Abstract
We performed experiments to characterize the inducibility of nucleotide excision repair (NER) in Caenorhabditis elegans, and to examine global gene expression in NER-deficient and -proficient strains as well as germline vs. somatic tissues, with and without genotoxic stress. We also carried out experiments to elucidate the importance of NER in the adult life of C. elegans under genotoxin-stressed and control conditions. Adult lifespan was not detectably different between wild-type and NER-deficient xpa-1 nematodes under control conditions. However, exposure to 6 J/m2/day of ultraviolet C radiation (UVC) decreased lifespan in xpa-1 nematodes more than a dose of 100 J/m2/day in wild-type. Similar differential sensitivities were observed for adult size and feeding. Remarkably, global gene expression was nearly identical in young adult wild-type and xpa-1 nematodes, both in control conditions and three hours after exposure to 50 J/m2 UVC. Neither NER genes nor protein activity were detectably inducible in young adults that lacked germ cells and developing embryos (glp-1 strain). However, expression levels of dozens of NER and other DNA damage response genes were much (5 to 30-fold) lower in adults lacking germ cells and developing embryos, suggesting that somatic and post-mitotic cells have a much lower DNA repair ability. Finally, we describe a refinement of our DNA damage assay that allows damage measurement in single nematodes.
Keywords: Caenorhabditis elegans, DNA damage response, nucleotide excision repair, global gene expression, gene-environment interaction
1. Introduction
Nucleotide excision repair (NER) is responsible for the removal of a wide range of DNA helix-distorting damage, including lesions caused by ultraviolet radiation, important environmental contaminants such as polycyclic aromatic hydrocarbons and mycotoxins, as well as some types of cisplatin-induced and oxidative damage [1,2]. The importance of this pathway is illustrated by the human disorder xeroderma pigmentosum (XP), which is caused by a mutation of one of several NER genes (XPA to XPG) that leads to a markedly (~1000-fold) increased susceptibility to skin cancer and, in some cases, neurological effects [3,4]. Furthermore, NER is very highly conserved and observed in all free-living organisms studied in all three domains of life [5–7]. However, in shorter-lived organisms including mice and Caenorhabditis elegans, very few phenotypes associated with NER deficiency have been observed in unstressed organisms. We therefore chose to study in more detail the phenotype of NER-deficient C. elegans both in control and DNA-damaging environments.
C. elegans is a useful model for DNA damage and repair studies because it is a simple organism sharing many homologs with human DNA damage responsive genes [8–10]. Experiments using C. elegans exposed to ionizing radiation or UV have provided powerful insights into the apoptotic and checkpoint pathways [11–14]. Several studies have focused on C. elegans XPA-1, a homolog of the human XP group A protein [15]. Most researchers have reported few obvious phenotypes in xpa-1 mutants or RNAi-treated nematodes in the absence of stress, except for an increase in germline apoptosis in xpa-1 mutants compared to wild-type animals [14] and a small increase in mutation accumulation [16]. However, Hyun et al. [17] reported that the xpa-1 strain had a significantly shorter lifespan under control conditions, in contrast to previous observations [18,19]. After genotoxic stress, in contrast, the xpa-1 phenotype is marked, constituting a striking example of a gene-environment interaction. Following UV exposures, xpa-1 mutants display decreased embryonic survival [15], lack of inducibility of apoptosis in the germline [14], larval growth arrest, decreased transcriptional competence, DNA damage-induced mutation accumulation [19], and reduced adult lifespan [17]. Earlier work with a rad-3 mutant, recently identified as allelic to xpa-1 [19], also demonstrated ultraviolet C (UVC) sensitivity [20–22].
Previously, we described a quantitative polymerase chain reaction (QPCR)-based assay to quantify DNA damage in the nuclear or mitochondrial genomes of C. elegans [23]. Using the QPCR DNA damage assay, we observed that after exposure to UVC, xpa-1 mutant L1s (first larval stage nematodes) failed to repair nuclear DNA damage after 6 or 24 h. Abolition of repair of UVC damage was also observed in xpa-1 and rad-3 mutants using other assays [17,24]. However, we observed no detectable difference in DNA damage between xpa-1 and wild-type L1s in the absence of UV exposure. Inducibility of NER (or other DNA repair pathways) has not been characterized in C. elegans.
