Summary
The identification of the cellular mechanisms responsible for the wide differences in species lifespan remains one of the major unsolved problems of the biology of aging. We measured the capacity of nuclear protein to recognize DNA double strand breaks (DSB) and telomere length of skin fibroblasts derived from mammalian species that exhibit wide differences in longevity. Our results indicate DNA DSB recognition increases exponentially with longevity. Further, an analysis of the level of Ku80 protein in human, cow, and mouse suggests that Ku levels vary dramatically between species and these levels are strongly correlated with longevity. In contrast mean telomere length appears to decrease with increasing longevity of the species, although not significantly. These findings suggest that an enhanced ability to bind to DNA-ends may be important for longevity. A number of possible roles for increased levels of Ku and DNA-PKcs are discussed.
Keywords: Species life-span, DNA-end binding activity, telomere length, DNA double-strand break repair, Ku protein
Introduction
In mammals, species lifespan can vary by more than 100 fold (shrew 2 years, bowhead whale 211 years). Despite considerable research, the cellular mechanisms that make this variation possible remain unclear. Assuming the simplest underpinning of these mechanisms, several predictions can be made. First, they likely impact fundamental processes. Second, they would be expected to be reflected by structural differences between species at the cellular level. Furthermore, these differences would be predicted to approximate the differences in observed lifespan in magnitude and to correlate independently with lifespan. As a tool to investigate these mechanisms, we have developed a series of skin fibroblast cell lines derived from mammalian species with a wide variation in lifespan. Using these lines, we have previously shown that the reported dependence of replicative capacity on longevity (Rohme, 1981) is most likely due to differences in body mass, which is itself correlated with longevity (Lorenzini et al., 2005). Therefore, comparative studies of longevity must address the influence of body mass. In the above and present analysis we used maximum species longevity and mean adult body mass from fully authenticated sources and followed published recommendations for comparative studies on longevity (Speakman, 2005). In the present study, we have examined these lines for their DNA break recognition capacity and telomere length. The results of this examination reveal a robust correlation between lifespan and DNA break recognition but little correlation with telomere length.
Materials and Methods
Origin and culture conditions of cell lines used
For DNA end binding assays biopsies was taken from the skin of each of the following animals: mouse, Mexican Free Tailed bat, rabbit, Big Brown bat, cow and cat to produce primary fibroblast cultures. For the dog fibroblasts cultures were derived from a Beagle and a Rottweiler. For human, DNA binding activity was determined on a fibroblast cell line derived from a biopsy taken from an adult man but also on WI 38 lung embryo fibroblasts and on HeLa cells (a commonly used cell line derived from a cervical cancer). For Rhesus monkey, horse and gorilla skin fibroblast cultures were obtained from the Coriell Institute for Medical Research (Camden, NJ). For Chinese Hamster, we used CHO cells, a cell line derived from ovary cells.
For telomere length assays the following skin fibroblast cultures were used: little brown bat (2 cultures from 2 different wild caught individuals), mouse (2 cultures, one from a wild mouse from Pennsylvania and one from a wild mouse from Idaho), rat (2 cultures from 2 laboratory animals), rabbit (2 cultures from a laboratory animal), cat (1 line from a house cat), Rhesus monkey (2 cultures from the Coriell Institute), dog (2 cultures from a laboratory beagle), human (2 cultures from adult individuals), horse (2 cultures from the Coriell Institute), cow (2 cultures from a biopsy obtained from a New Jersey slaughter house).
The exact age of the majority of individuals used was known. It ranged between young and early middle age but it was generally young adult. The two mice and all the bats, which were all caught in the wild, were estimated to be young adults. Skin fibroblasts were grown according to our standard procedures (Cristofalo et al., 1980) with the exception of the addition of antibiotics and antimycotic (100 IU/ml penicillin, 100 mg/ml streptomycin and 0.25 mg/ml amphotericin B). Bats skin samples were obtained from Dr Anja Brunet-Rossinni. Rottweiler skin samples were obtained from Dr David J. Waters and Deborah Schlittler (Purdue University, West Lafayette, IN).
Longevity and body mass data
Maximum longevity and adult body mass for the species analyzed were obtained from Dr Steven N. Austad’s personal database (The Sam and Ann Barshop Institute for Longevity and Aging Studies, San Antonio, TX) and from the online Longevity Records of the Max Planck Institute for Demographic Research (Rostock, Germany, http://www.demogr.mpg.de/). Both databases are compiled from fully authenticated sources. Note that the reported maximum human longevity is 122.5 years. In this study, human longevity is adjusted to 90 years to account for the fact that, for the others species, only small cohorts are used to determine maximum longevity, and 90 years is an estimate for a similar sized random sample of humans.
Telomere restriction fragment (TRF) length analysis
TRF analysis was performed essentially as previously described (Steinert et al., 2002). Briefly, 1 μg of genomic DNA was digested with a cocktail of AluI, HaeIII, HhaI, HinfI, MspI and RsaI, separated on a 0.5% agarose gel for 27 hours at 1 V/cm (or for PFGE at 6 v/cm with a ramped pulse from 1 to 15 secs for 20 hours), and the gel was then dried and probed using the 32P-end labeled telomere repeat oligonucleotide (CCCTAA)4. Complete DNA digestion was confirmed by ethidium staining of DNA run for four hours into the gel. The washed gel was visualized with a Molecular Dynamics Phosphoimager. Mean telomere length was calculated as the weighted average (ODi)/(ODi/Li), where ODi is the background-corrected intensity of telomere signal in interval i and Li is the average length of telomeres in interval i (each interval equal to a pixel), thus normalizing for the stronger signal emitted by longer telomeres. End-labeled full-length and HindIII-digested lambda DNA fragments were used as markers. Signals between 4 and 40 kb for standard gels (and between 5 and 65 kb for PFGE gels) were used for calculations. For little brown bat measurements, only telomere signals that were detected under non-denaturing conditions were used to estimate mean telomere length.
Fluorescence in situ hybridization
For telomere repeat fluorescence in situ hybridization (FISH), a Cy3-conjugated peptide nucleic was used. Slide preparation, acid probe (CCCTAA)3 hybridization, and detection were performed as described in Unit 18.4, Current Protocols in Cell Biology Online, 2006 [www.interscience.wiley.com.]
