Introduction
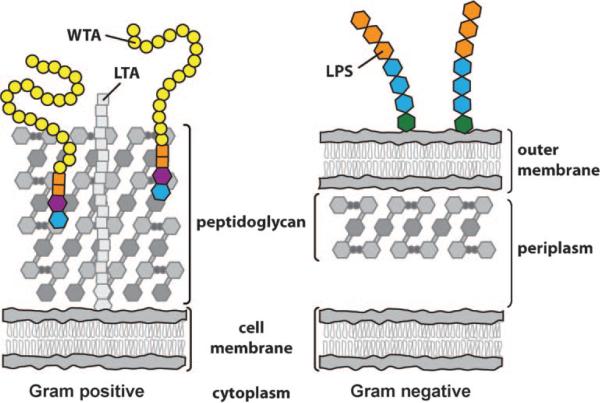
One of the major differences between Gram-negative and Gram-positive organisms is the presence or absence of an outer membrane (Figure 1). In Gram-negative organisms, the outer membrane protects the organism from the environment. It filters out toxic molecules and establishes a compartment, the periplasm, which retains extracytoplasmic enzymes required for cell-wall growth and degradation. It also serves as a scaffold to which proteins and polysaccharides that mediate interactions between the organism and its environment are anchored.[1] In addition, in ways that are not completely understood, the outer membrane functions along with a thin layer of peptidoglycan to help stabilize the inner membrane so that it can withstand the high osmotic pressures within the cell.[2]
Figure 1.
Simplified depiction of Gram-positive and Gram-negative bacterial cell envelopes. Gram-negative organisms have a distinct periplasm; Gram-positive organisms do not, but recent studies have suggested that they have a periplasmic-like compartment between the plasma membrane and the base of the peptidoglycan layers.[3] Proteins are omitted from the depictions for clarity. Membrane-embedded, membrane-anchored, and peptidoglycan-associated proteins are abundant in the cell membranes of both Gram-positive and Gram-negative organisms. LTA: lipoteichoic acid; LPS: lipopolysaccharide; WTA: wall teichoic acid.
Gram-positive organisms, in contrast, lack an outer membrane and a distinct periplasm (Figure 1). The peptidoglycan layers are consequently very thick compared to those in Gram-negative organisms.[4] These thick layers of peptidoglycan stabilize the cell membrane and also provide many sites to which other molecules can be attached. Gram-positive peptidoglycan is heavily modified with carbohydrate-based anionic polymers that play an important role in membrane integrity.[5] These anionic polymers appear to perform some of the same functions as the outer membrane: they influence membrane permeability, mediate extracellular interactions, provide additional stability to the plasma membrane, and, along with peptidoglycan, act as scaffolds for extracytoplasmic enzymes required for cell-wall growth and degradation.
A major class of these cell surface glycopolymers are the teichoic acids (TAs), which are phosphate-rich molecules found in a wide range of Gram-positive bacteria, pathogens and nonpathogens alike. There are two types of TAs: the lipo-TAs (LTAs), which are anchored to the plasma membrane and extend from the cell surface into the peptidoglycan layer;[6] and the wall TAs (WTAs), which are covalently attached to peptidoglycan and extend through and beyond the cell wall (Figure 1).[7] Together, LTAs and WTAs create what has been aptly described as a “continuum of negative charge” that extends from the bacterial cell surface beyond the outermost layers of peptidoglycan.[5] Neuhaus and Baddiley comprehensively reviewed both LTAs and WTAs in 2003.[5] Since then, however, new functions for WTAs in pathogenesis have been uncovered and it has been suggested that the biosynthetic enzymes that make these polymers are targets for novel antibacterial agents.[8,9] Indeed, the first WTA-active antibiotic has just been reported.[10] This review will focus primarily on recent developments in the study of WTAs in Bacillus subtilis and Staphylococcus aureus, and will include a discussion of strategies for the discovery of WTA inhibitors and prospects for these inhibitors as antibiotics.
Wall Teichoic Acid Structure
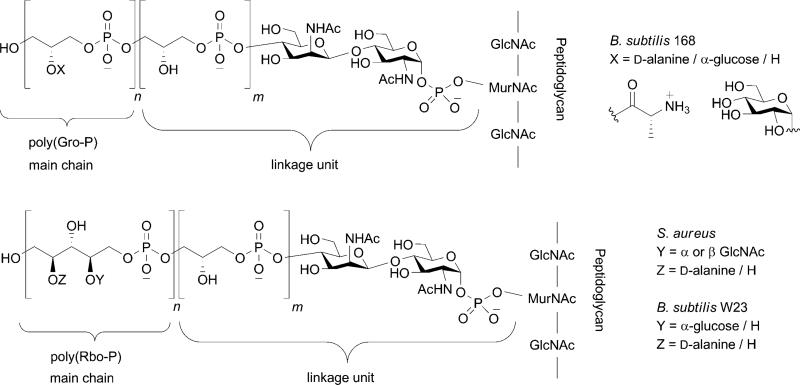
WTAs are anionic glycopolymers that are covalently attached to peptidoglycan via a phosphodiester linkage to the C6 hydroxyl of the N-acetyl muramic acid sugars.[5] They can account for as much as 60 % of the total cell wall mass in Gram-positive organisms. The chemical structures of WTAs vary among organisms, as described in detail by Neuhaus and Baddiley,[5] but the most common structures are composed of a ManNAc(β1→4)GlcNAc disaccharide with one to three glycerol phosphates attached to the C4 hydroxyl of the ManNAc residue (the “linkage unit”) followed by a much longer chain of glycerol- or ribitol phosphate repeats (the “main chain”; Figure 2).[11–18] B. subtilis, the Gram-positive model organism, makes poly(glycerol phosphate) or poly(ribitol phosphate) WTAs depending on the strain,[19] while S. aureus strains primarily make poly(ribitol phosphate) WTAs.[20–23] The hydroxyls on the glycerol- or ribitol phosphate repeats are tailored with cationic D-alanine esters and monosaccharides, such as glucose or N-acetylglucosamine.[24,25] The presence of WTAs and the particular tailoring modifications that are found on them have profound effects on the physiology of Gram-positive organisms, and impact everything from cation homeostasis to antibiotic susceptibility to survival in a host.
Figure 2.
Representative chemical structures of wall teichoic acids from different Gram-positive bacteria (m = 1–3 and n = 20–40).
