Abstract
Studies of the genetics of gene expression reveal expression SNPs that explain variation in transcript abundance. Here we address the robustness of eSNP associations to environmental geography and population structure in a comparison of 194 Arab and Amazigh individuals from a city and two villages in southern Morocco. Gene expression differed between pairs of locations for up to a third of all transcripts, with notable enrichment for ribosomal biosynthesis and oxidative phosphorylation. Robust associations were observed in the leukocyte samples with cis-eSNPs (P < 10−08) for 346 genes, and trans-eSNPs (P < 10−11) with 10 genes. All of these were consistent across the three sample locations and after controlling for ethnicity and relatedness. No evidence for large-effect trans-acting mediators of the pervasive environmental influence was found and instead genetic and environmental factors acted in a largely additive manner.
Keywords: Peripheral blood, eSNP, GWAS, ethnicity, relatedness, environmental geography
The human transition from pastoral and rural to urban lifestyles has been accompanied by increased incidence of numerous chronic diseases such as asthma, diabetes and cancer1. Environmental contributors, likely including dietary shifts, pollution, and psychological factors, are the subject of ongoing epidemiological research. It is equally interesting to ask whether genetic influences on disease susceptibility change across environments. Since disease risk is commonly thought to often involve differential gene expression2, we have assessed the robustness of transcript abundance to environmental variation by performing a genome-wide association study on leukocyte gene expression profiles across two ethnicities in three locations. Our previous work had demonstrated a substantial effect of environmental geography3 on gene expression in Moroccan Amazigh, and here we additionally add the contrast with Arabs, allowing us to test whether geography and/or ethnicity affect each of several hundred robust associations between genotypes and transcript abundance.
The Souss region in southern Morocco is home to several million people of two dominant ethnicities, living either in cities, or rural villages (Fig. 1). The Amazigh Berbers are descendant of the first modern humans who populated North Africa 35,000 or more years ago4, and many still live in traditional villages in the low Atlas Mountains. The Arabs by contrast moved into southern Morocco between the 7th and 11th centuries and tend to occupy lowland villages, while both groups inhabit the cities, often retaining their linguistic and cultural identities.
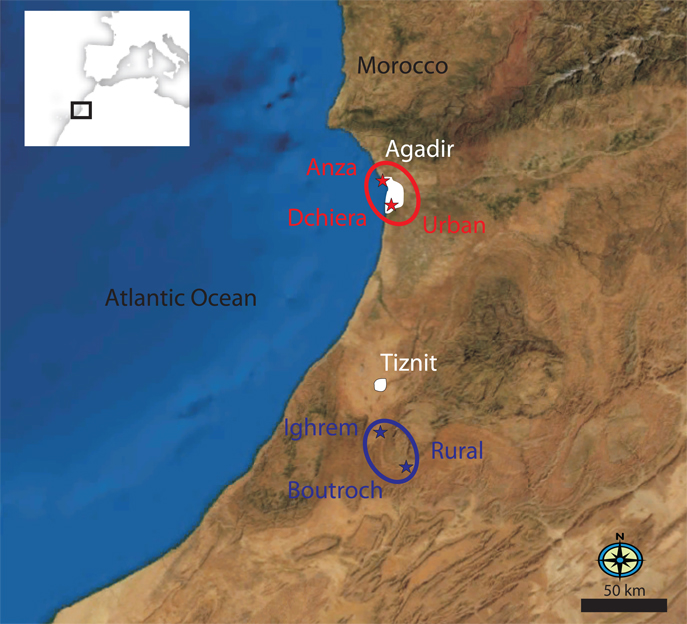
Figure 1. Map of the Souss region of southern Morocco.
showing the location of the two rural villages, Boutroch and Ighrem, near the town of Tiznit, relative to the urban locations Anza and Dchiera north and south of the city of Agadir, respectively.
We collected peripheral blood samples from 284 healthy adults in June and July of 2008 from four locations, including approximately equal numbers of men and women, and of Amazigh and Arabs. Half the sample was from two high density, low to middle-income, urban communities, Anza and Dchiera, on either side of the city of Agadir. The other half was from two rural villages near Tiznit, 120 km to the south. Boutroch is predominantly Amazigh and remains quite isolated, while Ighrem is predominantly Arab and (based on self-reported information and our observations at the collection site) many of the men, in particular, commute into the cities.
Leukocytes were isolated from serum, platelets and erythrocytes at the time of blood sampling by depletion filter technology5 and fixed in RNALater® solution within a few minutes of blood collection. Gene expression profiles were obtained from 208 high quality RNA samples using Illumina HumanHT12 bead arrays that include 48,804 probes, of which 22,300 RefSeq probes in 16,738 genes were deemed to have signal above background. In order to minimize batch effects, all samples were processed in the same week, and the extraction, labeling and hybridization steps were all performed according to a randomized block design. Whole genome genotypes were obtained from whole blood samples using Illumina Human 610-Quad arrays. After quality control filters, 516,972 SNPs were available for 194 of the individuals who also had gene expression profiles.