The goal of the current study was to study the expression and function of NER genes in C. elegans under control conditions, and to elucidate the importance of NER in genotoxin-stressed adult somatic tissues. We hypothesized that subtle phenotypic differences would be observed between xpa-1 and wild-type nematodes upon careful observation, and therefore examined lifespan, adult growth, and adult feeding, in the presence and absence of genotoxin exposure (single or daily doses of UVC). Since many previous studies have focused on the consequences of ionizing radiation and UV exposures on the germline of exposed animals (e.g., [11]), we focused here on the effects of UVC on adult C. elegans. All of these phenotypes were indistinguishable under control conditions, but greatly impaired after UVC. We further employed microarray analysis as a particularly powerful way to test for differences between N2 and xpa-1 nematodes, since it offers the ability to examine mRNA levels of most genes in the nematode genome. We hypothesized that basal global gene expression, or early DNA damage-responsive signaling-related gene expression, would be different in N2 and xpa-1 young adults under control conditions and 3 hours after exposure to 50 J/m2 UVC. Instead, global gene expression was highly similar between N2 and xpa-1 nematodes.
Finally, we carried out experiments to test the inducibility of NER in adult C. elegans, and to address the potentially different responses of germline and somatic cells to UVC damage. For these experiments, we used a glp-1 strain, which is a temperature-sensitive mutant deficient in germline proliferation at 25°C [25]. While we did not find evidence for inducibility of NER in C. elegans, the constitutive levels of expression of most DNA damage response genes were markedly higher in N2 and xpa-1 than the germline (and embryo)-deficient glp-1 young adults. This suggests that transcription of DNA repair genes falls off very dramatically after germ cell differentiation and early embryogenesis in C. elegans.
2. Materials and Methods
2.1. Nematode culture
All strains of C. elegans were obtained from the Caenorhabditis Genetic Center (Minneapolis, MN) and cultured as previously described [23]. Bristol N2 (wild-type) and RB684 (xpa-1) were maintained at 20°C; JK1107 (glp-1) was raised at 25°C to prevent germline proliferation [25]. C. elegans embryos were prepared as previously described to yield age-synchronized adult nematodes [26].
2.2. UVC exposures
UV radiation exposures were performed in a CL-1000 Ultraviolet Crosslinker (UVP, Upland, CA, USA) with an emission peak at 254 nm (referred to in this manuscript as 'UVC') as previously described [23]. After exposures, nematodes were rinsed from agar plates and either frozen for QPCR-based DNA damage assays or measured for growth or feeding as described below. Single UVC exposures were administered once, on day 0. Chronic UVC exposures were administered every 24h, beginning on day 0. Sampling of nematodes for growth, feeding, and QPCR receiving chronic UVC exposures was performed every 24h with all remaining nematodes then receiving the next UVC exposure. Nematodes were frozen by dripping pelleted nematodes suspended in about 500 µl K-medium [27] into liquid nitrogen; frozen pellets were stored at −80°C.
2.3. Lifespan assay
Survival assays were performed at 20°C for N2 and xpa-1 adult lifespans and at 25°C for glp-1 lifespans. One day post-L4 molt was counted as "day 0”. A total of 8–10 replicate plates per treatment group were monitored; four separate (in time) experiments were carried out. Plates had between 26 and 42 non-censored nematodes each. Censored nematodes were those that crawled onto the sides of the petri dishes and dessicated, were lost during transfer, or disappeared but were not observed to be dead. Nematodes were transferred by washing on a daily basis during the first 7–8 days and every 1–3 days thereafter.