Functions of Teichoic Acids in Bacterial Physiology
The functions of TAs in bacterial physiology are incompletely understood, but evidence for their importance is overwhelming. B. subtilis and S. aureus mutants deficient in LTA biosynthesis can be obtained but only if grown under a narrow range of conditions; they are temperature sensitive and exhibit severe growth defects.[26,27] Mutants deficient in WTA biosynthesis are also compromised and manifest increased sensitivity to temperature and certain buffer components, including citrate; they also tend to aggregate in culture.[26–31] In addition, B. subtilis strains that do not express WTAs show profound morphological aberrations. Bacterial strains in which both LTA and WTA expression are prevented are not viable, an observation suggesting that these polymers have overlapping functions and can partially compensate for one another.[26,27] Indeed, this might be expected for some functions since both polymers contain phosphate-linked repeat units with similar tailoring modifications. One of the tailoring modifications, D-alanylation, is accomplished by the same machinery, so there is even some overlap in the biosynthetic pathways. This fact makes dissecting the functions of the individual anionic glycopolymers difficult, but is consistent with the idea that LTAs and WTAs are partially redundant. Some of the functions attributed to WTAs are described in the following paragraphs. LTAs are beyond the scope of this review, but will be mentioned in cases where it is relevant to the discussion of WTAs. Morath et al. and Rahman et al. have each written recent reviews on LTA structure and biosynthesis.[6,32]
Cation binding functions
WTAs form a dense network of negative charges on Gram-positive cell surfaces. To alleviate the resulting electrostatic repulsive interactions between neighboring phosphates, TAs bind cationic groups, including mono- and divalent metal cations. Networks of WTA-coordinated cations affect the overall structure of the polymers, and this in turn influences the porosity and rigidity of the cell envelope. WTAs are proposed to be important for cation homeostasis in Gram-positive organisms,[33,34] and provide a reservoir of ions close to the cell surface that might be required for enzyme activity. In addition, the gradient of ions could in some way mitigate the osmotic pressure change between the inside and outside of the cell. The amount of bound cations can be modulated by d-alanylation, a tailoring modification that introduces positively charged amines.[35] WTAs that lack d-alanyl esters can bind up to 60 % more Mg2+ ions than analogous polymers that contain this modification.[36] The importance of cation binding is highlighted by the observation that B. subtilis strains up-regulate their production of TAs in the presence of low Mg2+ concentrations, and produce other negatively charged polymers (teichuronic acid) in the presence of limiting phosphate concentrations.[37] Recent structural studies have been focused on elucidating modes of cation binding by WTA polymer phosphate groups, and researchers have suggested that a clear understanding of the three-dimensional structure of WTAs and their bound cation groups might provide insights that facilitate the design of novel antimicrobials.[38]
Scaffolding roles
In addition to providing binding sites for cations, WTAs serve as scaffolds or receptors for a wide range of other molecules. In S. aureus, for example, they function as receptors that are required for phage infection.[39] Depending on their tailoring modifications (see below) they might also promote adhesion by lytic enzymes produced by neutrophils.[40] They are additionally thought to serve as scaffolds for endogenously produced cell wall hydrolases (autolysins) involved in cell growth and division.[41] In general, the molecular interactions between WTAs and other biomolecules are not well understood but could provide crucial insights into cell envelope function.
Tailoring modification-dependent functions
The main chain hydroxyl groups on both glycerol- and ribitol phosphate WTA polymers are subject to further derivatization by tailoring enzymes (Figure 2). There are two classes of tailoring enzymes: those that catalyze the addition of d-alanyl esters, and those that append glycosyl groups. The extent to which these modifications occur on the TA polymers is strain dependent and can also be affected by environmental conditions. Efforts have been made to understand the role(s) of these modifications in bacterial physiology, and some of these studies are highlighted below.
The d-alanylation tailoring modification has been more extensively investigated than glycosylation and is far better understood at this point. Perego et al. were the first to characterize the genetic pathway responsible for this modification (dlt operon) in B. subtilis.[42] Briefly, the biosynthetic pathway begins intracellularly with the activation of d-alanine to its corresponding aminoacyl adenylate by DltA. This molecule is then covalently attached, as a thioester, to a cofactor bound to the d-Ala carrier protein, DltC. Although the precise roles of DltB and DltD have not been confirmed, it is believed that they facilitate the transport of DltC through the membrane and the incorporation of d-Ala onto both LTAs and WTAs.[43] It has been found that d-alanylation is affected by several factors, including growth media, pH and temperature.[5] The attachment of d-alanyl esters to the hydroxyls on TAs alters the net charge of the polymer by adding positively charged amines. This modification reduces the electrostatic repulsion between neighboring TA chains and possibly facilitates stabilizing ion-pair formation between the cationic esters and the anionic phosphate groups.[38]
The d-alanine modification modulates interactions between the cell envelope and the environment and has been implicated in many of the known scaffolding/receptor functions of WTAs.[5,44] For example, it has been shown that the absence of d-alanyl esters on the TA polymers increases susceptibility to cationic antimicrobial peptides, possibly by increasing the negative charge density on the cell surface.[45,46] Removing the alanine residues also increases bacterial sensitivity to glycopeptide antibiotics and to the lytic activity of enzymes produced by neutrophils during host infection.[40,41] In contrast, the activity of autolytic enzymes is decreased, suggesting a role for TAs in scaffolding and/or activating bacterial enzymes involved in the processes of cell-wall synthesis and degradation.[41] Removal of d-alanyl esters from TAs has also been shown to attenuate the binding of S. aureus to artificial surfaces as well as host tissue. A recent study has illustrated the importance of the charge balance of WTAs in adhesion to artificial surfaces, such as glass and polystyrene.[44]
Since d-alanylation promotes better adhesion to host tissue and confers some resistance to lytic enzymes produced by the host, mutant strains lacking this modification have been studied in animal infection models. For example, in a mouse tissue cage infection model, bacterial strains lacking d-alanylation were more susceptible to Toll-like receptor 2-dependent host defenses;[46] in a septicemia model, such strains were attenuated in their ability to establish an infection, possibly because they were more readily killed by neutrophils.[40] Based on these and other studies, it was proposed that the d-alanine modification is a putative target for novel antimicrobials that function by attenuating virulence. In 2005, May et al. reported the synthesis and evaluation of a nonhydrolysable analogue of d-Ala aminoacyl adenylate as the first designed inhibitor of DltA, the enzyme that activates d-Ala. The compound enhanced the activity of vancomycin against B. subtilis.[43] This result is consistent with inhibition of DltA, and supports the idea that small molecules that interfere with d-alanylation might provide a novel strategy for antimicrobials.
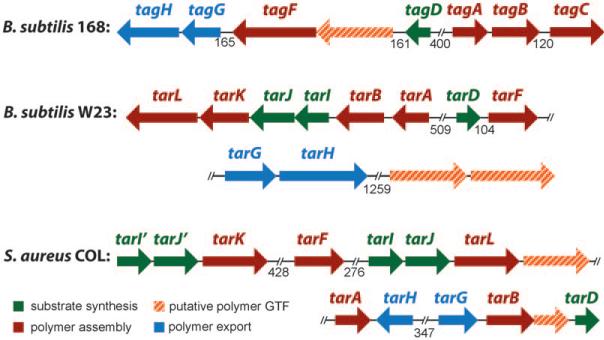
Glycosylation is a ubiquitous tailoring modification of WTAs but its functions are not well understood. Glucose is commonly added to the WTA polymers in B. subtilis, whereas N-acetyl glucosamine (GlcNAc) is added in S. aureus (Figure 2).[5] Depending on the bacterial strain, the stereochemistry of the glycosidic linkage may be β-, α-, or a mixture of the two anomers. All sequenced B. subtilis and S. aureus strains contain one or more putative glycosyltransferase genes clustered with the WTA biosynthetic genes (Figure 3). For example, B. subtilis 168 contains a gene for a putative retaining glycosyltransferase that might add a-Glu to the glycerol phosphate polymers. S. aureus strains contain two genes encoding putative inverting glycosyltransferases that might transfer β-GlcNAc to the poly(ribitol phosphate) polymers. Although some S. aureus strains have been shown to contain α-glycosidically linked WTAs, there are no genes yet identified for any glycosyltransferases that can carry out this tailoring modification. Furthermore, no studies have confirmed the enzymatic functions of any of the putative WTA glycosyltransferases or have explored the effects of preventing WTA glycosylation on bacterial cell growth, division, intercellular interactions, or pathogenesis. In fact, as far as we know there is only one piece of data pertaining to the functions of WTA glycosyltransferases in the literature: a transposon mutant in a putative glycosyltransferase in the S. aureus strain Newman showed attenuated virulence in a nematode killing assay, suggesting that glycosylation might play a role in pathogenesis in S. aureus.[47] If glycoslyation proves important for bacterial pathogenesis, the glycosyltransferase tailoring enzymes, like the enzymes involved in d-alanylation (see above) would be possible targets for antimicrobials.