DNA end binding activity assay
Nuclear protein extracts were prepared from nuclei using the method of (Dignam et al., 1983) using 0.2% (v/v) NP-40 to lyse the cells. Extraction buffer contained 5mM MgCl2 to prevent clumping of the nuclei. Protein concentration of the nuclear extracts was determined using the Bradford method (Bio-Rad laboratories, Hercules, CA) and bovine serum albumin as a standard. DNA end binding activity was determined using our established protocol (Getts et al., 1994), briefly, for each species increasing amounts of nuclear extracts (0.01–20 μg) were incubated with 1ng of a 144bp 32P-labeled DNA probe for 15min at room temperature in the presence of 1μg of supercoiled circular pUC18 plasmid (1000 fold ratio of competitor to probe) in a final volume of 20μl in binding buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 150 mM NaCl, 5 mM DTT, 200μM PMSF, 2.5 mg/ml leupeptin, 1 mg/ml pepstatin A, and 10% (v/v) glycerol). The 144 bp probe was obtained from a Pvu II and Eco-RI digest of the pUC18 plasmid. Since the probe was derived from pUC18 plasmid, those proteins that bind to internal DNA sequences will be bound by the excess circular plasmid and will not bind to the probe. Also, since the plasmid does not contain DNA ends, only proteins that bind to DNA ends will bind to the probe. The reaction mixtures were electrophoretically separated in 5% polyacrylamide gels. The gels are then dried and the fraction of probe bound at each protein concentration is determined from phosphorimager scans of DNA end binding patterns. To estimate DNA binding activity, for each species we calculated the amount of protein required to bind to 50% of the probe.
Immunoblotting
Nuclear extracts prepared as reported above were size-fractionated on 5–20% gradient SDS-polyacrylamide gels (Cambrex Biopharmaceuticals, Baltimore, MD) and electrotransferred on nitrocellulose membranes (Schleicher & Schuell BioScience Inc, Keene, NH) using a BioRad mini Protean electrophoresis system. Abundance of the proteins of interest was assayed using antibodies that react with sequences that are 100% conserved across mouse, cow and human: DNA dependent protein kinase catalytic subunit (cat# IMG-534) and Ku80 (cat# IMG-4174) antibodies were purchased from Imgenex (San Diego, Ca); they are both rabbit polyclonal antibody. For DNA-PKcs the antibody was raised against a synthetic peptide corresponding to amino acids 4118–4128 (GRTWEGWEPWM) of human DNA-PKcs. For Ku80 the antibody was raised against a synthetic peptide corresponding to amino acids 323–338 (FSKVDEEQMKYKSEGK) of the 80 kDa human Ku protein. The DNA Ligase IV antibody was made against a peptide (CELQEENQYLI) at the carboxyl terminus of the Human DNA Ligase IV protein (Bryans et al., 1999). Antibodies against serum response factor (SRF, G-20 cat# sc-335) and histone H3 (N-20 cat# sc-8653) were from Santa Cruz Biotechnology (Santa Cruz, CA). SRF G-20 is a polyclonal antibody raised against a peptide mapping within the C-terminus of SRF of human origin; SRF is more then 95% conserved across mouse, cow and human. Histone H3 N20 is an affinity purified goat polyclonal antibody raised against a peptide mapping at the N-terminus of histone H3 of human origin; the N-terminus of histone H3 is 100% conserved across mouse, cow and human. To control for loading, all membranes were stained with 0.5% w/v Ponceau S prior to antibody hybridization and gels were stained with Coomassie Brilliant Blue G-250 immediately after transfer.
Mass spectrometry
Ten μg of the above cow and human nuclear protein extracts were separated in adjacent lanes on a 10% gel by SDS-PAGE. Proteins present in a series of horizontal 1mm wide gel samples across the sample lanes were digested with trypsin and the resulting peptides analyzed by high resolution tandem time of flight MALDI-(TOF/TOF) mass spectrometry. Peptides similarities were searched based on the human sequences information. Then identified proteins were arrayed in descending order of abundance based on their total peptide ion current. Thus the protein whose peptides produced the highest total ion current is number 1. Total ion current is a function of a protein’s abundance, the theoretical number of trypsin cleavable peptide fragments in its amino acid sequence, and their probability of cleavage. Relative abundance of human and cow DNA-PKcs, Ku80 and Ku70 was estimated by comparing total ion current and position in a list of proteins ordered by total ion current (Table 2).
Table 2. Mass spectrometry results.
A semi quantitative estimate of the relative abundance of protein from nuclear extracts used for DNA end binding and Western analysis was obtained by comparing total peptide ion current in Mass Spectrometry. The Mass Spectrometry analysis was performed on trypsin digests of two regions of a polyacrylamide gel. One region was from the bottom of the well to 97 kDa and in this region 349 proteins were detected in cow and 381 in human. Another region was from 97 kDa to 65 kDa and in this region 366 proteins were detected in cow and 314 in human. Filamin A and HSP90α are shown here as controls for the capacity of the Mass Spectrometer to detect abundant proteins in both species.
| Region of polyacrylamide gel analyzed | Protein Name | COW | HUMAN |
|---|---|---|---|
| Position in descending list of proteins ordered by Total Ion Current | Position in descending list of proteins ordered by total Ion Current (Total Ion Current) | ||
| > 97 kDa | Filamin A | 1st of 349 (7.6e8) | 1st of 381 (2.4e8) |
| DNA-PKcs | No Peptides detected | 6th of 381 (7.6e7) | |
| Between 65 kDa and 97 kDa | HSP90ƒ¿ | 4th of 366 (1.5e7) | 1st of 314 (2.7e7) |
| Ku 80 | 274th of 366 (1.4e5) | 3rd of 314 (2.1e7) | |
| Ku 70 | 355th of 366 (5.3e4) | 2nd of 314 (2.5e7) |
DNA double strand break repair
Cells growing exponentially as monolayers in flasks were irradiated on ice using a Sheppard irradiator at approximately 12.1 Gy/min. and then incubated varying times at 37°C. Cells were removed from the flasks, washed with phosphate-buffered saline (PBS) at 0°C and resuspended at a density of 0.5–1.0 × 107 cells/ml in PBS containing 1.0% low melting point agarose (Incert, FMC), and agarose plugs were prepared by casing into 5 mm × 6mm × 1.0 mm inserts using a BRL mold cooled to 0–4°C. Cells in agarose plugs were placed in 50°C lysis solution containing; 0.5 M EDTA pH 7.9 (Sigma), 1.0% Sarkosyl (Sigma), 1.0 mg/ml proteinase K (Boeringer Manheim). After 18–24 h of digestion, the plugs were dialyzed twice against 10 volumes of TE (10 mM Tris, pH 7.8, 1 mM EDTA), and the RNA hydrolyzed by digestion in 1 volume of TE with 0.1 mg/ml RNAse A (Sigma) for 2 h at 37°C. The agarose plugs were subjected to AFIGE electrophoresis as described previously (Denko et al., 1989; Stamato et al., 1993; Stamato et al., 1990), the gel stained with ethidium bromide, photographed, and agarose sections containing the DNA in the lane and the DNA in the plug were excised. The percentage of DNA released from the plug into the lane was determined by counting the radioactivity in agarose sections.
