neg | |||||
| Rectal | Neg | Neg | Neg | Neg | |||||||
| Rh10 | Oral | Neg | Neg | +/– | Neg | Neg | |||||
| Rectal | Neg | Neg | Neg | Neg | |||||||
| Cy1 | Oral | Neg | Neg | Neg | Neg | Neg | Neg | Neg | |||
| Rectal | +/– | Neg | +/– | Neg | Neg | Neg | Neg | ||||
| Cy2c | Oral | Neg | Neg | Neg | −/− | −/− | +/– | Neg | |||
| Rectal | +/– | +/– | −/− | +/– | −/− | +/– | +/– | ||||
| Cy3c | Oral | Neg | −/− | +/– | −/− | −/− | −/− | Neg | Neg | ||
| Rectal | Neg | +/– | Neg | Neg | Neg | Neg | Neg | ||||
| Cy4c | Oral | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | ||
| Rectal | −/− | Neg | +/– | Neg | −/− | Neg | −/− | ||||
| Cy5 | Oral | Neg | Neg | +/– | Neg | Neg | +/– | Neg | |||
| Rectal | +/– | +/– | +/– | +/– | +/– | Neg | Neg | ||||
| Cy6 | Oral | Neg | Neg | Neg | Neg | ||||||
| Rectal | –/+ | –/+ | –/+ | –/+ | |||||||
Rh, rhesus macaque; Cy, cynomolgus macaque. Blank cells indicate that culture was not performed.
HMV K. pneumoniae-positive isolates indicated by rmpA/magA status
This animal was the index case and was euthanized after initial positive culture.
Animal had variable PCR results over time.
RAPD analysis.
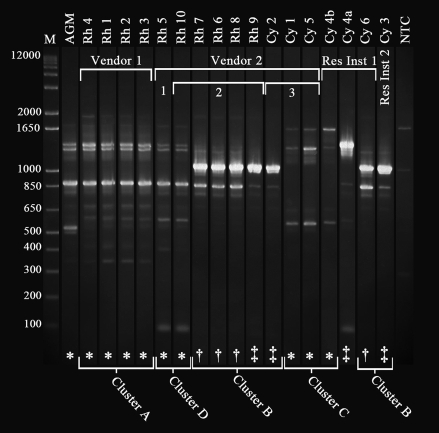
A subset of 18 isolates, identified as HMV K. pneumoniae in light of bacteriology and phenotypic analysis, was characterized by RAPD typing in an attempt to confirm genetic variability suggested by the PCR results. Visual inspection readily distinguished multiple RAPD types among the HMV K. pneumoniae isolates (Figure 5). Patterns were considered to be distinct if 2 or more bands were present or absent between 2 isolates. According to this criterion, 4 clusters (A, B, C, D) were identified. There was no apparent association of RAPD patterns with animal species, given that rhesus macaques were infected with 3 different types (A, D, B), and cynomolgus macaques were infected with 2 different types (C and B). Likewise, RAPD pattern was not associated with rmpA/magA status. However, RAPD pattern related loosely to vendor or vendor shipment cohort. For example, cluster A was uniquely associated with vendor 1. However, these associations were not absolute: shipment cohort 2 from vendor 2 contained animals infected with HMV K. pneumoniae of both clusters B and D.
Figure 5.
RAPD-PCR banding pattern of a subset of isolates from NHP infected with HMV K. pneumoniae. Isolates were identified as being HMV K. pneumoniae based on Vitek analysis and a positive string test. “M” indicates a 1 kb plus marker, and NTC indicates a negative control. The isolate labeled AGM was an HMV K. pneumoniae culture from a fatal abdominal abscess in an African green monkey in 2006. All other isolates were from oropharyngeal or rectal swabs from asymptomatic macaques. Animal identifications correlate with those in Tables 1 and 2. Isolates labeled Cy4a and Cy4b represent 2 rectal samples taken from NHP Cy4 about 3 wk apart (Cy 4a, 2008 Apr 28; Cy4b, 2008 May 20). Animals from Vendor 1 comprised a single shipment, whereas NHP that came from Vendor 2 arrived in 3 separate shipments, as indicated. The remaining 2 macaques were transferred from 2 different Department of Defense research centers, indicated as Research Institute 1 (Res Ins 1) and Research Institute 2 (Res Ins 2). PCR results were correlated with the RAPD-PCR banding patterns. Isolates were determined to be rmpA+/magA− (*), rmpA−/magA+ (†), or rmpA−/magA− (‡). Clusters A through D indicate distinct RAPD types, which were determined based on the absence or presence of 2 or more bands.