RESULTS
Population Structure of southern Morocco
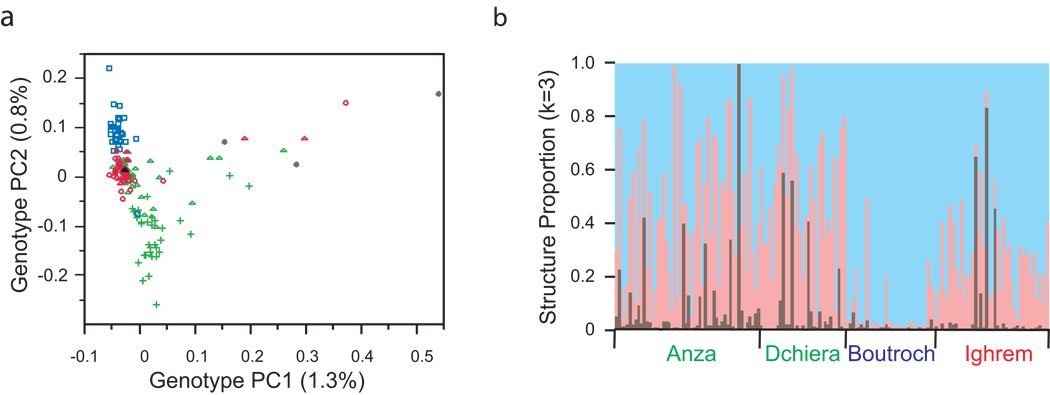
Population structure was assessed by examining the principal components (PC) of the variance of the genotype profiles, using Eigenstrat software6. Initial examination revealed several clusters of siblings and other close relatives (cousins or similar) whose similarity skewed the axes; where data were available, these identities were in agreement with participant records. After removal of these relatives, analysis of 163 unrelated individuals revealed seven significant eigenvectors. None of these explain more than 5% of the variance, and PC3 through PC7 are heavily weighted by large clusters of SNPs on one or a few chromosomes. As described by others, such axes are commonly observed and do not provide reliable genome-wide estimates of population structure7,8, but it is interesting to note that PC3 distinguishes Ighrem from the other locations (Supplementary Fig. 1a online).
A plot of the first two eigenvectors (Fig. 2a) highlights the major historical influences on population structure in southern Morocco. PC1 separates just a dozen individuals, and we inferred that this axis represents a sub-Saharan African contribution, consistent with expected levels of admixture in Morocco, by performing an analysis including 21 Yoruban individuals (Supplementary Fig. 1b online). PC2 is highly correlated with both location and self-reported ethnicity, so is inferred to capture the major component of Arab-Amazigh ancestry.
Figure 2. Population structure in southern Morocco.
(a)Eigenstrat principal component analysis of 579,144 SNPs reveals 7 significant eigenvectors, the first two of which, explaining just 1.3 and 0.8 % of the genotypic variance respectively, are plotted here. By self-report, Boutroch Amazigh are blue squares, Agadir Amazigh green triangles, Agadir Arabs green plus symbols, Ighrem Arabsred circles, and Ighrem Amazigh red triangles. 3 individuals with uncertain ethnicity possibly including sub-Saharan heritage, are indicated as gray spots, and have high values of PC1, which is characteristic of Yoruban ancestry as shown in Supplementary Figure 1b online. (b) Structure analysis of 16,000 autosomal SNPs, with k=3 and employing the admixture model with correlated allele frequencies, highlights the same individuals with large PC1 values (brown bars) and shows that Boutroch Amazigh are predominantly derived from one population group (pale blue) while all other samples are a mixture of the two populations represented by pale red and blue bars.
A surprising aspect of this analysis is the positioning of Ighrem Arabs between Boutroch Amazigh and half of the Agadir Arabs along PC2. This was confirmed by Structure analysis9 of 16,000 randomly chosen autosomal SNPs assuming admixture of two ancestral populations (Fig. 2b), which indicates that Ighrem residents tend to be a mixture, while most Amazigh are derived from one population, and only a handful of Agadir Arabs represent the other. There has thus likely been considerable admixture between these two groups over an extended period of time, possibly with movement of Arabs from other locations into Agadir recently. A slight shift of Ighrem Arabs toward the Amazigh pole of PC2, relative to Agadir Arabs, would also be consistent with some genetic exchange between the villages over 50 generations. Further sampling of villages in the region may reveal subtle population structure across southern Morocco10–13.
Regional Differentiation in Gene Expression
Next we asked whether region, location and ethnicity impact gene expression profiles, and if they do so in a gender-specific manner. Since location and ethnicity are confounded in the villages, several parallel analyses were undertaken to tease apart these influences. Transcript abundance data was transformed by median centering on the log base 2 scale (Supplementary Fig. 2 online), which results in maximal overlap of profiles without altering their variance.
Gene specific analysis of variance14 with expression as a function of region, gender, and their interaction discovered 1,521 probes significant at a 1% false discovery rate (FDR; P < 0.0007). Region, namely the rural (Boutroch plus Ighrem) versus city (Anza plus Dchiera) comparison is by far the main effect in this joint analysis. Approaching 7% of all expressed genes differentiate these individuals by this conservative criterion, whereas considerably fewer than 1% of the probes show gender differences. A full list of genes is provided in Supplementary Table 1 online. Among several classes of over-represented genes for this lifestyle comparison, small nucleolar RNA genes stand out: 5 of the top 8 overall and 15 of 29 members of the SNORD family are in the highly significant list, compared with just 1 of 10 SNORA genes. There is little in the literature to indicate why this is the case, or what the physiological consequences may be, but it is interesting to note that epigenetic modification has been observed for many small nucleolar RNA genes15.