Statistical and phylogenetic analysis
Statistical analyses were performed using the software GraphPad InSat 3 and non linear regression analysis was performed with Graph Pad Prism 4 software (both from GraphPad Software, Inc. San Diego, CA). The phylogeny used for the phylogenetically independent comparisons was derived from Adkins and Murphy (Adkins et al., 2003; Murphy et al., 2001). Phylogenetic independent comparisons were performed using the “Comparative Analysis by Independent Contrasts” (CAIC) statistical program of Purvis and Rambault (Purvis et al., 1995).
Results
DNA double strand break (DSB) recognition and longevity
We tested the capacity of nuclear extracts from skin fibroblasts and cell lines from different mammalian species to bind to a linear DNA probe using a mobility shift method that we have previously developed (Getts et al., 1994). Displayed in Fig. 1 are mobility shift patterns for varying amounts of nuclear extract isolated from human (panel A), cow (panel B), and mouse (panel C) and their quantitation from phosphoimager scans (panel D). The analysis of our data indicates that this capacity increases dramatically with species lifespan (Fig. 2A & B; Table 1). Across the species examined, the ability to recognize DNA DSB increased 100 fold between the species with the shortest lifespan, Chinese hamster, and that with the longest, human. Using the data for adult skin fibroblasts, regression analysis of the logarithm of DNA-end binding activity versus maximum longevity produced a linear relationship (Fig. 2 panel A; blue diamonds) with significant correlation (Pearson correlation coefficient r=0.903, p < .0001, n=12). Further, the statistical significance of this correlation increased (Pearson correlation coefficient r=0.953, p< .0001, n=15) when data from the Chinese hamster, Hela and WI38 cells (red dots) were included in the analysis. This linear relationship strongly suggests that that there is an exponential relationship between DNA-end binding activity and maximum longevity. In contrast the logarithm of DNA-end binding activity was not significantly correlated with body mass (r=0.097, p=0.5824, n=12) Fig. 2, panel C & D). Further, when the contribution of body mass to the correlation was statistically removed by partial correlation, the correlation of the logarithm of DNA-end binding activity and maximum longevity was not significantly altered (not shown). In contrast, when DNA end binding and body mass were compared by removing the influence of longevity, no significant correlation was observed (not shown). Species in the same phylum may share particular traits through decent from a common ancestor rather than through independent evolution (Speakman, 2005). To examine this possibility, phylogenetically independent comparisons of DNA-end binding activity, maximum longevity, and body mass were performed using approaches described by Felsenstein and Garland (Felsenstein, 1985; Garland et al., 2005). In the 10 phylogenetically independent species comparisons available here (Fig. 3a), DNA end binding activity differed in the same direction as maximum longevity in 9 instances (p = 0.01 one-tailed, p = 0.025 two-tailed with binomial test, Fig. 3b) compared with 7 instances for body mass (Fig. 3b, p=0.35 two tailed, not significant).
Figure 1. Assay for determination of Nuclear DNA-end binding activity.
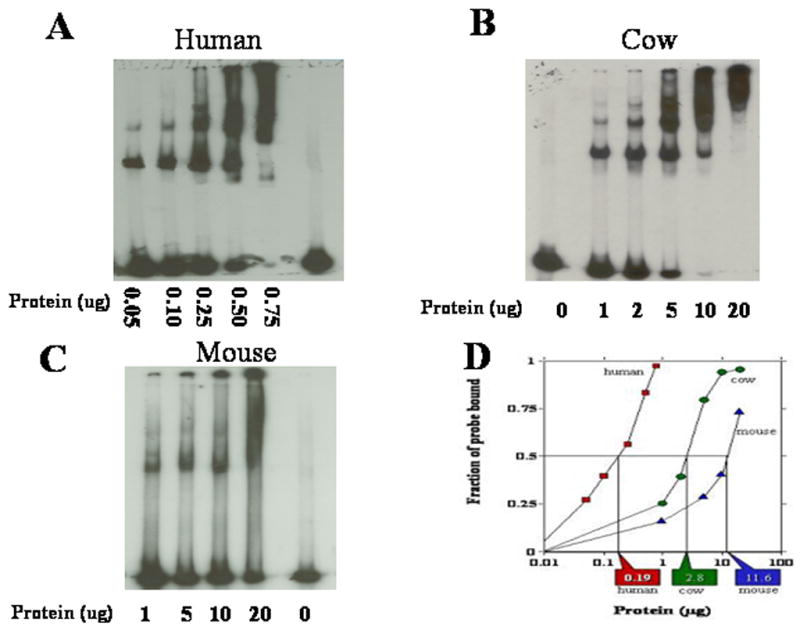
Typical mobility shift assays for the determination of DNA-end binding activity are shown for nuclear protein extracts isolated from human (A), mouse (B) and cow (C) fibroblasts. (D) Quantification of phosphorimager scans of DNA probe binding for each mobility shift. Vertical lines and rectangular callouts represent that amount of protein necessary to bind 50% of the 32P-labeled probe of linear DNA.
Figure 2. Exponential correlation of Nuclear DNA-end binding activity with maximum longevity but not with adult body mass in mammals.
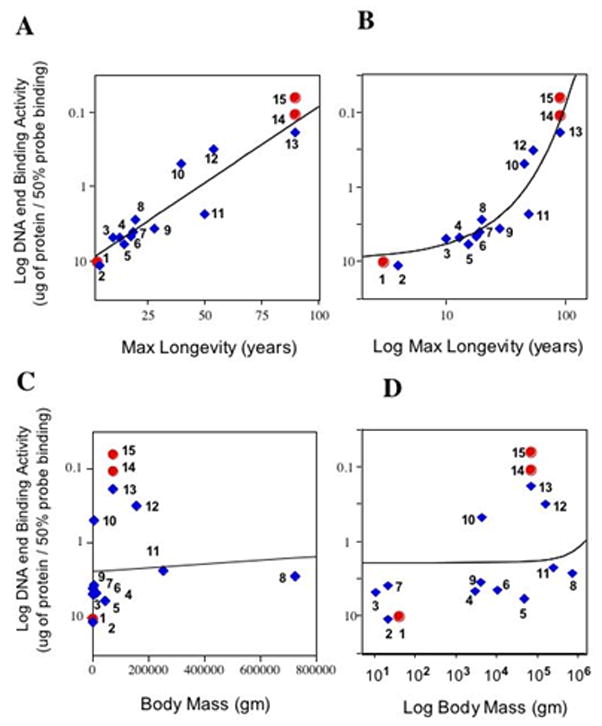
Plots of the Log of DNA end binding activity versus species maximum longevity or versus the Log of species maximum longevity are displayed in panels (A) and (B), respectively. Displayed in panels (C) and (D) are plots of the Log of DNA end binding activity versus species body mass or versus the Log of species body mass, respectively. Note that the “Y” axis values are in reverse order because higher binding activity means that less nuclear protein is necessary to bind the same amount of linear DNA probe. Each data point is identified by number, species name, DNA-end binding activity (ug protein to bind 50% of the probe), maximum longevity in years, and average adult body mass in grams are contained in Table 1. Statistical determinations were performed using cultured adult skin fibroblasts (blue diamonds), and thus did not include Chinese hamster CHO lines, human WI 38 fibroblast or human HeLa cells (round red circles). The line in each of the panels represents a regression analysis fit of the data to an exponential function. For the majority of the skin fibroblast cultures the population doublings (PDs) of the culture was 25 and cells had not immortalized. Mouse and Mexican Free Tailed bat (TB) skin fibroblasts had immortalized. TB was used at PDs=40.