A single animal, Cy4 from research institute 1, yielded 2 different strains isolated from rectal swabs taken about 3 wk apart. The first isolate appeared similar to cluster C bacteria, but the second isolate was unique and did not correlate with any other identified cluster. The HMV K. pneumoniae isolated from an abscess in 2005 from an AGM with fatal multisystemic disease appeared to be a minor variant of cluster A from vendor 1 (Figure 5).
Environmental testing.
In the first test method, 252 environmental swabs were taken from predetermined locations in NHP rooms over 6 wk, and none of those samples cultured positive for HMV K. pneumoniae. Six additional environmental swabs were added during weeks 5 and 6. HMV K. pneumoniae was cultured during week 5 from the cage front of an infected macaque in the room housing known infected animals, and HMV K. pneumoniae was cultured from feces in pans of 3 infected macaques during week 6.
The second test method, which used a grid system of MacConkey culture plates affixed to the walls and cages in the room housing known infected animals, yielded only 1 positive culture. The plate that was placed in position 8 (Figure 3) during spray-down of cage bank 1 yielded HMV K. pneumoniae.
The final test method, which involved uncovering culture plates for different times after completion of spray down of organic material from cages in the room housing known infected NHP, yielded no samples that were positive for K. pneumoniae.
Discussion
Although HMV K. pneumoniae has emerged as an important pathogen of humans, causing pyogenic liver abscesses and other metastatic lesions, this organism previously caused fatal abdominal abscesses in AGM at our facility.35 To our knowledge, these cases reflect the only documented cases of clinical disease attributed specifically to HMV K. pneumoniae in NHP. After a subclinically infected rhesus macaque was identified in March 2008, subsequent screening and testing of 307 NHP resulted in the collection of 2297 oropharyngeal and rectal swabs and 326 environmental swabs over 7 mo. The HMV phenotype was found in 81 samples, representing a total of 19 subclinically infected macaques. Three samples, obtained from animals that were moved into biocontainment, were unavailable for further analysis, leaving a total of 78 samples. In addition, 2 of the 326 environmental swabs tested positive for HMV K. pneumoniae.
Fifteen of the 19 infected animals were isolated and monitored clinically with monthly evaluations, which included physical exam, hematology and serum chemistry, and oropharyngeal and rectal culture. The infected macaques, regardless of species, remained asymptomatic from the time they were diagnosed through early September 2008, when they were used on a separate research protocol to further characterize HMV K. pneumoniae.
Asymptomatic carriers of Klebsiella are common in both animals and humans. In 1 study,24 prevalence of fecal shedding in healthy adult dairy cows was over 80%. In humans, fecal carriage of Klebsiella is also common, ranging in prevalence from 5% to 38% in the general population to 77% in hospital settings.30 Rates of Klebsiella-positive cultures from the human nasopharynx range from 1% to 6% in the general population to 19% in hospital settings.30 In our study, the index case was identified by positive results from an oropharyngeal swab, which was the standard sampling site before the detection of infection within the colony. Once colony screening began, both oropharyngeal and rectal swabs were submitted for culture. This procedural change likely altered the sensitivity of testing, perhaps resulting in improved detection of subclinical carriers of HMV K. pneumoniae.