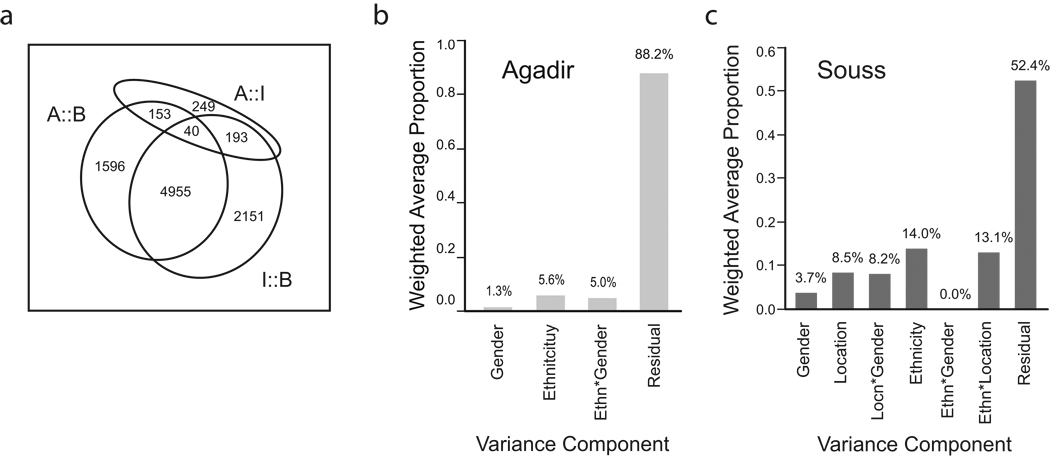
Even more differentiation was observed when we fit analysis of variance models including location, gender, and their interaction. Since exploratory analyses indicated that the Anza and Dchiera samples are indistinguishable either for gene expression or genotypes, these were combined into a single location, Agadir, in all subsequent analyses. In the three-way comparison, 8,459 probes (38%) were significant at the 1% FDR threshold for location (Supplementary Table 2 online). Boutroch differs from both Ighrem and Agadir at over seven thousand probes each, with a high degree of overlap (Fig. 3a; Table 1). Ighrem and Agadir are much more similar to one another, in part because there is considerably more diversity within the Ighrem sample that reduces the significance of the location contrast. We also noted that women are much more differentiated among locations than men (Table 1). These results confirm our previous report2 of substantial differentiation between Bedouin nomads, urban Anza, and another remote Amazigh village, Sebt Nabor.
Figure 3. Location impacts gene expression transcriptome-wide.
(a) Venn diagram of the number of genes significant at 1% FDR for ANOVA of the three pair-wise comparisons indicated. Variance components of expression variation (b) just in the 118 residents of Agadir (excluding 9 individuals with strongly positive gPC1 scores, and including reassignment of ethnicity according to gPC2 for just 11 individuals relative to self-report, Supplementary Table 5),where Ethnicity is modeled as the PC2 of the genotype variation as shown in Figure 1a, or (c) for all 22,300 probes in the full sample of 208 individuals.
Table 1.
Number of transcripts significant at 1% FDR
| Location | Gender | Interaction | ||||||
|---|---|---|---|---|---|---|---|---|
| ANOVA | ANCOVA | ANOVA | ANCOVA | ANOVA | ANCOVA | |||
| 3-way | 8459 | 7057 | Male : Female | 151 | 233 | Location*Gender | 133 | 203 |
| Aga : Bou | 6744 | 4974 | In Agadir | 24 | 24 | Fem (Aga : Bou) | 4830 | 3791 |
| Aga : Igh | 635 | 651 | In Boutroch | 13 | 14 | Fem (Aga : Igh) | 1451 | 1467 |
| Bou : Igh | 7339 | 6286 | In Ighrem | 589 | 890 | Mal (Aga : Bou)† | 407 | 806 |
| Aga : Rural | 1521 | 607 | Mal (Aga : Igh) | 8 | 8 | |||
ANOVA includes terms for Location, Gender, and Location*Gender interaction. The False Discovery Rate was evaluated using the conservative Benjamin and Hochberg method. The left hand columns show the number of genes significant at the 1% FDR threshold for Location effects (either in the 3-way comparison of Agadir (Aga), Boutroch (Bou) and Ighrem (Igh); between pairs of locations, or between Agadir (Aga) and the two rural sites combined). The central columns contrast gender (male versus female) effects, either in the total sample or each location individually. The right hand columns show interaction effects, either in the total sample or showing the indicated contrast between Agadir and either village, for females or males separately. ANCOVA is the same model with an additional continuous covariate for ethnicity, genotypic PC2.
Significance of this contrast was reduced by the small sample of Boutroch males (12, cf 26 females).
In order to evaluate the possible independent contribution of ethnicity more carefully, variance component analysis of the expression variation was performed. Within Agadir alone, neither ethnicity (modeled as the second eigenvector of the genotype data, gPC2) nor gender have a noteworthy impact on the principal components of the expression variation, as shown by the bar charts in Figure 3b. However, in the total dataset there is some evidence for a contribution: Figure 3c shows that when fit jointly with location, the ethnicity, ethnicity-and gender-by-location interaction terms make a substantial contribution to the expression profiles.