Table 1.
Maximum Longevity, DNA-End binding activity, and body mass for species in Fig. 2.
| Data point number in Figure 2 | Species | Maximum Longevity (years) | DNA-End Binding activity* | Average adult body mass (grams) |
|---|---|---|---|---|
| #1 | Chinese hamster ovary cell | 3 | 10.45 | 42 |
| #2 | House mouse | 4 | 11.6 | 22.5 |
| #3 | Mexican Free Tailed bat | 10 | 4.9 | 11 |
| #4 | Rabbit | 13 | 4.8 | 3000 |
| #5 | Dog, Rottweiler | 15 | 6.0 | 45,000 |
| #6 | Dog, Beagle | 18 | 4.6 | 10,800 |
| #7 | Big Brown bat | 19 | 4.1 | 22 |
| #8 | Cow | 20 | 2.8 | 725,000 |
| #9 | House cat | 28 | 3.7 | 4000 |
| #10 | Rhesus monkey | 40 | 0.5 | 8500 |
| #11 | Horse | 50 | 2.4 | 250,000 |
| #12 | Low Land gorilla | 54 | 0.32 | 155,000 |
| #13 | Human adult Fibroblast | 90 | 0.19 | 70,000 |
| #14 | Human WI 38 lung embryo fibroblasts | 90 | 0.064 | 70,000 |
| #15 | Hela human ovary tumor cell | 90 | 0.11 | 70,000 |
Amount of protein (ug) required to bind 50% of the probe
Figure 3. Co-variation of longevity and body mass with DNA end binding activity across the available independent comparisons.
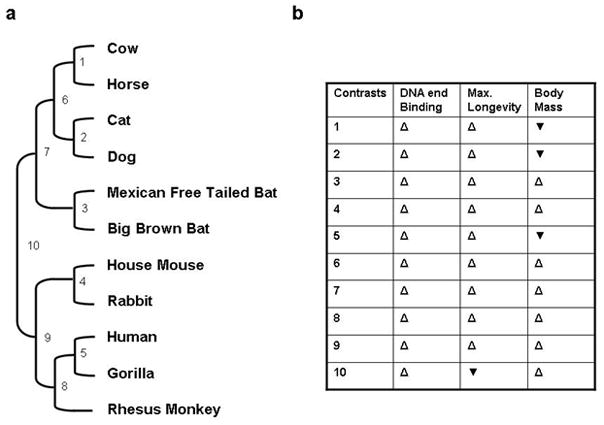
(a) Phylogeny for the relationships between maximum longevity, DNA end binding and body mass. Branch lengths are not drawn to scale. (b): Direction of variations in maximum longevity, DNA end binding and body mass in the available independent comparisons were determined using the CAIC software package (Purvis et al., 1995). Open, upward-pointed triangles indicate that the two parameters vary in the same direction, while filled, downward-pointing triangles indicate that the parameters vary in opposite directions.
We, and others, have previously demonstrated that the highly evolutionary conserved Ku80/70 heterodimer is the primary protein responsible for the DNA end binding activity of nuclear extracts (Downs et al., 2004; Getts et al., 1994; Marangoni et al., 2000). After the Ku80/70 heterodimer binds to DNA DSB the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is recruited and activated by the Ku complex. The Ku80/70 heterodimer together with the DNA-PKcs constitute the DNA-PK complex. DNA-PKcs through phosphorylation of many downstream targets acts as a DNA damage signal enhancer. The level of Ku80 and DNA-PKcs protein in nuclear extracts isolated from mouse, cow, and human cells was examined by Western analysis using antibodies raised against peptide sequences that are 100% conserved in these three species. This analysis indicated that the level of Ku80 and DNA-PKcs proteins are much more abundant in long-lived species (human, 90 years) compared to species with intermediate lifespan (cow, 20 years) or the species with the shortest lifespan (mouse 4 years) (Fig. 4). These results are similar to differences in DNA-PKcs kinase activity and Ku levels previously observed in human and mouse cells (Lees-Miller et al., 1992; Wang et al., 1993a) and now have been extended to the cow, a species with intermediate lifespan. These observations are consistent with the correlation we have observed between DNA-end binding activity and levels of DNA-PKcs and Ku proteins in these species (Fig. 1. & Table 1).
Figure 4. Comparison of Ku 80 and DNA-PKcs protein abundance in human, cow and mouse.
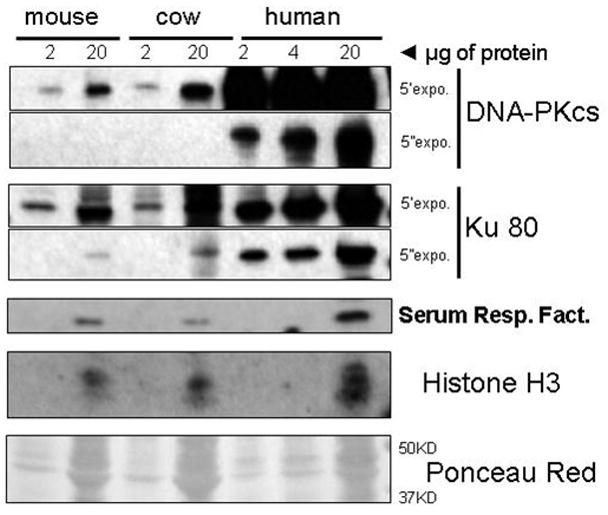
Nuclear extracts from mouse, cow, and human were probed for DNA-PKcs, Ku80, serum response factor (SRF) and Histone H3 using antibodies that recognize protein regions that are 100% conserved between these species. The maximum lifespan for these species are 4 years (mouse), 20 years (cow) and 90 years (human). For DNA-PKcs and Ku80 two different film exposures of 5 min (5′) and 5 sec (5′) are shown. The abundance of these proteins related with the capacity to bind DNA ends (Ku80 and DNA-PKcs) reflects the DNA binding capacity of the species. SRF, Histone H3 and the Ponceau Red staining of the membrane (shown only in the region between 37 and 50 kDa) are shown as loading controls.