Figure 3.
Genetic organization of wall teichoic acid biosynthetic genes; tag: teichoic acid glycerol; tar: teichoic acid ribitol. Adapted from Qian et al.[48] (//: number of nucleic acids between genes if > 120 base pairs).
Roles in cell elongation and division
Recent studies have implicated LTAs and WTAs in cell growth, division, and morphogenesis. In the rod-shaped organism B. subtilis, TAs have been shown to play distinct roles in bacterial morphogenesis. Preventing WTA expression results in the production of round, severely defective progeny, while preventing LTA biosynthesis causes major defects in septum formation and cell separation.[27,49] It is known that there are separate multiprotein complexes involved in septation and elongation in B. subtilis, and Errington and co-workers have suggested (based on localization studies using fluorescently tagged enzymes) that the WTA biosynthetic enzymes associate with the machinery involved in elongation, while the LTA enzymes might associate with machinery involved in septation and cell division.[27,50] It was suggested that the spatial distribution of these two anionic glycopolymers determines their specific functions. Defects in S. aureus upon deletion of WTAs are less pronounced than in B. subtilis, and no specific roles in cell growth and division for WTAs in this organism have been proposed. However, Oku et al. recently reported that S. aureus strains devoid of LTAs show major defects in septal formation and cell separation and grow only under a restricted range of conditions, including reduced temperatures.[26]
Functions in biofilm formation and host tissue adhesion
As major components of the cell envelope, WTAs influence the interactions of bacterial cells with their environment in many ways. We have already mentioned that S. aureus mutants lacking WTAs show reduced initial adherence to artificial surfaces, including glass and polystyrene;[44] they are also impaired in their ability to form biofilms. It has been shown that WTA null mutants that are impaired in biofilm formation do not have a reduced production of the exopolysaccharide poly-N-acetylglucosamine (PNAG), which has been identified as an important factor for biofilm formation.[30] This finding highlights the independent role that WTAs play in biofilm formation.
S. aureus WTAs are also required for adhesion to host tissue. Peschel and co-workers have shown that S. aureus strains that do not express WTAs are severely impaired in their ability to adhere to nasal epithelial cells and are unable to colonize the nasal passages of cotton rats.[8] They have also shown that WTA-null mutants cannot colonize endothelial tissues derived from kidney and spleen.[9] The d-alanylation machinery was not impaired in these strains, and d-alanylation could still have occurred on LTAs; therefore, these results implicate WTAs as independent factors involved in cell adhesion. Since WTAs are required for host infection and play important roles in biofilm formation, it was suggested that they are virulence factors, that is, factors required for the establishment and spread of infection in a host. Therefore, the enzymes involved in WTA biosynthesis were suggested to be targets for novel antimicrobials that impede host colonization by S. aureus.[7]
Biosynthesis of Wall Teichoic Acids
Poly(glycerol phosphate) WTA biosynthesis in B. subtilis 168
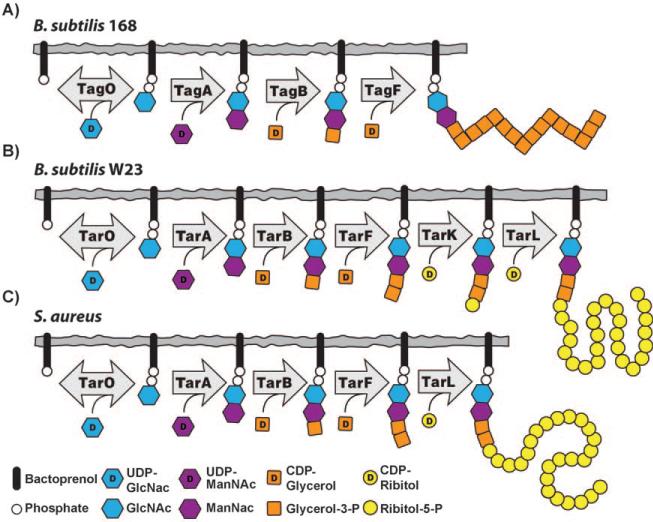
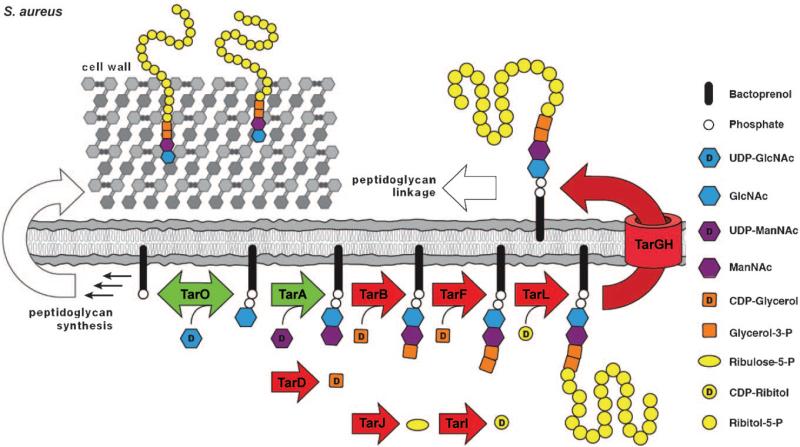
The pathway for WTA biosynthesis was first characterized in B. subtilis 168, which makes poly(glycerol phosphate) WTAs (Figure 4 A).[51] The genes involved in the synthesis of these WTAs are known as tag genes (for teichoic acid glycerol). The pathway starts in the cytoplasm with the transfer of GlcNAc phosphate to an undecaprenyl phosphate (also known as bactoprenyl phosphate) carrier anchored in the bacterial membrane. The enzyme that catalyzes this reaction, TagO, is reversible and is related to a large family of phosphosugar transferases that includes the first enzyme in the dolichol pathway for N-linked glycosylation in eukaryotes, GPT, as well as MraY, an essential bacterial enzyme involved in peptidoglycan biosynthesis.[52,53] Following formation of the GlcNAc-pp-lipid by TagO, an N-acetylmannosaminyl transferase, TagA, transfers ManNAc from UDP-ManNAc to the C4 hydroxyl of the GlcNAc residue to form a β-linked disaccharide, which is the substrate for the next enzyme in the pathway, TagB.[54,55] TagB is a glycerophosphate transferase that transfers a single phosphoglycerol unit from CDP-glycerol to the C4 hydroxyl of ManNAc to complete the synthesis of the linkage unit (Figure 2).[54,56] The next enzyme in the B. subtilis 168 pathway, TagF, is a polymerizing cytidylyl transferase that attaches 35 or more glycerol phosphates to the linkage unit to form the anionic polymer.[57–59] The catalytic domains of TagB and TagF share significant sequence identity and belong to a group of phosphotransferases that are apparently unique to WTA biosynthesis. Other members of this family include TarB, F, K, and L (see below). Once assembled, the lipid-linked WTA polymer is putatively modified by a glycosyltransferase (TagE) and then exported to the external surface of the bacterial membrane by a two-component ABC (ATP binding cassette) transporter, TagGH.[60] The polymer is coupled to peptidoglycan through the anomeric phosphate of the GlcNAc residue. The transferase responsible for carrying out this reaction has not been identified. D-Alanine ester formation occurs outside of the cell as described above.[5]
Figure 4.