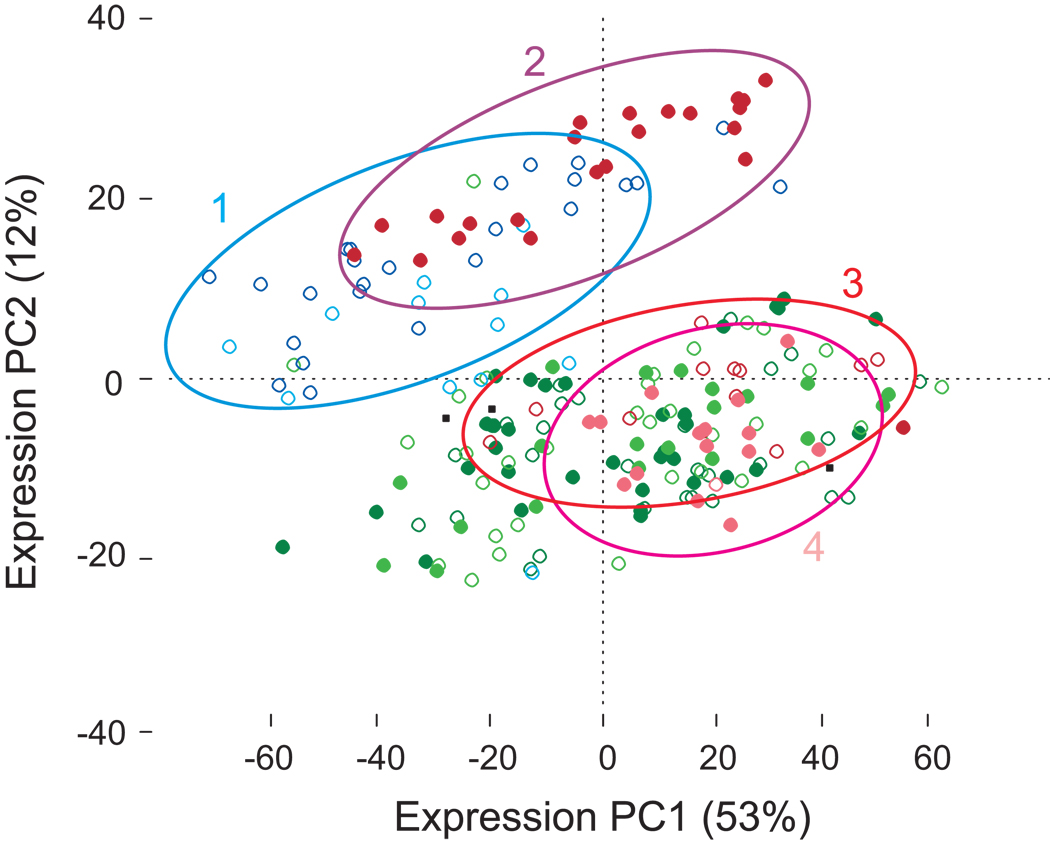
Although gender and ethnicity affect the expression of fewer genes than location, the plot of expression PC1 by PC2 for the most differentially expressed 1,500 genes in Figure 4 indicates that for many genes the interaction between these three factors is quite complex. This can also be seen in the expression profiles of characteristic individual genes (Supplementary Fig. 3 online). Boutroch and Ighrem villagers in general separate along PC1, while high values of PC2 are obtained for all Boutroch residents (cluster 1) and for Arab women in Ighrem (cluster 2). Amazigh women from Ighrem (cluster 3) and the Ighrem men (cluster 4) have lower values of PC2 similar to those observed for all Agadir residents. The simplest interpretation is that cultural or behavioral differences, likely including time spent outside the village, contribute strongly to the observed gender and ethnicity effects. Deeper sampling would be required to firmly establish whether intrinsic biological differences between the sexes and/or populations also make significant contributions to expression divergence in lymphocytes, as they appear to do for lymphoblast cell lines grown in culture16–19.
Figure 4. Principal component plot for the most differentially expressed genes.
The two major principal components of the expression of the 1,500 most significant genes shows significant separation of individuals by location (PC1 and PC2) and gender (PC2) (all P < 0.0001) as described in the text. Individuals from Boutroch are blue, Ighrem red, and Agadir green. Arabs are indicated with solid spots, Amazigh open circles, and males are lighter symbols for each color. Boutroch and Arab women from Ighrem (clusters 1 and 2) separate from Amazigh women and Arab men from Ighrem (clusters 3 and 4) who are closer to Agadir residents. If Boutroch residents and Ighrem Arab women are grouped and contrasted with Agadir residents, Ighrem Amazigh women, and Ighrem men, 8,239 genes are significantly differentially expressed at the 1% FDR rate, more than any pair-wise comparison of locations. A similar plot for all genes is shown in Supplementary Figure 11.
Two classes of genes stand out as significantly differentially-expressed among locations. These are ribosomal proteins of both the small and large subunits as well as the cytoplasmic and mitochondrial compartments, and proteins involved in oxidative phosphorylation, which are highly up-regulated in half of the Agadir residents (Supplementary Fig. 4a online). All of the transcripts encoding these proteins form a module of co-regulated genes, but as shown in Supplementary Figure 4b online, it is noteworthy that this module is not co-expressed with the SNORDs, which tend to be relatively down-regulated in Agadir but are particularly high in the Arab women from Ighrem. Regulation of ribosomal biosynthesis may be related to response to viral infection, and it also seems to be involved in tumorigenesis in conjunction with mitochondrial activity20,21. Oxidative phosphorylation is correlated with renal health and the production or disposal of free radicals22, so our data suggests that deeper evaluation of health risks associated with lifestyle transitions may be revealing.