Because the Western blot examination was limited to mouse, cow, and human proteins due to the limited availability of antibodies raised against a peptide sequence that is 100% conserved across multiple species, the differences in DNA-PKcs and Ku80 levels between cow and human cells was confirmed by mass spectrometry. The mass spectrometric analysis showed that while some abundant proteins such as Filamin A and HSP90 are present in similar amounts in both human and cow, DNA-PKcs, Ku80 and additionally Ku70 are expressed much more abundantly in human cells (Table 2). As with our Western blot analysis, these results are consistent with the differences in DNA-Pkcs kinase activity and Ku levels previously observed in human and mouse cells (Lees-Miller et al., 1992; Wang et al., 1993b). Because DNA-Pkcs and Ku play keys roles in DNA DSBs repair by non-homologous recombination (Lieber et al., 2003), we also examined the kinetics of repair of DNA DSBs induced by ionizing radiation in WI 38 human fibroblasts and the CHO Chinese hamster cell line, see Fig. 5A. Although the above results suggest that there are significantly greater amounts of Ku and DNA-Pkcs in human cells than rodent cells, the kinetics of repair in these two cells appear to be similar. To determine whether other proteins involved in NHEJ are differentially expressed in species with differing lifespans, we examined the levels of DNA ligase IV using an antibody raised against a peptide that is conserved between human and rodents. We find that human, hamster, and mouse cells express similar levels of DNA ligase IV (Figure 5B & C) suggesting that the species differences we observed in Ku and DNA-PKcs levels do not extend to all components of the NHEJ pathway.
Figure 5. Comparison of kinetics of DNA double strand break repair in CHO Chinese hamster and WI 38 human fibroblast cells and Western analysis of DNA ligase IV levels.
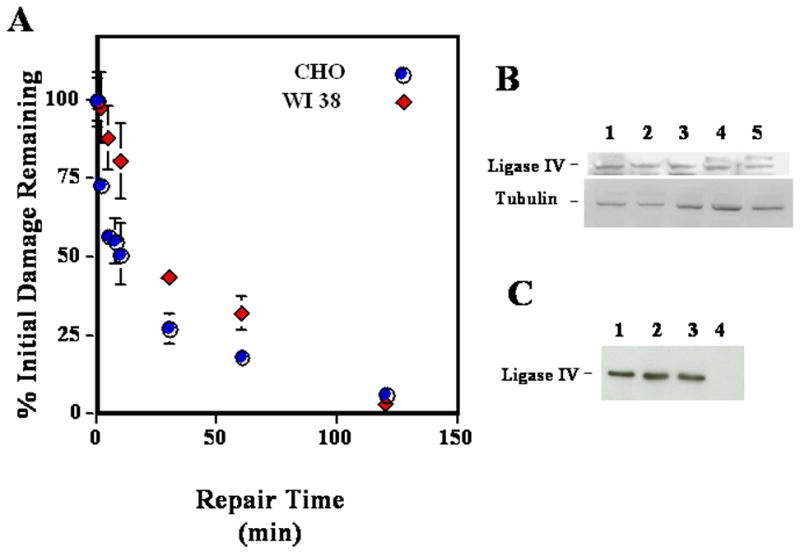
In panel A CHO Chinese hamster (blue circles) and WI-38 human fibroblast cells (red diamonds) growing exponentially in monolayers were irradiated with 20 or 15 Gy of γ-rays, respectively, incubated various times at 37°C in growth medium for repair, and analyzed by asymmetric field inversion gel electrophoresis (Denko et al., 1989; Stamato et al., 1993). Panel B contains a Western blot analysis of the levels of DNA ligase IV in total cellular extracts isolated from hamster (lanes 1 and 2), human fibroblasts (lane 3), LN229 human glioblastoma cells (lane 4), and primary mouse fibroblasts (lane 5). The levels of tubulin are shown as a loading control. Panel C contains a Western blot analysis for DNA ligase IV using nuclear protein extracts from human (lane 1) and hamster cells (lanes 2 and 3). Lane 4 contains extracts from XR1, a negative control hamster CHO-derived cell line that does not express DNA ligase IV (Bryans et al., 1999; Lee et al., 2003).
Telomere length and longevity
Since Ku80/70 has been shown to be part of the telomeric complex we also examined mean telomere length for its potential correlation with longevity. Telomere restriction fragment (TRF) length analysis was performed essentially as previously described (Steinert et al., 2002). We first used standard gel electrophoresis and quantified signals between 4 and 40 kb (Fig. 6a). Then we used pulse field gel electrophoresis (PFGE) to quantify the telomere length of the species with the longer telomeres (Fig. 6b). With PFGE we were able to quantify fragment length between 5 and 65 kb.
Figure 6. Telomere length across mammalian species with different longevity.
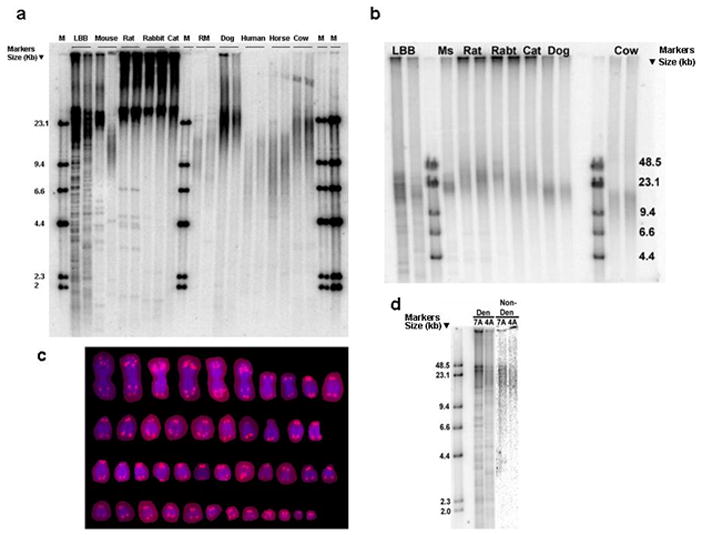
(a) Digested genomic DNA was resolved on a 0.5% agarose gel and probed with an end-labeled (CCCTTA)4 oligonucleotide. Species are ordered by increasing body mass (in grams). DNA marker lengths are shown in kilobases. The first line of the mouse is derived from a wild caught mouse from Pennsylvania, the second line is derived from a wild caught mouse from Idaho. RM = Rhesus monkey. M = DNA ladder. (b) Pulse field gel electrophoresis was used to resolve the telomeres that were too long to be measured in (a), above. Ms = Pennsylvania wild caught mouse, Rabt = rabbit. (c) Fluorescence in situ hybridization (FISH) with a Cy3-conjugated peptide nucleic acid probe (CCCTAA)3 showing that little brown bat telomeres contain internal telomeric repeats. (d) Little brown bat (LBB) telomeres were probed under denaturing conditions (left) or non-denaturing condition (right), only the telomere signals that were detected under non-denaturing conditions were used to estimate mean telomere length. For the majority of the lines the population doublings of the culture were 25 and cells had not immortalized. Mouse and rat cells had spontaneously immortalized.