Differences in wall teichoic acid biosynthesis for: A) B. subtilis 168, B) B. subtilis W23, and C) S. aureus.
Poly(ribitol phosphate) WTA biosynthesis in B. subtilis W23
B. subtilis W23 makes a poly(ribitol phosphate) WTA rather than a poly(glycerol phosphate) WTA (Figure 2). A pathway for the biosynthesis of B. subtilis W23 WTA was proposed by Lazarevic et al., who designated the genes involved as tar genes (for teichoic acid ribitol).[19] The first three steps of the proposed pathway, mediated by TarO, TarA, and TarB are identical to those in B. subtilis 168, but the pathways then diverge (Figure 4 B). The TagF homologue in B. subtilis W23, TarF, functions not as a polymerase but as a primase, and adds one additional glycerol phosphate unit to the 168-type linkage unit. The catalytic domains of TagF, which is a polymerase, and TarF, which is a primase, share significant sequence identity and the structural features in each enzyme that determine whether one or many glycerol phosphate units is transferred to the linkage unit have not been identified. Once the W23 linkage unit is complete, the poly(ribitol phosphate) main chain is assembled. Lazarevic et al. proposed that the assembly of this poly(ribitol phosphate) chain requires two enzymes: TarK, which transfers a single ribitol phosphate residue to the linkage unit, and TarL, which carries out the polymerization of the ribitol phosphate chain.[19] TarK and TarL in B. subtilis W23 were thus suggested to function as a primase/polymerase pair, analogous to the primase/polymerase pair (TagB/TagF) that assembles the poly(glycerol phosphate) chain in strain 168. Meredith et al. used a genetic approach to verify that tarK and tarL from B. subtilis W23 are both required for the assembly of poly(ribitol phosphate) WTAs;[61] this is consistent with the proposed primase/polymerase model for biosynthesis. Pereira et al. subsequently confirmed this finding.[62] Once the poly(ribitol phosphate) WTA polymer is assembled, the remaining steps are thought to be similar to those in strain 168. That is, the WTA polymer is glycosylated, transported through the bacterial membrane by a two-component transporter, TarGH, attached to peptidoglycan by an unidentified transferase, and esterified with d-alanine residues.
Poly(ribitol phosphate) WTA biosynthesis in S. aureus
Like B. subtilis W23, S. aureus also makes a ribitol phosphate WTA polymer. Except for the length of the polymer chain and the nature of the appended sugar residues, the structures of the poly(ribitol phosphate) WTAs are thought to be the same in B. subtilis W23 and S. aureus (Figure 2). The assembly of the linkage unit in S. aureus is identical to its synthesis in B. subtilis (TarO, TarA, TarB, TarF catalyze the same reactions), but the main chain is assembled not by a primase/polymerase pair, but by one of two bifunctional poly(ribitol phosphate) primase/polymerases (currently designated TarK and TarL although their functions are different from TarK/TarL in B. subtilis; Figure 4 C). It has been proposed that S. aureus TarL makes a primary WTA polymer (L-WTA) while S. aureus TarK makes a secondary WTA polymer (K-WTA).[61] As outlined in the following section, however, there are still a number of questions about the cellular roles of the two bifunctional poly(ribitol phosphate) polymerases in S. aureus. Once the ribitol phosphate polymer is completed in the cytoplasm, glycosylation occurs and the polymer is flipped to the external surface of the membrane by an ABC-dependent transporter complex (TarGH) before ligation to the cell wall by unidentified enzyme(s) and d-alanylation.
Gene Cluster Duplication in S. aureus
As noted in the previous section, S. aureus contains two bifunctional poly(ribitol phosphate) polymerases that have similar enzymatic functions rather than a pair of enzymes containing separate primase and polymerase activities. Qian et al. were the first to note that S. aureus may differ from B. subtilis in how it accomplishes poly(ribitol phosphate) polymerization. In a genomic analysis of six sequenced S. aureus strains (Figure 3)[48] these authors noted that all of the strains contained an apparent duplication of the region of the chromosome containing the putative ribitol phosphate polymerase gene and the two genes involved in the synthesis of its CDP-ribitol substrate (tarI,J,L). These two similar gene clusters have since been designated in the literature as tarI,J,L and tarI′,J′,K. The tarK gene is highly homologous to the tarL gene, which suggests it might have the same enzymatic function. Walker and co-workers provided the first experimental evidence for a unique S. aureus-specific poly(ribitol phosphate) WTA biosynthetic pathway. By utilizing an approach previously developed to study peptidoglycan biosynthesis[63,64] and later applied to validate part of the B. subtilis WTA pathway,[54] the Walker group reconstituted the biosynthesis of S. aureus poly(ribitol phosphate) WTA in vitro.[65] Through the use of synthetic substrates, it was shown that S. aureus TarL is, in fact, a bifunctional enzyme that combines both primase and polymerase activities (Figure 4).[65] They demonstrated that a dedicated ribitol phosphate primase was not required for WTA polymer synthesis in S. aureus as it is in B. subtilis.
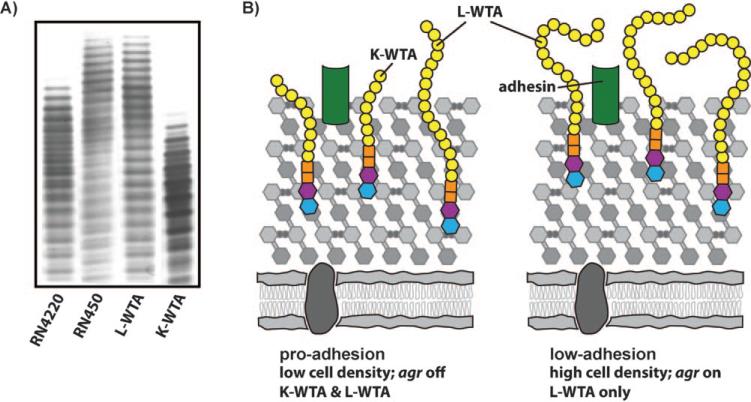
The in vitro studies of WTA biosynthesis established the enzymatic function of TarL in S. aureus, but did not answer the question of why S. aureus contains an additional gene, tarK, that is homologous to tarL. Meredith et al. and Pereira et al. used genetics to probe the cellular roles of tarK and tarL.[61,62] Under certain conditions, it was shown that tarK can compensate for the loss of tarL, and both groups have concluded that TarK, like TarL, is a bifunctional enzyme that combines ribitol phosphate primase and polymerase activities. However, Meredith et al. and Pereira et al. have proposed alternative explanations for the cellular roles of tarK and tarL. Pereira et al. have suggested that tarK is a redundant gene resulting from duplication, and have argued that its function is decaying. Meredith et al. reached a different conclusion based on an in-depth analysis of tarK and tarL expression in cells. Analysis of extracted WTAs from strains that produce only TarL or TarK showed that these two enzymes produce electrophoretically distinct poly(ribitol phosphate) WTAs, designated L-WTA and K-WTA (Figure 5 A). K-WTA is significantly shorter than L-WTA and is presumed, based on PAGE banding patterns, to contain subtle differences in composition. Furthermore, K-WTA biosynthesis is negatively regulated (directly or indirectly) by the agr (accessory gene regulator) quorum sensing system.[61] Since tarK expression can cause the polymer to shorten by up to 50 %,[61] it was suggested that regulation of tarK by agr allows S. aureus to dynamically control WTA chain length as a function of cell density. WTA polymer length might affect exposure of surface adhesins, and it was proposed that dynamic regulation of WTA polymer length allows S. aureus to cycle between a pro-adhesion state and a low-adhesion state, perhaps to promote adhesion and dissemination at appropriate times during the infection process (Figure 5 B). Determination of the exact structures of K-WTA and L-WTA and their potential roles in virulence remain to be addressed.