Genome-wide association with gene expression variation
The genetic contribution to expression variation was evaluated by genome-wide association with expression of all 22,300 probes. Starting with a simple test of the correlation between each transcript abundance and each genotype, and filtering to retain only eSNPs with a minor allele frequency greater than 0.05, we observed 3,430 associations at P < 10−8. Further filtering of eSNPs to retain only autosomal associations with annotated genes, and imposing the additional stringency of P < 10−11 for putative trans associations between an eSNP on one chromosome and a probe on another chromosome, reduced this to 1,636 associations. 1,569 (96%) of these are intra-chromosomal linkages, the vast majority within 50 kb and hence cis-acting(Supplementary Fig. 5 online), and only 3 clearly in different chromosomal intervals. Facsimile associations were observed for 39 of the target genes represented by a second probe (37 cis, 2 trans). Reducing the dataset further to exclude linked associations within haplotype blocks leaves 346unique cis and 10 unique trans associations at the stringent genome-wide 5% significance level. These proportions are in good agreement with most other GWAS expression studies on blood or lymphocyte cell lines16,17,23–26, and a 30-fold or greater excess of cis over trans associations is also supported by 1% FDR estimates of 600 and 20 genes respectively. Complete lists of peak cis and trans associations are provided in Supplementary Table 3 online.
Given the high degree of population structure for gene expression, we addressed the possibility that differentiation of eSNP allele frequencies may contribute to the observed associations by calculating FST estimates for each pair-wise comparison of location for the 516,972 SNPs and 16,500 of the genes. No fixed differences were observed and plots of the FST comparisons (Supplementary Fig. 6a online) indicate only moderate overall genetic differentiation, with occasional SNPs having FST values between 0.12 and 0.3. There was no tendency for these outliers to have elevated expression differentiation and in fact almost all of the top 10% most differentially expressed genes are among the least genetically differentiated. Nor was there any correlation between FST and significance of gene expression divergence (Supplementary Fig. 6b online), confirming that the observed expression differences between locations are for the most part not attributable to gene-specific allelic frequency differences between locations.
The robustness of the 3,430 associations to environmental sources of variance and population structure was further evaluated by fitting two additional linear trend models to the data. The first included location, gender and the interaction between them. The second included two measures of ethnicity (the first three genotype eigenvectors and a four-way categorical ethnicity cluster: see methods), a matrix of relatedness based on an identity by descent measure27, as well as gender interactions with ethnicity cluster and genotype. Figures 5a and 5b show the Manhattan plot of associations by chromosomal location for the second of these models, and the cis-trans plot of target against eSNP location, respectively. Figure 5c and Supplementary Fig. 7 online show that the logarithm of the genotype significance term is highly correlated (r > 0.95) between both of these models and the original correlation test. Furthermore, Figure 5d shows that there is no evidence for significant genotype-by-location interactions in any of the association trend tests. Neither the ethnicity nor the relatedness variance components explain an appreciable amount of the expression variation for any of the transcripts (Supplementary Fig. 8 online).
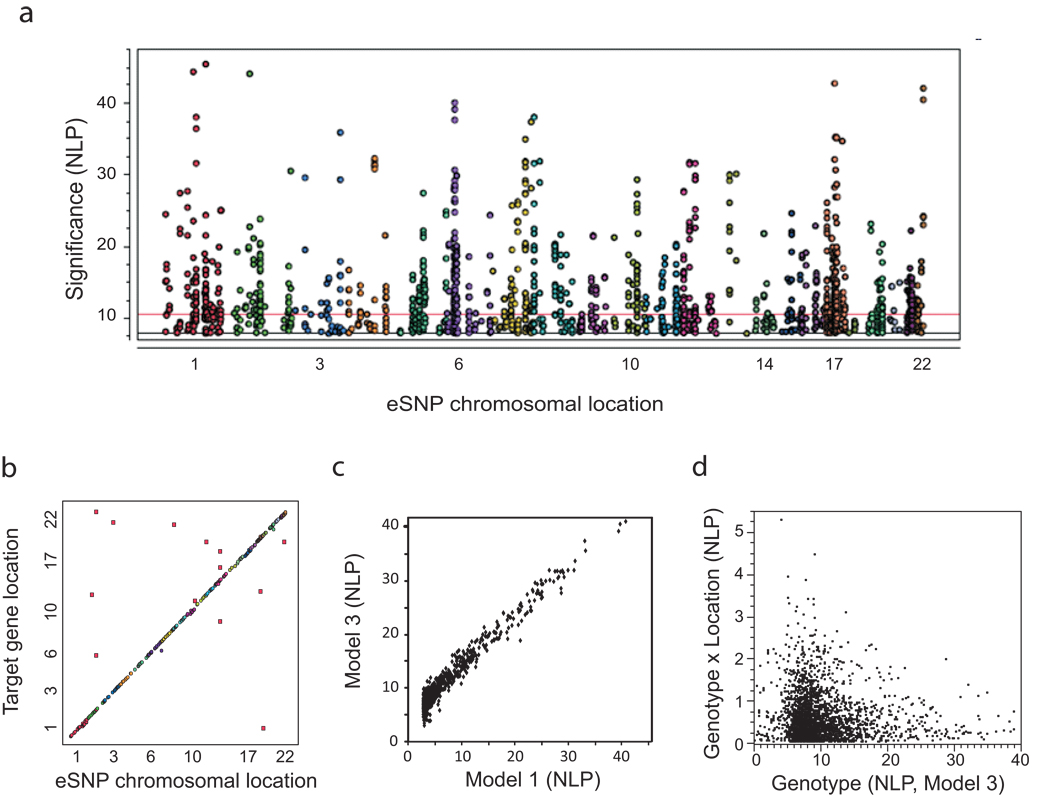
Figure 5. Genome-wide association with transcript abundance.