2.4. Growth assay
Adult nematodes were rinsed from plates, allowed to settle, and the pellet was transferred to the sample cup of a COPAS Biosort [28] (Union Biometrica Inc., Somerville, MA, USA) and diluted to approximately 1 nematode/µL. The COPAS Biosort was used as previously described to measure the optical extinction of individual nematodes (EXT), which takes into account nematode length as well as optical density (width and opacity) [29,30]. In this manuscript, we use the generic terms “size” to refer to optical extinction and “growth” to refer to age-related increases in EXT. After the nematodes were measured, they were dispensed onto fresh E. coli-seeded agar plates and incubated at 20° C until the following day.
2.5. Feeding assay
Feeding assays were modified from Boyd et al. [31]. Nematodes were transferred to the sample cup of the COPAS Biosort and diluted to approximately 1 nematode/µL. Twenty-five adults were then dispensed into each well of a 96-well plate, containing a mixture of K-medium and OP50 E. coli at a final volume of 50µL. After 1–4 h, 5 µL aliquots of Fluoresbrite® polychromatic red microspheres (0.5 µm diameter; diluted 1:20 in K-medium) (Polysciences, Inc., Warrington, PA) were pipetted into each well and plates were rotated on a nutator mixer for 5 min. The nematodes were allowed to ingest the microspheres for 10 additional min (15 min total) and were then anesthetized using 5 µL of sodium azide (10 mM, final concentration), inhibiting further bead ingestion. Nematodes were aspirated from each well with the COPAS Biosort ReFLx; the level of red fluorescence for individual C. elegans were measured and referred to as “feeding”.
2.6. Analysis of growth and feeding data
Dose responses for xpa-1 and N2 were plotted together for single and chronic UVC exposures and at all time points. The dose responses were significantly different (p < 0.05) as indicated by non-overlapping standard error bars (Fig. 3 and Fig.4). The mean responses +/− standard errors for starved wild-type nematodes were included for reference.
Figure 3. NER is required for normal growth in UVC-exposed adult C. elegans.
Wild-type (left) and xpa-1 (right) adult growth after single (upper) or chronic (lower) exposures to 0, 6, 12, 25, 50, or 100 J/m2 UVC irradiation. Mean size measurements (optical extinction) ± standard error of the mean for one representative experiment is presented. Optical extinction measurements of individual nematodes were performed on a COPAS Biosort.
Figure 4. NER is required for normal feeding in UVC-exposed adult C. elegans.
Wild-type (left) and xpa-1 (right) adult feeding after single (upper) or chronic (lower) exposures to 0, 6, 12, 25, 50, or 100 J/m2 UVC irradiation. Mean feeding measurements (red fluorescence) ± standard error of the mean for one representative experiment is presented. Red fluorescence measurements of individual nematodes were performed on a COPAS Biosort.
2.7. Quantitative PCR (QPCR)
Previously, we described the adaptation of a QPCR-based DNA damage assay [32,33] to C. elegans [23]. This assay is based on the ability of DNA damage to block the progression of the DNA polymerase used for PCR amplification. Thus, damaged samples are amplified less well than control samples when the assay is carried out using quantitative conditions. The degree of inhibition of amplification of large (~10 kb) genomic fragments is used to quantify the number of lesions. Small (~200 bp) fragments are also amplified from the mitochondrial and nuclear genomes to permit, respectively, normalization to mitochondrial copy number and verification of equal starting amount of nuclear DNA.