Figure 5.
Proposed role for the distinct TarK and TarL WTA polymers. A) TarK and TarL make electrophoretically distinct WTA polymers. PAGE analysis of WTAs extracted from various S. aureus strains shows that poly(ribitol phosphate) polymer length corresponds to gene regulation. The agr system directly or indirectly represses the expression of TarK, leading to an increase in WTA polymer length. RN4220 has a partial defect in agr while RN450 has a fully functional agr system (agr+). TarK makes a short secondary polymer, K-WTA, while TarL makes a longer primary polymer, L-WTA. The L-WTA was extracted from a ΔtarK mutant of S. aureus RN4220. K-WTA was extracted from an S. aureus RN4220 mutant over-expressing tarK in a ΔtarL background.[61] B) Expression of the shorter K-WTA at low cell density might allow for increased accessibility of adhesins, leading to a pro-adhesion state; whereas, the expression of the longer L-WTA at high cell density (mediated by the activation of agr) might lead to a low-adhesion phenotype.
Disagreement about the functions of tarK and tarL extends to other genes within the two tarI′J′K/IJL clusters. It has been reported that tarI′ and tarI are both nonessential,[66] that only tarI is essential,[67] or that tarI is only essential under a certain set of growth conditions in vitro but is nonessential in an in vivo infection model.[68] Furthermore, in S. aureus Newman, seven viable transposants were isolated within tarI′J′K but none was isolated in tarIJL,[47] suggesting that the gene duplications are not redundant. Recently, Chaudhuri et al. have reported that tarI, tarJ, and tarL are essential in S. aureus.[69] These discrepancies can be collectively resolved by suggesting that there are differences in tarI′J′K expression, which depend on culture conditions and strain backgrounds. The fact that all fourteen sequenced S. aureus strains retain both tarI′J′K and tarIJL intact, argues against simple functional redundancy and suggests a selective pressure for maintaining both clusters.
Characterization of the Wall Teichoic Acid Biosynthetic Genes in S. aureus
WTA biosynthesis exhibits a mixed gene dispensability pattern
It was first observed in the 1960s that WTAs are not essential for the survival of S. aureus in vitro,[39] and a number of studies on genetically uncharacterized WTA null mutants were reported in subsequent years.[70–74] In 2004, Peschel and co-workers characterized a defined WTA-null S. aureus strain lacking tarO, the first gene in the WTA biosynthetic pathway.[8] This ΔtarO strain was reported to have a similar in vitro growth rate to the wild-type strain, but was greatly impaired in its ability to adhere to epithelial and endothelial tissues. The adhesion-impaired mutant was unable to colonize nasal passages, leading to the suggestion that WTAs might be virulence factors since they are required for host infection. Brown and co-workers subsequently reported that tarA, like tarO, can also be deleted.[75] The ΔtarA WTA-null strains are viable in vitro and are phenotypically identical to the ΔtarO strains; however, many of the genes downstream of tarA in the S. aureus WTA pathway (depicted in red in Figure 6) cannot be deleted unless tarO (or tarA) is deleted first.[67] These downstream genes are “conditionally essential”: that is, they are required for viability in a strain background containing a functional WTA pathway, but are not required in a WTA-null background. This mixed gene dispensability pattern implies that blocking late-acting WTA biosynthetic enzymes after flux into the pathway has been initiated is deleterious to bacterial growth.
Figure 6.
Depiction of the primary Staphylococcus aureus wall teichoic acid (L-WTA) biosynthetic pathway. Nonessential WTA pathway enzymes are colored green and their deletion leads to an avirulent phenotype. Conditionally essential enzymes are colored red and their deletion is lethal in a wild-type background but permitted in a ΔtarO or ΔtarA background. Following intracellular assembly, the poly(ribitol phosphate) polymer is transported to the outside by a two-component ABC transporter, TarGH, and then covalently linked through a phosphodiester bond to the MurNAc sugars of peptidoglycan by an unidentified enzyme the biological and genetic properties of which have not been established. In addition to TarI, J, and L, all S. aureus strains contain a homologous set of enzymes (designated TarI′,J′ and K; Figure 3) that directs the synthesis of a distinct WTA polymer (K-WTA); their cellular functions remain incompletely understood.
Possible explanations for conditional essentiality
The mixed gene dispensability pattern observed for WTA biosynthesis has also been reported for several other nonessential biosynthetic pathways in which a cell surface glycopolymer is assembled on an undecaprenyl phosphate carrier lipid.[76–80] Such pathways exist in virtually all bacteria. For example, they are involved in the synthesis of O-antigens, capsular polysaccharides, and exopolysaccharides. In pathogenic bacteria, these cell surface polymers, like WTAs, play roles in virulence. Two explanations have generally been considered for the apparent toxicity caused by blocking late steps in these pathways. One explanation attributes toxicity to depletion of undecaprenyl phosphate-linked peptidoglycan precursors and the resulting effects on peptidoglycan biosynthesis. Undecaprenyl phosphate is used as the carrier lipid in the peptidoglycan biosynthetic pathway; however, only small amounts of this carrier lipid are produced and the cell's capacity to increase these levels is limited. Therefore, any metabolic block that leads to accumulation or sequestration of undecaprenyl phosphate-linked intermediates is potentially harmful to cells. Indeed, a number of cell-wall active antibiotics, including bacitracin, vancomycin, and ramoplanin, function by sequestering peptidoglycan precursors.[81–83] An alternative explanation attributes the observed toxicity upon blocking nonessential bactoprenol-dependent pathways to an accumulation of bactoprenol-linked intermediates that are somehow directly harmful to cells.[84] Evidence for and against both mechanisms has been presented, but a consensus has not yet been reached. Since these possibilities are not mutually exclusive, it is possible that both play a role.
Brown and co-workers have proposed that peptidoglycan substrate depletion is the mechanism for toxicity when WTA biosynthesis is blocked in B. subtilis.[85] Microarray analysis was used to identify genes up-regulated in B. subtilis upon depletion of TagD, the cytidylyltransferase that provides activated glycerol phosphate for poly(glycerol phosphate) synthesis. The promoters for ten highly up-regulated genes were then fused to the lux operon and the luminescence signal upon tag gene depletion was evaluated. One of the promoters, PywaC, gave a particularly robust luminescent signal when WTA biosynthesis was disrupted at a late step. The PywaC reporter was activated by cell-wall active antibiotics that sequester peptidoglycan precursors as well as by depletion of genes involved in undecaprenol biosynthesis. It was not activated by depletion of tagO. Since the PywaC reporter strain responded to perturbations known to affect pool levels of bactoprenol-linked peptidoglycan intermediates and to late-acting tag gene depletion, it was suggested that blocking late-acting WTA enzymes is toxic because it affects levels of bactoprenol-linked peptidoglycan substrates. Brown and co-workers have speculated that their reporter assay can be used in a high-throughput screen to identify compounds that target either the cell wall or wall teichoic acid biosynthetic pathways.