(a)Manhattan plot of all 1,636 genome-wide associations at P < 10−8(NLP > 8) for model 3, which includes control for genotype-determined ethnicity, location, relatedness, and gender. Each chromosome is indicated by a different color. The horizontal red line indicates the genome-wide significance threshold (NLP > 11.4) for trans associations. Note the excess of peaks at the MHC complex on chromosome 6 due to multiple cis-eSNPs. (b) Cis-Trans plot showing target transcript location against eSNP location indicating that most eSNPs are in cis to the regulated transcript, while just 13 trans associations at NLP > 11.4are visible. (c) High correlation of significance measures for all eSNPs detected by simple correlation of genotype with expression (model 1) or robust control for ethnicity, gender and location (model 3). (d) Absence of genome-wide significance for the Genotype-by-Location interaction effect, which is not correlated with the Genotype effect.
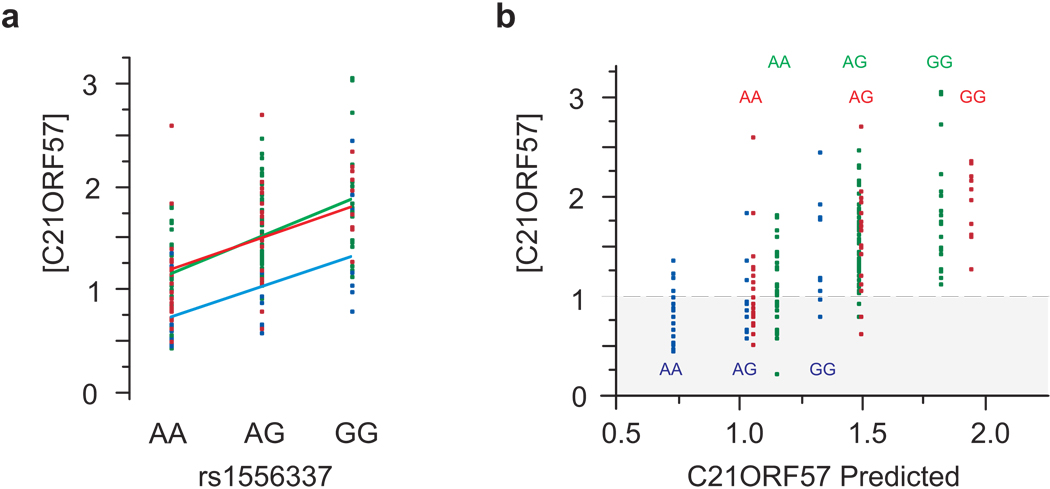
The absence of interaction effects is readily visualized by plotting expression as a function of genotype with color coding of each location, for each association. An example of a trans association in Supplementary Figure 9 online shows the clear trend of increased expression of AMY1A (chromosome 1) in homozygotes for the A allele of ACTG1 gamma actin (chromosome 17), consistently across the three locations despite slight overall location effects. Expression of AMY1A is highly correlated with that of AMY1B (r > 0.8) as well as of dozens of other genes in a co-expression module, but the eSNP only regulates AMY1A, because it increases expression of the gene two-fold in an additive manner. A similar plot for a representative gene that shows highly significant location and genotype effects in cis, C21ORF57, is provided in Figure 6a and further discussed below, and further examples can be seen in Supplementary Figure 9c online.
Figure 6. The relationship between genotype, expression, and phenotype.
(a) A typical example of a transcript (encoding C21ORF57, a putative metallo-proteinase) that shows both a significant difference between locations (P < 10−5) and a cis-eSNP association, with rs1556337 (P < 10−13) but no interaction effect in an additive model on the log scale. Expression is lower in Boutroch (blue points and line), while genotype has a consistent effect across all three locations (Ighrem, red; Agadir, green). (b) The Actual vs Predicted plot separates the genotypes by location for clarity. Suppose that a disease or phenotype is only seen in individuals with transcript abundance less than 1.0 (on a relative log2 scale), indicated by the gray area. Then in Agadir and Ighrem (green and red respectively) almost all affected are AA homozygotes, whereas in Boutroch (blue) heterozygotes and some GG homozygotes are also affected. There is thus a G×E interaction for the phenotype in the absence of a G×E interaction for transcription, because the environment shifts more individuals into the susceptible zone. Similar arguments would apply for phenotypes with high expression values, and for graded rather than threshold-dependent traits.