The presence of multiple discrete short bands in electrophoretically separated little brown bat (LBB) genomic DNA that hybridized to the telomere probe in Fig. 6a, led us to hypothesize the presence of intra-chromosomal telomeric repeats. This was confirmed by the appearance of several interstitial regions on metaphase spreads that hybridized to a telomere repeat probe, particularly within the longer chromosomes (Fig. 6c). LBB telomeres were probed under denaturing conditions (Fig. 6d, left) or non-denaturing conditions (Fig. 6d, right). Only the telomeric 3′ single strand overhang is detected under non-denaturing conditions, and thus the longer signals appear to correspond to true telomeres, while the shorter signals seen only under denaturing conditions likely correspond to interstitial repeats. For LBB, then, only the telomere signals that were detected under non-denaturing conditions were used to estimate mean telomere length.
With logarithmic data transformation, species mean telomere length appeared to decrease with increasing longevity. However, this negative correlation did not reach statistical significance (Pearson r= −0.622, p=0.0547, n=10, Fig 7a). In addition when the influence of body mass was removed with partial correlation, we also found a negative correlation (still not significant r=−0.453, two tailed p=0.2221). We also observed a negative trend between telomere length and adult body mass, although the correlation was never significant, before (Fig. 7b) or after the removal of longevity by partial correlation. Additionally, among the nine available phylogenetically independent comparisons (Fig. 8a) telomere length differed in the same direction as maximum longevity in 5 instances and in 3 instances with body mass (Fig. 8b); these associations were both not significant (exact binomial test). Thus, of the two parameters we examined, DNA end binding and mean telomere length, only DNA end binding showed a positive correlation with species longevity that is both highly significant and supported by phylogenetic analysis.
Figure 7. Correlations of average telomere length to maximum longevity and adult body mass in mammals.
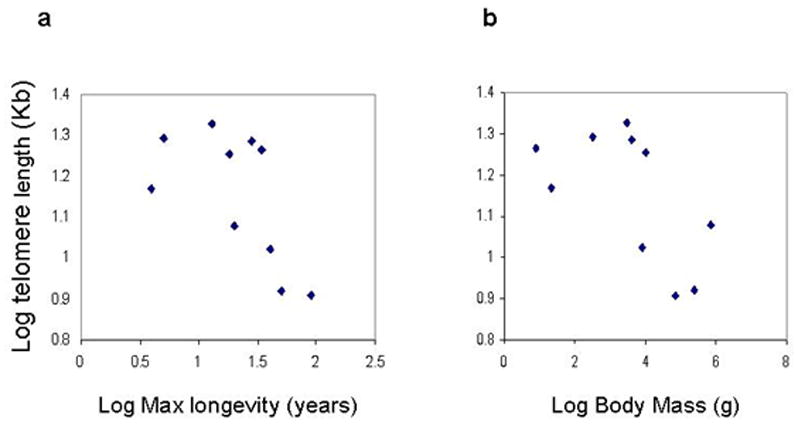
(a) Log average telomere length versus Log maximum longevity. (b) Log average telomere length versus Log adult body mass. All the determinations were from cultured adult skin fibroblasts. The species analyzed here are shown in Fig. 6 and 8.
Figure 8. Co-variation of longevity and body mass with telomere length across the available independent comparisons.
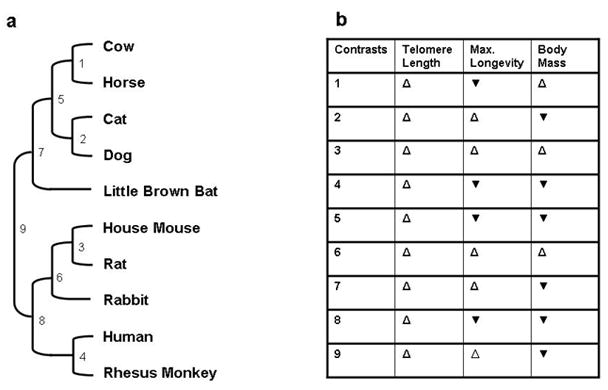
(a) Phylogeny for the relationships between telomere length, maximum longevity and body mass. Branch lengths are not drawn to scale. (b): Direction of variations in telomere length, maximum longevity and body mass among the available independent comparisons. Open, upward-pointed triangles indicate that the two parameters vary in the same direction, while filled, downward-pointing triangles indicate that the parameters vary in opposite directions.
Discussion
We have observed a novel exponential relationship between the capacity of nuclear proteins to bind DNA ends and the longevity of mammalian species. This activity increases 100 fold from mouse to man and appears to be correlated with increased levels of Ku and DNA-PKcs. There are a number of possible interpretations for these results:
Improved DNA repair
The most obvious interpretation of these data is that higher levels of these proteins in long-lived species provides them with increased capacity for DNA repair. This interpretation is consistent with reports showing that DSBs (Herbig et al., 2006) and mutations (Martin et al., 1996) accumulate exponentially during aging, and with the fact that humans are less sensitive to oxygen (Parrinello et al., 2003) and that they are estimated to be 1×105 fold more tumor resistant than mice (Austad, 1997). However, when the kinetics of DSB repair in WI38 human fibroblasts and Chinese hamster cells were compared, no significant differences were observed (Figure 5). These results are consistent with published observations on the rates of DSB repair in rodent and human cells as measured by neutral filter elution, comet assay, or other pulsed field gel electrophoresis approaches (Evans et al., 1993; Flentje et al., 1993; Gauter et al., 2002; Olive et al., 1994). It could be argued that human Ku and DNA-PKcs proteins are about 100 times less effective in repairing DSBs than the mouse proteins, and thus higher levels are required in humans to support equal levels of non-homologous end-joining (NHEJ) repair. If this is a tenable explanation, we would expect that the specific protein kinase activity of human DNA-PK (Ku80/70, DNA-PKcs) would be a 100 fold less that rodent DNA-PK; however DNA-PK kinase activity in human and rodent cells is consistent with the level of the DNA-PKcs and Ku80/70 proteins in these cells (Finnie et al., 1995; Lees-Miller et al., 1992). In contrast, we find that human, hamster, and mouse cells express similar levels of DNA ligase IV (Figure 5B & C) which suggests that not all NHEJ factors are elevated in human cells and is consistent with the similar rates of DNA DSB repair in human and rodent cells. While one cannot rule out the possibility that there are subtle differences in DSB DNA repair that are not detected by the techniques used, it seems that the differences in levels of Ku and DNA-PKcs do not reflect differences in global DNA repair per se.