Inhibitors of Wall Teichoic Acid Biosynthesis
Methicillin-resistant S. aureus infections have become a major problem in the United States, recently surpassing HIV/AIDS as a cause of death. Although there are still a handful of effective anti-MRSA antibiotics, clinical resistance is inevitable and, indeed, has already been observed for the most recently introduced antibiotics.[86] Thus, an urgent need exists for the exploration of new strategies to battle resistant S. aureus infections.
The WTA biosynthetic pathway has been speculated to be an antibiotic target for many years, but only one specific inhibitor has recently been reported. The mixed gene dispensability pattern implies that there are two distinct types of possible antimicrobial targets within the pathway: antivirulence targets (TarO and TarA; depicted in green in Figure 6) and antibiotic targets (the conditionally essential downstream enzymes; depicted in red). Small molecule inhibitors of the former are expected to impede colonization and the spread of infection, while inhibitors of the latter have been shown to prevent bacterial growth (see below). The known inhibitors of WTA biosynthesis are described below.
Inhibitors of antivirulence targets in the WTA pathway
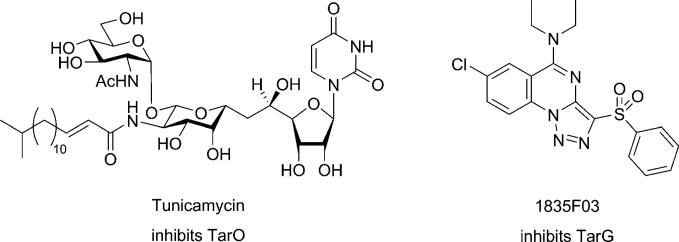
Peschel and co-workers were the first to suggest that WTA biosynthesis is an antivirulence target in S. aureus.[8] This possibility has attracted considerable attention because it is speculated that resistance to nonessential targets involved in pathogenicity (virulence factor targets) will not develop as readily as it does to more traditional antibiotic targets.[87,88] May et al. have reported a small molecule that inhibits the d-alanine tailoring modification in both LTAs and WTAs, and the activity of this compound in preliminary studies supports the possibility that inhibiting d-alanylation could attenuate the virulence of pathogenic organisms.[43] In addition to this compound, there is a very potent natural product inhibitor of WTA biosynthesis, the uridine-containing antibiotic tunicamyin (Figure 7).[72,89] Tunicamycin is a promiscuous inhibitor of the large family of enzymes that couple sugar phosphates to membrane-embedded lipid phosphates.[52] Its well-known antibacterial activity derives from its ability to inhibit MraY, an essential phosphosugar transferase in the peptidoglycan biosynthetic pathway; however, tunicamycin also inhibits TarO.[10,52] In fact, tunicamycin is selective for TarO over MraY by a factor of at least 100. Its selectivity for TarO makes it a useful tool for shutting off WTA expression in vitro without affecting bacterial growth rates. Unfortunately, it cannot be used in animals to assess whether inhibiting TarO is a viable strategy for treating S. aureus infections because it is toxic to eukaryotes. It inhibits an essential eukaryotic phosphosugar transferase involved in the dolichol pathway for N-linked glycosylation (GPT), which catalyzes the same chemical transformation as TarO. Nontoxic, selective inhibitors of TarO (or TarA) remain to be discovered.
Figure 7.
Chemical structures of currently known inhibitors of wall teichoic acid synthesis and export in Staphylococcus aureus. An inhibitor of DltA, which is involved in modification of both WTAs and LTAs, has also been reported.[43]
Inhibitors of antibiotic targets in the WTA pathway
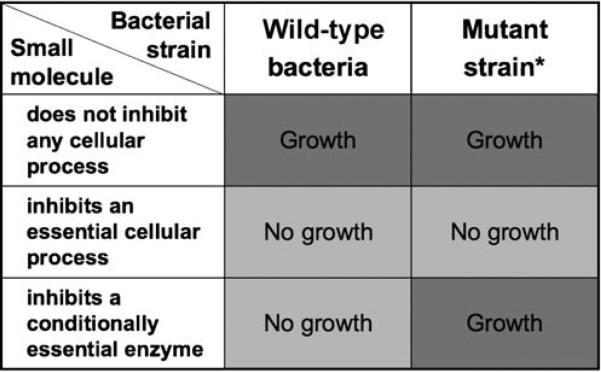
The first inhibitor of a putative antibiotic target in the WTA biosynthetic pathway was recently reported by Swoboda et al.[10] It was discovered by using a general cell-based screening approach that exploits the conditional essentiality of the late-acting enzymes (Figure 8).[10] The screening strategy developed to discover WTA inhibitors is applicable, in principle, to any nonessential biosynthetic pathway containing conditionally essential genes. It involves screening a compound library against a pair of bacterial strains, one a wild-type strain and the other a null mutant that does not express the polymer of interest (e.g., WTAs). Compounds that inhibit the growth of the polymer-expressing wild-type strain, but not of the mutant, are expected to target the conditionally essential enzymes in the desired biosynthetic pathway. Screening paired strains ensures target specificity while eliminating compounds that inhibit essential cellular processes or are toxic for other reasons (Figure 8). Since antibiotic discovery is challenging, a cell-based screen that ensures cellular activity is critical, but designing a screen to report on a particular pathway is typically difficult. The reported strategy combines two important features that are not often found together in high-throughput screens: it is both cell-based and pathway specific.
Figure 8.
General screening strategy for the identification of small-molecule inhibitors of conditionally essential enzymes/targets in nonessential biosynthetic pathways; *: the mutant strain is incapable of initiating polymer synthesis. In the described screen, the paired strain lacked the first enzyme involved in WTA biosynthesis (TarO).
Swoboda et al. used the paired strain screening strategy to identify WTA inhibitors with antibiotic activity.[10] The growth of S. aureus RN4220 and the corresponding ΔtarO strain were monitored in the presence of a library of 55 000 small molecules. Three inhibitors were found to inhibit the wild-type strain without affecting the mutant. The most active of the three compounds (1835F03; Figure 7) was found to have a minimum inhibitory concentration of 1–2 μg mL–1 (2.5–5 μm) against all S. aureus strains examined, including clinical MSSA and MRSA isolates. A comprehensive set of genetic and biochemical experiments have shown that the target of the compound is TarG, the transmembrane component of the dedicated, two-component ABC transporter that exports WTAs from the cytoplasm to the cell surface.