Four of the 15 infected NHP had PCR results that varied from culture to culture. A study using RAPD typing to characterize fecal Klebsiella shedding in dairy cows showed marked variation in strains within and between fecal samples collected from individual cows on multiple consecutive days.25 Two cows were found to harbor multiple strains of Klebsiella in fecal samples taken on a single day. In our study, a single cynomolgus macaque (Cy4) had 2 different strains, as determined by RAPD typing, isolated from rectal samples taken 3 wk apart. Although the study evaluating dairy cows25 examined Klebsiella species in general and not HMV K. pneumoniae specifically, its findings of coinfection with multiple strains may still help to explain the variable PCR results in our current study. Here, pure culture isolates were generated by picking a single bacterial colony based on phenotype and performing a boil preparation for PCR. If various strains coinfect a single animal, several phenotypically similar colonies from those strains may grow on a plate from which only a single colony would have been selected. Depending on the genotype of the colony selected, PCR results would vary from culture to culture. An additional consideration is that, once diagnosed as positive for HMV K. pneumoniae, each animal was moved into a single room that housed all infected macaques, regardless of genotype. The potential for coinfection or superinfection with different strains was increased during that period.
A persistent carrier status did not appear to exist in dairy cows; instead a transient presence of Klebsiella was evidenced by the large variety of Klebsiella strains found within samples and between samples collected from individual animals on multiple consecutive days.25 The suggestion that infection in dairy cows is transient differs from literature on Klebsiella spp. in other species, which indicates that the organism colonizes the mucosal surfaces of mammals including humans, horses, and swine.30 HMV K. pneumoniae infections in macaques do not appear to be as transient as those in the dairy cows previously described.25 Most macaques in the current study maintained infection with at least 1 strain of K. pneumoniae for weeks to months; however, some animals appeared to clear the organism, testing positive once or twice and then subsequently testing negative.
In our facility, animals from an individual quarantine group (shipment cohort) generally stay grouped together until they are assigned to an experimental protocol. There appear to be some broad (but not absolute) cohort associations with regard to rmpA and magA gene status (Table 2). However some building renovation projects in our facility in the months preceding identification of the index case necessitated moving NHP into different rooms. As a result, identifying all NHP that an individual animal might have been exposed to during this time was impossible. Consequently, an NHP may have had unknown exposure to an NHP with a subclinical infection of HMV K. pneumoniae.
Table 2.
Cohort associations between macaques infected with HMV K. pneumoniae
| Initial PCR results |
||||
| Animal | rmpA | magA | Date of arrival | Source |
| Rh1a | + | – | July 2007 | Vendor 1 |
| Rh2 | + | — | July 2007 | Vendor 1 |
| Rh3 | + | — | July 2007 | Vendor 1 |
| Rh4b | + | — | July 2007 | Vendor 1 |
| Cy1 | + | — | January 2008 | Vendor 2 |
| Cy2b | + | — | January 2008 | Vendor 2 |
| Cy5 | + | — | January 2008 | Vendor 2 |
| Cy4b | — | — | January 2008 | Research institute 1 |
| Cy6 | — | + | May 2008 | Research institute 1 |
| Cy3b | — | — | June 2007 | Research institute 2 |
| Rh5 | + | — | May 2007 | Vendor 2 |
| Rh6 | — | + | August 2007 | Vendor 2 |
| Rh7 | — | + | August 2007 | Vendor 2 |
| Rh8 | — | + | August 2007 | Vendor 2 |
| Rh10 | — | — | August 2007 | Vendor 2 |
| Rh 9 | — | — | August 2007 | Vendor 2 |
This animal was the index case and was euthanized after initial positive culture.
Animal had variable PCR results over time.