Improved control of cell cycle arrest and/or apoptosis
The above observations support the possibility that functions of Ku and DNA-PKcs other than those involved in NHEJ may be an important component of species-specific functions of Ku and DNA-PKcs. For example abundant levels of Ku80/70 and DNA-PKcs could allow for the rapid recruitment of other proteins to the DSB damage site (Mauldin et al., 2002). Thus, even if correct repair is not accomplished, a signal originating from the damage site could rapidly induce apoptosis and/or cell cycle arrest, thus eliminating damaged cells or preventing proliferation of potentially malignant ones. The recently demonstrated extension of lifespan in mice with additional copies of loci encoding the DNA damage response factors ARF and p53 is consistent with this idea (Matheu et al., 2007), as is the observation that foci of DNA damage persist in mice deficient for Ku (Busuttil et al., 2003). In addition to binding to double strand DNA ends, the Ku protein recognizes a variety of DNA structures including single strand DNA gaps and hairpin loops (Blier et al., 1993; Falzon et al., 1993; Tuteja et al., 1994). Like DNA ends, both of these DNA structures are able to activate DNA-PKcs (Dip et al., 2005; Morozov et al., 1994). Thus, it is possible that high levels of Ku and DNA-PKcs may be needed to detect errors in DNA replication by recognizing these abnormal DNA structures especially in repetitive DNA sequences. Activation of DNA-PK would signal cell cycle arrest and/or apoptosis in cases of harmful replication abnormalities.
Improved telomere maintenance
Another plausible explanation for the high levels of Ku and DNA-PKcs in long-lived mammals is that these proteins function in maintaining telomere stability rather than NHEJ repair. Data from a number of published articles support this explanation. First, targeted gene disruption of Ku80 and Ku70 in human Nalm-6 pre-B ALL cells was used to generate lines heterozygous for Ku deficiency alleles (Ku80+/− or Ku70+/−) (Fattah et al., 2008; Uegaki et al., 2006). The level of both Ku subunits was reduced approximately 50% in these cell lines, but no or minimal increases in x-ray sensitivity or reduction in growth rate was observed, inconsistent with a requirement for high levels of Ku to support NHEJ. Telomere length increased in one study and decreased slightly in the other, suggesting that the high level of telomerase in Nalm-6 cells (Uegaki et al., 2006) was able to compensate for loss of telomere capping by Ku. Second, targeted gene disruption was used to generate heterozygous deficiencies for Ku80, Ku70 or DNA-PKCS in HCT116 colon cancer cells (Fattah et al., 2008; Li et al., 2002; Ruis et al., 2008), which have lower levels of telomerase than Nalm-6 cells (Uegaki et al., 2006). The cells had 20–50% of normal protein levels, and had only a “slight” (less than two to three fold) increase in sensitivity to X-rays or etoposide. However, their telomeres shortened considerably, indicating that the lower levels of telomerase in these cells was not able to compensate for loss of telomere capping by the reduced levels of Ku. Very similar results were obtained by Jaco et al (Jaco et al., 2004), who used RNAi to decrease Ku80 levels in moderately telomerase-positive HeLa cells or in telomerase-negative ALT cell lines U2OS and Saos2. Importantly, the mild levels of genome instability observed by Li et al. (Li et al., 2002) and by Jaco et al. (Jaco et al., 2004) could be secondary to telomere defects, rather than NHEJ defects, as the authors suggested in these studies. These observations are consistent with our hypothesis that Ku proteins function in maintaining telomere stability in human cells while only a small fraction of the Ku proteins cells are necessary for repair of DNA DSBs.
In a recent publication Wang et al. (Wang et al., 2009) generated a human cell line containing a conditional null allele of the Ku80 gene. Upon induction of the homozygous knockout, they observed a dramatic loss of telomeres and cell death. This observation is consistent with our suggestion that increased levels of Ku and DNA-PKcs function in long-lived species by stabilizing (capping) telomere ends rather than by supporting a general DNA DSB break repair mechanism, and indeed the main conclusion of the authors was that higher levels of DNA-PK in humans are needed for telomere maintenance and not to support NHEJ. Furthermore, we note that the trend toward shorter telomeres among longer-lived mammalian species that we observed in our study, which is consistent with previously published studies (Argyle et al., 2003; Cherif et al., 2003; Kozik et al., 1998; McKevitt et al., 2003; Nasir et al., 2001; Steinert et al., 2002), together with higher levels of DNA-PK in longer-lived species and a key role for DNA-PK in telomere integrity raises the possibility that increased levels of DNA-PK might provide a more ideal mechanism for capping telomeres (employed in long-lived mammals) than simply having long telomeres (employed in short-lived organisms).
In conclusion our findings suggest that DNA end-recognition may be an important component of the mechanisms that cells use to maintain genome stability, and reinforce the idea that loss of genome stability contributes to the aging process. Together with other recent studies, they further suggest that the processes involved in genome stability related to aging are likely to be more complex than an enhancement in simple NHEJ kinetics or telomere length, but may be related to maintenance of telomere capping.
Acknowledgments
We thank Dr Anja Brunet-Rossinni and the Coriell Institute for Medical Research for providing bats skin samples and gorilla, Rhesus monkey and horse lines, respectively. We thank Dr. Edward J. Gracely of Drexel University for statistical support. We are also grateful to Ulana Prociuk for help with karyotyping. We also wish to acknowledge the support of the Lankenau Institute for Medical Research.