Novel associations with potential disease alleles
GWAS-expression associations detected in one tissue can identify regulatory variants that may be active in other tissues that are directly engaged in the etiology of disease23,25,26. One example is the cis-linkages in peripheral blood with the T1D susceptibility locus at chromosome 12q13. The strongest expression association is with transcription of the RPS26 ribosomal protein gene, and network analyses have been employed to argue that this is the more likely diabetes candidate gene than the initially reported ERBB328. However, the strongest T1D association involves a different SNP than that associated with expression and/or splicing24 of RPS26. We further find that the same linkage group of eSNPs, centered on the rs10876864 in the SUOX gene 35kb from RPS26, is also associated in trans with half a dozen other RP26 paralogs (probably due to cross-hybridization), and with CCDC4 on chromosome 4, albeit at the suggestive significance level ofP = 3.5×10−10. Intriguingly, expression of RPS26 is only weakly correlated with that of the module of ribosomal proteins that differentiate locations (Supplementary Fig. 4b online), so this association does not contribute to the environmental effect on transcription of ribosomal protein genes.
Another trans-association of interest involves rs11987927 in MYOM2 at 8p23 with ZNF71 at 19q13, but also with its own MYOM2 transcript. Logic suggests that the cis-association likely affects the abundance of the MYOM2 myomesin protein, which in turn regulates ZNF71, but the trans association is actually significantly stronger and conditional dependence analysis29,30 points in the opposite direction, namely that the MYOM2 regulatory site influences ZNF71, which then feeds back on the MYOM2 transcript (Supplementary Fig. 10 online). This example may be a cautionary tale concerning the interpretation of conditional dependence results. It is worth mentioning that four of the seven strongest trans associations involve regulation by loci that include genes that encode structural proteins, the others being the LAMA5 laminin (20q13) with OSBPL2, and the PLEKHM1 plekstrin homology domain protein (17q21) with MAPK8IP1.
One further trans-association is of particular interest. Prolongation of fetal gamma hemoglobin expression in adults is often observed in thalassemia patients. We found association of two probes that detect both HBG1 and HBG2 transcripts from 11p15 with rs766432 in the second intron of the BCL11A zinc finger proto-oncogene at 2p16. This same SNP has previously been associated with the fraction of erythrocytes that contain measurable fetal hemoglobin31, and alteration of BCL11A activity was recently shown drive differences in globin switching between mice and humans32. Another SNP in BCL11A, rs4671393 has been associated with abundance of two BCL11A transcript isoforms in CEU and YRI HapMap lymphoblast cell lines33, but is not associated with BCL11A transcript abundance in our leukocyte data, suggesting that regulation of BCL11A translation or protein activity is more likely to be affecting HBG expression in our sample.
Numerous cis-associations are also likely to be of interest. We scanned the GWAS association database for overlap between our study and established disease associations at p<10−5. Of 1,628 entries, 10 involve cis associations observed in our dataset that explain between 15 and 55% of the transcript variance (Supplementary Table 4 online). Five of the associations are with disease conditions (rheumatoid arthritis, celiac disease, T1D, ulcerative colitis, and SLE) and five are with endophenotypes (PAFAH1B2 and ICAM-1 protein levels, triglycerides, LDL cholesterol, and hip bone mineral density). The two serum protein associations34,35 are with the same SNPs as we detect and hence suggest that protein abundance is largely regulated at the transcriptional level.
DISCUSSION
The genetic and environmental contributions to expression variation
Our geographical genomic survey of gene expression variation in southern Morocco has highlighted two parallel and for the most part non-overlapping insights. On the one hand, it is evident that as much as half of the transcriptome is influenced by the environment in a highly coordinated manner such that where a person lives explains up to a quarter of the variation for a substantial fraction of the transcripts. The environmental influences are likely a combination of biotic and abiotic factors, as well as cultural and behavioral ones, while genetic differences between the two North African ethnicities are relatively minor. On the other hand, the genome is littered with strong genetic associations, mainly in cis, that explain between 15 and 60 percent of the variance of 5% of the transcripts. Impressive as these associations are, particularly since they are discovered in a sample of just under 200 individuals, they have essentially no bearing on the vast majority of the transcriptional variation, and are not informative of the genetic basis of the environmental response.
The robustness of the observed associations to the environmental effect raises the issue of whether genotype-by-environment interactions influence the peripheral blood transcriptome at all. Genome-wide significant interaction effects are generally unlikely to occur in the absence of significant main genotype effects36. The only circumstances in which they will are if the genotype effect is in the opposite direction in two locations, and if the genetic effect in these locations is at least the same magnitude as the main effects detected in this GWAS, namely explaining over 30% of the variance of a particular transcript. While a few such interactions may exist, it would take a study comparing several thousand individuals from each location to reveal weaker genotype-by-environment interactions. If the genetic architecture of transcription is generally similar to that of visible phenotypes like height and body mass37,38, even such a study will be underpowered to explain the vast majority of transcriptional variance.