The RAPD technique has been used frequently to study genetic variability in populations. Unlike PCR, which requires well-defined and specific DNA sequences as primers, the RAPD technique uses primers of arbitrary nucleotide sequence to access random segments of genomic DNA to reveal polymorphisms. In epidemiologic studies, although pulsed-field gel electrophoresis generally is considered to be the ‘gold standard’7, it is technically demanding, requires specialized equipment, and is time-consuming.14 Both pulsed-field gel electrophoresis and RAPD typing have proved useful for differentiating between groups of epidemiologically related extended-spectrum–β-lactamase-producing K. pneumoniae.14 Whereas pulsed-field gel electrophoresis was more discriminating, RAPD typing proved to be more efficient for screening of clonally related isolates.14 In our current study, the RAPD technique was applied to a subset of HMV K. pneumoniae-positive cultures to further elucidate some of the variability seen in PCR results. Cohort associations identified on PCR appeared to hold relatively consistent with RAPD banding patterns. Two different isolates from the same cynomolgus macaque (Cy4) underwent RAPD testing. During PCR analysis, the 28 April 2008 positive rectal culture from Cy4 was rmpA−/magA−, whereas that on 20 May 2008 was rmpA+/magA− (Table 1). The RAPD genotypes of those 2 isolates showed banding patterns that differed from each other: 1 of the isolates had a banding pattern consistent with cluster C, but the other isolate appeared unique and did not fit into any other cluster grouping (Figure 5).
In the initial stages of this epidemiologic investigation, a great deal of attention was given to the potential for environmental spread of this organism within the NHP colony and to concerns about human health. K. pneumoniae is ubiquitous in nature, existing in surface water, sewage, and soil and on plants,30 and it is well known to exist in biofilms.22,23 In addition, K. pneumoniae is a common cause of nosocomial infections in the human healthcare systems.30 It is an opportunistic pathogen that has a survival time of 4 to 27 d (median, 7.5 d) in common hospital materials.11
Because of these characteristics, requirements regarding personal protective equipment were enhanced at our facility, and investigators, caretakers, technicians, and veterinary staff working with NHPs were trained about the human health risks associated with HMV K. pneumoniae. Concerns about the zoonotic potential of this known human pathogen led to the decision to initiate environmental surveillance for Klebsiella. Because we were able to culture HMV K. pneumoniae from the feces of 3 infected macaques, a potential for fecal–oral transmission existed. Screening involved taking cultures from areas in 6 different NHP rooms that we considered most likely to support the growth of K. pneumoniae. The results of these screenings revealed that environmental contamination was not a major factor in transmission of this agent between NHP rooms. As a primary respiratory pathogen, K. pneumoniae is known for droplet or aerosol transmission.1,32 Accordingly, 2 experiments were initiated to determine how readily K. pneumoniae was dispersed both during and after high-pressure spraying of cages within the room housing known infected NHP. The results of each experiment failed to support a theory of widespread droplet or aerosol transmission of the bacteria either during or immediately after high-pressure cage wash by animal caretakers.
Although HMV K. pneumoniae has been documented to cause abdominal abscesses in AGM35 and pyogenic liver abscesses (sometimes with bacteremia, endophthalmitis, and meningitis) in humans in Taiwan and other Asian countries,6,17-19,21,26,34 the organism was maintained as a subclinical infection in a colony of macaques at our facility. Despite initial concerns about clonal spread of the organism through environmental, aerosol, or droplet contamination within the colony, environmental testing did not suggest widespread contamination in the environment. In addition, PCR and RAPD testing verified the existence of multiple genotypes which could broadly be associated with vendor cohort groups. Infection with multiple strains appeared likely in cohoused, infected macaques through a fecal–oral transmission pattern. As a result of this study, our institute has initiated procedures to increase physical separation between AGM and macaque species, including designation of room entry order and changes of personal protective equipment between species.
Future research efforts should focus on immunologic changes that result in macaques from subclinical infection with HMV K. pneumoniae and the potential for use as a research model for human disease. In addition, further characterization of the organism in AGM, a species that appears particularly susceptible to clinical disease resulting from infection with HMV K. pneumoniae, may prove to be valuable by providing insight into virulence factors and potential immune-evading measures used by this pathogen in infected AGM and humans.
Acknowledgments
We are grateful to Laurie Hartman, Dave Kulesh, Jeanne Geyer, and Mark Wolcott (Diagnostic Systems Division, US Army Medical Research Institute of Infectious Diseases) for developing the rmpA and magA assays and for providing technical expertise and support during this project. We also thank the technicians from the Nonhuman Primate Section and the technicians and staff from Clinical Laboratory for plating, subculturing, and reading results from cultures during a 6-mo timeframe that saw the microbiologic workload triple from that of the entire previous year.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Army. The authors do not have a commercial or other association that would pose a conflict of interest. This work was presented in part at the Association of Primate Veterinarians 36th Annual Workshop, Indianapolis, IN, 6–8 November 2008.