This work was supported by USPHS grants AG20955-03 (VJC), CA877144 (TS) and 5R01AG021521 (FBJ) and AG022443 (CS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 2003;26:409–20. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- Argyle D, Ellsmore V, Gault EA, Munro AF, Nasir L. Equine telomeres and telomerase in cellular immortalisation and ageing. Mech Ageing Dev. 2003;124:759–64. doi: 10.1016/s0047-6374(03)00104-0. [DOI] [PubMed] [Google Scholar]
- Austad SN. Why we age. John Wiley & Sons, Inc; New York, NY: 1997. pp. 140–145. [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–601. [PubMed] [Google Scholar]
- Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–8. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2003;2:287–94. doi: 10.1046/j.1474-9728.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- Cherif H, Tarry JL, Ozanne SE, Hales CN. Ageing and telomeres: a study into organ- and gender-specific telomere shortening. Nucleic Acids Res. 2003;31:1576–83. doi: 10.1093/nar/gkg208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofalo VJ, Charpentier R. A standard procedure for cultivating human diploid fibroblastlike cells to study cellular aging. Journal of Tissue Culture Methods. 1980;6:117–121. [Google Scholar]
- Denko N, Giaccia A, Peters B, Stamato TD. An asymmetric field inversion gel electrophoresis method for the separation of large DNA molecules. Anal Biochem. 1989;178:172–6. doi: 10.1016/0003-2697(89)90375-8. [DOI] [PubMed] [Google Scholar]
- Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dip R, Naegeli H. More than just strand breaks: the recognition of structural DNA discontinuities by DNA-dependent protein kinase catalytic subunit. Faseb J. 2005;19:704–15. doi: 10.1096/fj.04-3041rev. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–78. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- Evans HH, Ricanati M, Horng MF, Jiang Q, Mencl J, Olive P. DNA double-strand break rejoining deficiency in TK6 and other human B-lymphoblast cell lines. Radiat Res. 1993;134:307–15. [PubMed] [Google Scholar]
- Falzon M, Fewell JW, Kuff EL. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268:10546–52. [PubMed] [Google Scholar]
- Fattah KR, Ruis BL, Hendrickson EA. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair (Amst) 2008;7:762–74. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci U S A. 1995;92:320–4. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentje M, Asadpour B, Latz D, Weber KJ. Sensitivity of neutral filter elution but not PFGE can be modified by non-dsb chromatin damage. Int J Radiat Biol. 1993;63:715–24. doi: 10.1080/09553009314552111. [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Bennett AF, Rezende EL. Phylogenetic approaches in comparative physiology. J Exp Biol. 2005;208:3015–35. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- Gauter B, Zlobinskaya O, Weber KJ. Rejoining of radiation-induced DNA double-strand breaks: pulsed-field electrophoresis analysis of fragment size distributions after incubation for repair. Radiat Res. 2002;157:721–33. doi: 10.1667/0033-7587(2002)157[0721:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Getts RC, Stamato TD. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–8. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- Kozik A, Bradbury EM, Zalensky A. Increased telomere size in sperm cells of mammals with long terminal (TTAGGG)n arrays. Mol Reprod Dev. 1998;51:98–104. doi: 10.1002/(SICI)1098-2795(199809)51:1<98::AID-MRD12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Lee SE, Pellicioli A, Vaze MB, Sugawara N, Malkova A, Foiani M, Haber JE. Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break. Mol Cell Biol. 2003;23:8913–23. doi: 10.1128/MCB.23.23.8913-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–9. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci U S A. 2002;99:832–7. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 2005;126:1130–3. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Marangoni E, Le Romancer M, Foray N, Muller C, Douc-Rasy S, Vaganay S, Abdulkarim B, Barrois M, Calsou P, Bernier J, Salles B, Bourhis J. Transfer of Ku86 RNA antisense decreases the radioresistance of human fibroblasts. Cancer Gene Ther. 2000;7:339–46. doi: 10.1038/sj.cgt.7700111. [DOI] [PubMed] [Google Scholar]
- Martin GM, Ogburn CE, Colgin LM, Gown AM, Edland SD, Monnat RJ., Jr Somatic mutations are frequent and increase with age in human kidney epithelial cells. Hum Mol Genet. 1996;5:215–21. doi: 10.1093/hmg/5.2.215. [DOI] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–9. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Mauldin SK, Getts RC, Liu W, Stamato TD. DNA-PK-dependent binding of DNA ends to plasmids containing nuclear matrix attachment region DNA sequences: evidence for assembly of a repair complex. Nucleic Acids Res. 2002;30:4075–4087. doi: 10.1093/nar/gkf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKevitt TP, Nasir L, Wallis CV, Argyle DJ. A cohort study of telomere and telomerase biology in cats. Am J Vet Res. 2003;64:1496–9. doi: 10.2460/ajvr.2003.64.1496. [DOI] [PubMed] [Google Scholar]
- Morozov VE, Falzon M, Anderson CW, Kuff EL. DNA-dependent protein kinase is activated by nicks and larger single-stranded gaps. J Biol Chem. 1994;269:16684–16688. [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–51. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Nasir L, Devlin P, McKevitt T, Rutteman G, Argyle DJ. Telomere lengths and telomerase activity in dog tissues: a potential model system to study human telomere and telomerase biology. Neoplasia. 2001;3:351–9. doi: 10.1038/sj.neo.7900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL, Banath JP, MacPhail HS. Lack of a correlation between radiosensitivity and DNA double-strand break induction or rejoining in six human tumor cell lines. Cancer Res. 1994;54:3939–46. [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrasts (CAIC): an Apple Macintosh application for analysing comparative data. Comput Appl Biosci. 1995;11:247–51. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Rohme D. Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc Natl Acad Sci U S A. 1981;78:5009–13. doi: 10.1073/pnas.78.8.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–95. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan--two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–75. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Stamato T, Guerriero S, Denko N. Two methods for assaying DNA double-strand break repair in mammalian cells by asymmetric field inversion gel electrophoresis. Radiat Res. 1993;133:60–6. [PubMed] [Google Scholar]
- Stamato TD, Denko N. Asymmetric field inversion gel electrophoresis: a new method for detecting DNA double-strand breaks in mammalian cells. Radiat Res. 1990;121:196–205. [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp Cell Res. 2002;272:146–52. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Tuteja R, Ochem A, Taneja P, Huang NW, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, et al. Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen. Embo J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uegaki K, Adachi N, So S, Iiizumi S, Koyama H. Heterozygous inactivation of human Ku70/Ku86 heterodimer does not affect cell growth, double-strand break repair, or genome integrity. DNA Repair (Amst) 2006;5:303–11. doi: 10.1016/j.dnarep.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Wang J, Chou CH, Blankson J, Satoh M, Knuth MW, Eisenberg RA, Pisetsky DS, Reeves WH. Murine monoclonal antibodies specific for conserved and nonconserved antigenic determinants of the human and murine Ku autoantigens. Mol Biol Rep. 1993a;18:15–28. doi: 10.1007/BF01006891. [DOI] [PubMed] [Google Scholar]
- Wang J, Chou CH, Blankson J, Satoh M, Knuth MW, Eisenberg RA, Pisetsky DS, Reeves WH. Murine monoclonal antibodies specific for conserved and nonconserved antigenic determinants of the human and murine Ku autoantigens. Mol Biol Rep. 1993b;18:15–28. doi: 10.1007/BF01006891. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–5. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]