The discovery of a small molecule that inhibits a late-acting step in WTA biosynthesis and has growth inhibitory activity validates the WTA pathway as a possible antibacterial target, but the efficacy of this antibacterial strategy has yet to be determined. Resistance to the reported WTA inhibitor occurs at a high frequency in vitro (1 in 106) and two classes of resistant mutants have been identified. One class involves mutations in the target (TarG), a common theme for antibiotics. The other mutants contain changes in the tarO or tarA genes, which abolish WTA expression. The observation that this latter class of mutations occurs frequently is perhaps not surprising, as the pathway is not essential for growth in vitro. Under ordinary circumstances, obtaining a high frequency of resistant mutants in vitro would suggest that a particular pathway is not a viable antimicrobial target. However, the WTA biosynthetic pathway presents an unusual and previously unexplored paradigm. A large percentage of the resistant mutants do not express WTAs but, because Peschel and co-workers have reported that S. aureus strains lacking WTAs are incapable of colonizing a host, these resistant mutants are not expected to survive in vivo. If they do not, then the null mutants are not a factor for resistance in animals. As we have pointed out above, there are numerous other pathways that contain conditionally essential enzymes linked to virulence-factor expression. Many of these enzymes could be good antibiotic targets provided that the major mechanism for resistance involves deletion of the pathway, and results in the production of avirulent organisms. The recent discovery of a small molecule that inhibits a conditionally essential step in a virulence factor pathway provides a starting point for investigating this novel antibacterial strategy.
Outlook
Extensive work over several decades has illuminated many of the roles of TAs in Gram-positive bacteria and has firmly established their importance in bacterial physiology. A better understanding of the WTA biosynthetic pathway has been aided by both biochemical and genetic studies, and most of the steps in the B. subtilis and S. aureus WTA biosynthetic pathways have been reconstituted in vitro by using synthetic substrates. A small molecule antibiotic that targets WTA biosynthesis in S. aureus was recently discovered by utilizing the recent genetic and biochemical advances in this field, and will make possible studies to evaluate WTA biosynthesis as a pathway for therapeutic intervention. Positive outcomes from these studies would validate this class of virulence factors as antibacterial targets and provide further impetus for their study and exploitation.
Acknowledgements
This work was supported by the NIH (1P01AI083214 and 5R01M078477 to S.W., and F3178727 to J.G.S.), a Mary Fieser Postdoctoral Fellowship to J.C., and a training grant to T.C.M (T32-AI07061-30).
References
- 1.DiRienzo JM, Nakamura K, Inouye M. Annu. Rev. Biochem. 1978;47:481. doi: 10.1146/annurev.bi.47.070178.002405. [DOI] [PubMed] [Google Scholar]
- 2.Lugtenberg B, Van Alphen L. Biochim. Biophys. Acta. 1983;737:51. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- 3.Matias VRF, Beveridge TJ. Mol. Microbiol. 2005;56:240. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 4.Vollmer W, Blanot D, De Pedro M. FEMS Microbiol. Rev. 2008;32:149. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 5.Neuhaus F, Baddiley J. Microbiol. Mol. Biol. Rev. 2003;67:686. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morath S, Von Aulock S, Hartung T. J. Endotoxin Res. 2005;11:348. doi: 10.1179/096805105X67328. [DOI] [PubMed] [Google Scholar]
- 7.Weidenmaier C, Peschel A. Nat. Rev. Microbiol. 2008;6:276. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 8.Weidenmaier C, Kokai-Kun J, Kristian S, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond J, Peschel A. Nat. Med. 2004;10:243. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 9.Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR, Bayer AS. J. Infect. Dis. 2005;191:1771. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- 10.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach MT, Malhowski AJ, Kishony R, Gilmore MS, Walker S. ACS Chem. Biol. 2009;4:875. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endl J, Seidl PH, Fiedler F, Schleifer KH. Arch. Microbiol. 1984;137:272. doi: 10.1007/BF00414557. [DOI] [PubMed] [Google Scholar]
- 12.Araki Y, Ito E. Crit. Rev. Microbiol. 1989;17:121. doi: 10.3109/10408418909105745. [DOI] [PubMed] [Google Scholar]
- 13.Coley J, Tarelli E, Archibald AR, Baddiley J. FEBS Lett. 1978;88:1. doi: 10.1016/0014-5793(78)80594-8. [DOI] [PubMed] [Google Scholar]
- 14.Endl J, Seidl HP, Fiedler F, Schleifer KH. Arch. Microbiol. 1983;135:215. doi: 10.1007/BF00414483. [DOI] [PubMed] [Google Scholar]
- 15.Kojima N, Araki Y, Ito E. J. Biol. Chem. 1983;258:9043. [PubMed] [Google Scholar]
- 16.Kojima N, Araki Y, Ito E. J. Bacteriol. 1985;161:299. doi: 10.1128/jb.161.1.299-306.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradov E, Sadovskaya I, Li JJ, Jabbouri S. Carbohydr. Res. 2006;341:738. doi: 10.1016/j.carres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama K, Miyashita T, Araki Y, Ito E. Eur. J. Biochem. 1986;161:479. doi: 10.1111/j.1432-1033.1986.tb10469.x. [DOI] [PubMed] [Google Scholar]
- 19.Lazarevic V, Abellan FX, Moller SB, Karamata D, Mauel C. Microbiol. 2002;148:815. doi: 10.1099/00221287-148-3-815. [DOI] [PubMed] [Google Scholar]
- 20.Archibald AR, Baddiley J, Button D. Biochem. J. 1968;110:559. doi: 10.1042/bj1100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baddiley J, Buchanan JG, Hardy FE, Martin RO, Rajbhandary UL, Sanderson AR. Biochim. Biophys. Acta. 1961;52:406. doi: 10.1016/0006-3002(61)90699-0. [DOI] [PubMed] [Google Scholar]
- 22.Baddiley J, Buchanan JG, Martin RO, Rajbhandary UL. Biochem. J. 1962;85:49. doi: 10.1042/bj0850049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baddiley J, Buchanan JG, Rajbhandary UL, Sanderson AR. Biochem. J. 1962;82:439. doi: 10.1042/bj0820439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirelman D, Beck BD, Shaw DRD. Biochem. Biophys. Res. Commun. 1970;39:712. doi: 10.1016/0006-291x(70)90263-9. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama K, Mizuguchi H, Araki Y, Kaya S, Ito E. J. Bacteriol. 1989;171:940. doi: 10.1128/jb.171.2.940-946.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K. J. Bacteriol. 2009;191:141. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schirner K, Marles-Wright J, Lewis R, Errington J. EMBO J. 2009;28:830. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Elia M, Millar K, Beveridge T, Brown E. J. Bacteriol. 2006;188:8313. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaito C, Sekimizu K. J. Bacteriol. 2007;189:2553. doi: 10.1128/JB.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, Lasa I. Microbiol. 2008;154:865. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Götz F, Zähringer U, Peschel A. Mol. Microbiol. 2007;65:1078. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman O, Dover L, Sutcliffe I. Trends Microbiol. 2009;17:219. doi: 10.1016/j.tim.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Baddiley J. Acc. Chem. Res. 1970;3:98. [Google Scholar]
- 34.Marquis RE, Mayzel K, Carstensen EL. Can. J. Microbiol. 1976;22:975. doi: 10.1139/m76-142. [DOI] [PubMed] [Google Scholar]