The research described herein was sponsored by the US Army Medical Research Institute of Infectious Diseases under Research Plan Number 150874. It was supported in part by an appointment to the Student Research Participation Program at the US Army Medical Research Institute for Infectious Diseases administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and USAMRMC.
References
- 1.Bolister NJ, Johnson HE, Wathes CM. 1992. The ability of airborne Klebsiella pneumoniae to colonize mouse lungs. Epidemiol Infect 109:121–131 [PMC free article] [PubMed] [Google Scholar]
- 2.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan KS, Yu WL, Tsai CL, Cheng KC, Hou CC, Lee MC, Tan CK. 2007. Pyogenic liver abscess caused by Klebsiella pneumoniae: analysis of the clinical characteristics and outcomes of 84 patients. Chin Med J (Engl) 120:136–139 [PubMed] [Google Scholar]
- 4.Chang CM, Lu FH, Guo HR, Ko WC. 2005. Klebsiella pneumoniae fascial space infections of the head and neck in Taiwan: emphasis on diabetic patients and repetitive infections. J Infect 50:34–40 [DOI] [PubMed] [Google Scholar]
- 5.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654 [DOI] [PubMed] [Google Scholar]
- 6.Chung DR, Lee SS, Lee HR, Kim HB, Choi HJ, Eom JS, Kim JS, Choi YH, Lee JS, Chung MH, Kim YS, Lee H, Lee MS, Park CK. 2007. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect 54:578–583 [DOI] [PubMed] [Google Scholar]
- 7.Descheemaeker P, Lammens C, Pot B, Vandamme P, Goossens H. 1997. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int J Syst Bacteriol 47:555–561 [DOI] [PubMed] [Google Scholar]
- 8.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang FC, Sandler N, Libby SJ. 2005. Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J Clin Microbiol 43:991–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox JG, Rohovsky MW. 1975. Meningitis caused by Klebsiella spp. in two rhesus monkeys. J Am Vet Med Assoc 167:634–636 [PubMed] [Google Scholar]
- 11.Gastmeier P, Schwab F, Bärwolff S, Rüden H, Grundmann H. 2006. Correlation between the genetic diversity of nosocomial pathogens and their survival time in intensive care units. J Hosp Infect 62:181–186 [DOI] [PubMed] [Google Scholar]
- 12.Gibson SV. 1998. Bacterial and mycotic diseases, p 73–75. Bennett BT, Abee CR, Hendrickson R. Nonhuman primates in biomedical research: diseases. San Diego (CA): Academic Press [Google Scholar]
- 13.Gonzalo A, Montoyo E. 1991. Klebsiella pneumoniae infection in a New World nonhuman primate center. Lab Primate Newslett 30:13–20 [Google Scholar]
- 14.Gori A, Espinasse F, Deplano A, Nonhoff C, Nicolas MH, Struelens MJ. 1996. Comparison of pulsed-field gel electrophoresis and randomly amplified DNA polymorphism analysis for typing extended-spectrum–β-lactamase-producing Klebsiella pneumoniae. J Clin Microbiol 34:2448–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman LJ, Selby EE, Whitehouse CA, Coyne SR, Jaissle JG, Twenhafel NA, Burke RL, Kulesh DA. 2009. Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J Mol Diagn 11:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karama EM, Willermain F, Janssens X, Claus M, Van den Wijngaert S, Wang JT, Verougstraete C, Caspers L. 2008. Endogenous endophthalmitis complicating Klebsiella pneumoniae liver abscess in Europe: case report. Int Ophthalmol 28:111–113 [DOI] [PubMed] [Google Scholar]
- 17.Kawai T. 2006. Hypermucoviscosity: an extremely sticky phenotype of Klebsiella pneumoniae associated with emerging destructive tissue abscess syndrome. Clin Infect Dis 42:1359–1361 [DOI] [PubMed] [Google Scholar]
- 18.Keynan Y, Rubinstein E. 2007. The changing face of Klebsiella pneumoniae infections in the community. Int J Antimicrob Agents 30:385–389 [DOI] [PubMed] [Google Scholar]