A related question is whether or not genotype-by-environment interactions at the level of transcription are necessary to explain genotype-by-environment interactions for disease. It is possible the small interactions beneath the level of detection of GWAS are prevalent, or alternatively that disease arises primarily as a result of rare alleles of major effect, whose penetrance may be modulated in an environment-specific manner. However, transcriptional interactions are not required to explain the increased incidence of chronic disease. It is not difficult to imagine that individuals that fall into the major categories of transcriptome profiles (such as those implicated in Fig. 4 and Supplementary Fig. 4 online) have different distributions of disease susceptibility that alter the genotype-disease association matrix genome-wide, thereby inducing environment-by-genotype interactions for disease. Transcription of some genes that contribute to this expression component may also correlate with disease directly, effectively uncovering cryptic variation and resulting in environment-specific eSNP disease associations without any interaction effect at the level of transcription (Fig. 6)39. A corollary of this is that gene expression profiling might be used to stratify individuals at elevated risk for disease, thereby increasing the resolution of genome-wide association studies by focusing attention on the subset of individuals where genetic effects on disease are most pronounced.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank all of the study participants in Agadir, Ighrem and Boutroch, as well as numerous individuals who facilitated sample collection, in particular Malika, Aicha and Ahmed Idaghdour. Dongliang Ge and Alison Motsinger-Reif provided timely computational support, and we also thank Shameek Biswas and Josh Akey for providing HapMap genotypes. Funding for the study was provided by the University of Queensland, with YI supported by a Fulbright Fellowship and GG by an ARC Australian Professorial Fellowship.
Footnotes
ACCESSION NUMBERS
Accession codes. NCBI GEO: Gene expression data from this study have been deposited under series GSE17065.
AUTHOR CONTRIBUTIONS
YI collected the samples with the assistance of SJ and processed them with KS and DG; KM, HL, DG, PV and RW provided statistical and conceptual support for analysis of the data by YI, WC, HM and GG; and YI and GG conceived the study and wrote the paper.
COMPETING INTEREST: Co-authors Wolfinger, Czika and Miclaus are all employees of SAS, Inc, which are the producers of commercial JMP Genomics software used in the analysis of the data.
References
- 1.Abegunde DO, Mathers CD, Adam T, Ortegon M, Strong K. The burden and costs of chronic diseases in low-incomeand middle-income countries. Lancet. 2007;370:1929–1938. doi: 10.1016/S0140-6736(07)61696-1. [DOI] [PubMed] [Google Scholar]
- 2.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e52. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arredi B, Poloni ES, Paracchini S, et al. A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am. J. Hum. Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feezor RJ, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genom. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 6.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 7.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas S, Scheinfeldt LB, Akey JM. Genome-wide insights into the patterns and determinants of fine-scale population structure in humans. Am. J. Hum. Genet. 2009;84:641–650. doi: 10.1016/j.ajhg.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard J, Stephens M, P Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kéfir R, Stevanovitch A, Bouzaid E, Béraud-Colomb E. Diversité mitochondriale de la population de Taforalt (12.000 ans bp - Maroc): Une approche génétique à l'étude du peuplement de l'Afrique du nord. Anthropologie. 2005;43:1–11. [Google Scholar]
- 11.Coudray C, et al. Population genetic data of 15 tetrameric short tandem repeats (STRs) in Berbers from Morocco. Forensic Sci. Int. 2007;167:81–86. doi: 10.1016/j.forsciint.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Ennafaa H, et al. Mitochondrial DNA haplogroup H structure in North Africa. BMC Genet. 2009;10:8. doi: 10.1186/1471-2156-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch E, et al. Population history of North Africa: evidence from classical genetic markers. Hum. Biol. 1997;69:295–311. [PubMed] [Google Scholar]
- 14.Wolfinger RD, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 15.Royo H, Cavaillé J. Non-coding RNAs in imprinted gene clusters. Biol. Cell. 2008;100:149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 16.Dixon AL, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 17.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD, et al. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao CF, Chen SY, Lee YH. Activation of RNA polymerase I transcription by hepatitis C virus core protein. J. Biomed. Sci. 2004;11:72–94. doi: 10.1007/BF02256551. [DOI] [PubMed] [Google Scholar]
- 21.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 22.Shah SV, Baliga R, Rajapurkar M, Fonseca VA. Oxidants in chronic kidney disease. J. Am. Soc. Nephrol. 2007;18:16–28. doi: 10.1681/ASN.2006050500. [DOI] [PubMed] [Google Scholar]
- 23.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 24.Heinzen EL, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1000001. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 26.Heap GA, et al. Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genomics. 2009;2:1. doi: 10.1186/1755-8794-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes BJ, Visscher PM, Goddard ME. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 2009;91:47–60. doi: 10.1017/S0016672308009981. [DOI] [PubMed] [Google Scholar]
- 28.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2009;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LS, Emmert-Streib F, Storey JD. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome Biol. 2007;8:R219. doi: 10.1186/gb-2007-8-10-r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockman MV. Reverse engineering the genotype-phenotype map with natural genetic variation. Nature. 2008;456:738–744. doi: 10.1038/nature07633. [DOI] [PubMed] [Google Scholar]
- 31.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 32.Sankaran VG, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 34.Melzer D, et al. A Genome-Wide Association Study Identifies Protein Quantitative Trait Loci (pQTLs) PLoS Genet. 2008;4:1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paré G, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culverhouse R, Suarez B, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am. J. Hum. Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 38.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson G. Decanalization and the origin of complex disease. Nat. Rev. Genet. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visscher PM, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Idaghdour Y. Genetic and Environmental Components of Human Leukocyte Gene Expression Variation in Morocco. PhD thesis. available from the NCSU electronic library at http://www.lib.ncsu.edu/ETD-db/ETD-browse/browse?first_letter=I. [Google Scholar]
- 43.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda S, et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36(Web Server issue):W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.