- 35.Ellwood DC, Tempest DW. J. Gen. Microbiol. 1972;73:395. doi: 10.1099/00221287-73-2-395. [DOI] [PubMed] [Google Scholar]
- 36.Heptinstall S, Archibald AR, Baddiley J. Nature. 1970;225:519. doi: 10.1038/225519a0. [DOI] [PubMed] [Google Scholar]
- 37.Ellwood DC. Biochem. J. 1970;118:367. doi: 10.1042/bj1180367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickham JJR, Halye JL, Kashtanov S, Khandogin J, Rice CV. J. Phys. Chem. B. 2009;113:2177. doi: 10.1021/jp809313j. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee AN. J. Bacteriol. 1969;98:519. doi: 10.1128/jb.98.2.519-527.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins LV, Kristian S, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Götz F, Neumeister B, Peschel A. J. Infect. Dis. 2002;186:214. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 41.Peschel A, Vuong C, Otto M, Götz F. Antimicrob. Agents Chemother. 2000;44:2845. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. J. Biol. Chem. 1995;270:15 598. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 43.May JJ, Finking R, Wiegeshoff F, Weber TT, Bandur N, Koert U, Marahiel MA. FEBS J. 2005;272:2993. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 44.Gross M, Cramton SE, Götz F, Peschel A. Infect. Immun. 2001;69:3423. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. J. Biol. Chem. 1999;274:8405. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 46.Kristian S, Lauth X, Nizet V, Goetz F, Neumeister B, Peschel A, Landmann R. J. Infect. Dis. 2003;188:414. doi: 10.1086/376533. [DOI] [PubMed] [Google Scholar]
- 47.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. Proc. Natl. Acad. Sci. USA. 2004;101:12312. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian Z, Yin Y, Zhang Y, Lu L, Li Y, Jiang Y. BMC Genomics. 2006;7:74. doi: 10.1186/1471-2164-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollack JH, Neuhaus FC. J. Bacteriol. 1994;176:7252. doi: 10.1128/jb.176.23.7252-7259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Formstone A, Carballido-Lopez R, Noirot P, Errington J, Scheffers D. J. Bacteriol. 2008;190:1812. doi: 10.1128/JB.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward JB. Microbiol. Rev. 1981;45:211. doi: 10.1128/mr.45.2.211-243.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price NPJ, Tsvetanova B. J. Antibiot. 2007;60:485. doi: 10.1038/ja.2007.62. [DOI] [PubMed] [Google Scholar]
- 53.Soldo B, Lazarevic V, Karamata D. Microbiol. 2002;148:2079. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- 54.Ginsberg C, Zhang Y, Yuan Y, Walker S. ACS Chem. Biol. 2006;1:25. doi: 10.1021/cb0500041. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Ginsberg C, Yuan YQ, Walker S. Biochemistry. 2006;45:10895. doi: 10.1021/bi060872z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhavsar A, D'Elia M, Sahakian T, Brown E. J. Bacteriol. 2007;189:6816. doi: 10.1128/JB.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira M, Schertzer J, D'Elia M, Koteva K, Hughes D, Wright G, Brown E. ChemBioChem. 2008;9:1385. doi: 10.1002/cbic.200800026. [DOI] [PubMed] [Google Scholar]
- 58.Schertzer J, Brown E. J. Biol. Chem. 2003;278:18002. doi: 10.1074/jbc.M300706200. [DOI] [PubMed] [Google Scholar]
- 59.Schertzer J, Brown E. J. Bacteriol. 2008;190:6940. doi: 10.1128/JB.00851-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lazarevic V, Karamata D. Mol. Microbiol. 1995;16:345. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 61.Meredith TC, Swoboda JG, Walker S. J. Bacteriol. 2008;190:3046. doi: 10.1128/JB.01880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira MP, D'Elia MA, Troczynska J, Brown ED. J. Bacteriol. 2008;190:5642. doi: 10.1128/JB.00526-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Men H, Park P, Ge M, Walker S. J. Am. Chem. Soc. 1998;120:2484. [Google Scholar]
- 64.Ye X-Y, Lo M, Brunner L, Walker D, Kahne D, Walker S. J. Am. Chem. Soc. 2001;123:3155. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 65.Brown S, Zhang YH, Walker S. Chem. Biol. 2008;15:12. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zalacain M, Biswas S, Ingraham KA, Ambrad J, Bryant A, Chalker AF, Iordanescu S, Fan J, Fan F, Lunsford RD, O'Dwyer K, Palmer LM, So C, Sylvester D, Volker C, Warren P, McDevitt D, Brown JR, Holmes DJ, Burnham MKR. J. Mol. Microbiol. Biotechnol. 2003;6:109. doi: 10.1159/000076741. [DOI] [PubMed] [Google Scholar]
- 67.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. J. Bacteriol. 2006;188:4183. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Streker K, Schaefer T, Freiberg C, Broetz-Oesterhelt H, Hacker J, Labischinski H, Ohlsen K. Antimicrob. Agents Chemother. 2008;52:4470. doi: 10.1128/AAC.00548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. BMC Genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aly R, Shinefield HR, Litz C, Maibach HI. J. Infect. Dis. 1980;141:463. doi: 10.1093/infdis/141.4.463. [DOI] [PubMed] [Google Scholar]
- 71.Bracha R, Davidson R, Mirelman D. J. Bacteriol. 1978;134:412. doi: 10.1128/jb.134.2.412-417.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hancock IC, Wiseman G, Baddiley J. FEBS Lett. 1976;69:75. doi: 10.1016/0014-5793(76)80657-6. [DOI] [PubMed] [Google Scholar]
- 73.Ou LT, Chatterjee AN, Young FE, Marquis RE. Can. J. Microbiol. 1973;19:1393. doi: 10.1139/m73-225. [DOI] [PubMed] [Google Scholar]
- 74.Maki H, Yamaguchi T, Murakami K. J. Bacteriol. 1994;176:4993. doi: 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.D'Elia M, Henderson J, Beveridge T, Heinrichs D, Brown E. J. Bacteriol. 2009;191:4030. doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katzen F, Ferreiro DU, Oddo CG, Ielmini MV, Becker A, Puhler A, Ielpi L. J. Bacteriol. 1998;180:1607. doi: 10.1128/jb.180.7.1607-1617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xayarath B, Yother J. J. Bacteriol. 2007;189:3369. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuasa R, Levinthal M, Nikaido H. J. Bacteriol. 1969;100:433. doi: 10.1128/jb.100.1.433-444.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burrows LL, Lam JS. J. Bacteriol. 1999;181:973. doi: 10.1128/jb.181.3.973-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makela P, Stocker B. In: Handbook of Endotoxin. Rietschel E, editor. I. Elsevier; Amsterdam: 1984. p. 59. [Google Scholar]
- 81.Storm DR, Strominger JL. J. Biol. Chem. 1973;248:3940. [PubMed] [Google Scholar]
- 82.Walker S, Chen L, Hu YN, Rew Y, Shin DW, Boger DL. Chem. Rev. 2005;105:449. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- 83.Walsh TR, Howe RA. Annu. Rev. Microbiol. 2002;56:657. doi: 10.1146/annurev.micro.56.012302.160806. [DOI] [PubMed] [Google Scholar]
- 84.Danese PN, Oliver GR, Barr K, Bowman GD, Rick PD, Silhavy TJ. J. Bacteriol. 1998;180:5875. doi: 10.1128/jb.180.22.5875-5884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.D'Elia MA, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, Moreno-Hagelsieb G, Brown ED. Chem. Biol. 2009;16:548. doi: 10.1016/j.chembiol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Boucher HW, Corey GR. Clin. Infect. Dis. 2008;(Suppl 5):46, S344. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 87.Barczak AK, Hung DT. Curr. Opin. Microbiol. 2009;12:490. doi: 10.1016/j.mib.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clatworthy AE, Pierson E, Hung DT. Nat. Chem. Biol. 2007;3:541. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 89.Wyke A, Ward J. J. Bacteriol. 1977;130:1055. doi: 10.1128/jb.130.3.1055-1063.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]