- 19.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 8:160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler JE, Hutchens MP, Sadow PM, Modi BP, Tavakkolizadeh A, Gates JD. 2007. Klebsiella pneumoniae necrotizing fasciitis and septic arthritis: an appearance in the Western hemisphere. Surg Infect (Larchmt) 8:227–232 [DOI] [PubMed] [Google Scholar]
- 21.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, Wang LR, Ko WC. 2006. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med 259:606–614 [DOI] [PubMed] [Google Scholar]
- 22.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39 [DOI] [PubMed] [Google Scholar]
- 23.Matsen JM, Spindler JA, Blosser RO. 1974. Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl Microbiol 28:672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz MA, Ahlström C, Rauch BJ, Zadoks RN. 2006. Fecal shedding of Klebsiella pneumoniae by dairy cows. J Dairy Sci 89:3425–3430 [DOI] [PubMed] [Google Scholar]
- 25.Munoz MA, Zadoks RN. 2007. Short communication: Patterns of fecal shedding of Klebsiella by dairy cows. J Dairy Sci 90:1220–1224 [DOI] [PubMed] [Google Scholar]
- 26.Nadasy KA, Domiati-Saad R, Tribble MA. 2007. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis 45:e25–e28 [DOI] [PubMed] [Google Scholar]
- 27.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 57:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Research Council 1996. Guide for the care and use of laboratory animals. Washington (DC): National Academies Press [Google Scholar]
- 29.Pisharath HR, Cooper TK, Brice AK, Cianciolo RE, Pistorio AL, Wachtman LM, Mankowski JL, Newcomer CE. 2005. Septicemia and peritonitis in a colony of common marmosets (Callithrix jacchus) secondary to Klebsiella pneumoniae infection. Contemp Top Lab Anim Sci 44:35–37 [PubMed] [Google Scholar]
- 30.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. 2005. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J Med Microbiol 54:1111–1113 [DOI] [PubMed] [Google Scholar]
- 32.Theunissen HJ, Lemmens-den Toom NA, Burggraaf A, Stolz E, Michel MF. 1993. Influence of temperature and relative humidity on the survival of Chlamydia pneumoniae in aerosols. Appl Environ Microbiol 59:2589–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai FC, Huang YT, Chang LY, Wang JT. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsay RW, Siu LK, Fung CP, Chang FY. 2002. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med 162:1021–1027 [DOI] [PubMed] [Google Scholar]
- 35.Twenhafel NA, Whitehouse CA, Stevens EL, Hottel HE, Foster CD, Gamble S, Abbott S, Janda JM, Kreiselmeier N, Steele KE. 2008. Multisystemic abscesses in African green monkeys (Chlorocebus aethiops) with invasive Klebsiella pneumoniae: identification of the hypermucoviscosity phenotype. Vet Pathol 45:226–231 [DOI] [PubMed] [Google Scholar]
- 36.Yang CS, Tsai HY, Sung CS, Lin KH, Lee FL, Hsu WM. 2007. Endogenous Klebsiella endophthalmitis associated with pyogenic liver abscess. Ophthalmology 114:876–880 [DOI] [PubMed] [Google Scholar]
- 37.Yeh KM, Chang FY, Fung CP, Lin JC, Siu LK. 2006. Serotype K1 capsule, rather than magA per se, is really the virulence factor in Klebsiella pneumoniae strains that cause primary pyogenic liver abscess. J Infect Dis 194:403–404 [DOI] [PubMed] [Google Scholar]
- 38.Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, Chen TL, Chang FY, Koh TH. 2007. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol 45:466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ, International Klebseilla Study Group 2007. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 13:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. 2008. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and nonK1/K2 serotypes. Diagn Microbiol Infect Dis 62:1–6 [DOI] [PubMed] [Google Scholar]
- 41.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. 2006. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42:1351–1358 [DOI] [PubMed] [Google Scholar]