QPCR conditions were the same for these experiments as previously described [23], with one change and one major improvement. First, we amplified only one large nuclear target: the amplicon including most of the unc-2 gene [23]. Second, rather than extract genomic DNA from batches of several thousand pelleted and frozen nematodes as previously described, we carried out the PCR reactions on digests of a small number of individual nematodes. This procedure is an adaptation of a commonly used approach to PCR genotyping of C. elegans, and allows measurement of damage in a single nematode of any lifestage, corresponding to ~500–1000 cells. Briefly, adult nematodes in a ratio of 1 per 15 µl (or 1 per 10 µl in the case of glp-1 adults, which have significantly less DNA) were added by picking to lysis buffer (1× PCR buffer with proteinase K: using the rTth XL DNA polymerase (XL PCR) kit from Applied Biosystems, add 300 µl of 3.3× PCR buffer, 50 µl of Proteinase K (20 mg/ml; Qiagen), to 650 µl of dH2O). We experimented with adding detergents (per [34]) but found that they gave variable results and did not in general improve amplification. We also tried adding chitinase, but found that inclusion of chitinase in the lysis buffer nearly completely inhibited the PCR reaction. The nematodes in lysis buffer were immediately frozen and stored at −80°. Prior to QPCR, samples were processed as follows: heat to 65°C for 5 minutes; vortex at high speed for 4–5 seconds; heat for an additional 55 minutes at 65°C; heat to 95°C for 15 minutes to deactivate the Proteinase K; and finally briefly vortex and cool to 4°C. Samples can be stored at least several months at −80°C. At least two QPCR reactions with satisfactory 50% quality controls [33] were carried out per sample. In addition to permitting measurement of damage in a single nematode, this version of the QPCR assay reduces time (~2 h digestion vs. 1+-day extraction of DNA) and expense.
2.8. Analysis of DNA damage
All DNA damage assays were performed on 3–4 tubes of nematodes (3–6 nematodes/tube) picked from a single plate for each condition (dose, time point, presence/absence of pre-treatment). Values for the separate tubes were averaged to give a single value for each plate, which was considered an “n” of 1. In all cases the total “n” was between 4 and 8 (i.e., 4–8 plates obtained in experiments conducted at different times). The DNA lesion frequency data were compared by analysis of variance followed, when appropriate, by Fisher's Protected Least Significant Difference post hoc analysis.
2.9. Microarrays
Young adult (24 h post-vulval crescent L4 stage) N2, xpa-1, and glp-1 nematodes were reared at 25°C and exposed to 50 J/m2 UVC radiation on agar plates without food as described [23], placed on OP50-seeded plates for 2 h 45 minutes, washed off plates and rinsed with K-medium 3 times (~5 minutes each) by allowing settling, resuspending, settling again, etc. Nematodes were frozen by dripping 3,000–5,000 pelleted nematodes suspended in about 500 µl K-medium into liquid nitrogen, and stored at −80°C. The pellets were ground into fine powder with a liquid nitrogen-cooled mortar and pestle [35] and RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA, USA). RNA was quantified with a NanoDrop Fluorospectrometer (NanoDrop Technologies, Wilmington, DE, USA) and analyzed for integrity with a BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). These exposures were carried out 4 times, twice in 2007 and twice in 2008, for a total “n” of 4.
Gene expression analysis was conducted using Affymetrix /C. elegans/ GeneChip® arrays (Affymetrix, Santa Clara, CA). For the 2007 samples, 500 pg of total RNA was used to produce biotin-labeled cRNA with the Epicentre Biotechnologies TargetAmp 2-Round aRNA Amplification Kit 2.0 according to manufacturer's protocol, with the exception that 10 µl biotin-UTP was incorporated in place of 3.6 µl unlabeled UTP, and final product was eluted in 25 µl water. For the 2008 samples, 100 ng of total RNA was amplified as directed in the Affymetrix Two-Cycle cDNA Synthesis protocol. 15 µg (Epicentre) or 10 µg (Affymetrix Two-Cycle) of amplified cRNAs were fragmented and hybridized to the array for 16 h at 45°C in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Slides were stained and washed as indicated in the Antibody Amplification Stain for Eukaryotic Targets protocol using the Affymetrix Fluidics Station FS450. Arrays were then scanned with an Affymetrix Scanner 3000 and data was obtained using the GenechipÆ Operating Software (GCOS; Version 1.4.0.036).
2.10. Microarray data analysis
Data preprocessing, normalization, and error modeling was performed with Rosetta Resolver® (Gene Expression Analysis System version 6.0.0.0.311) after grouping within-year replicates. The resulting fold-changes and p-values were then used for GOMiner [36] and Cytoscape [37,38] analyses. For both analyses, we made the following comparisons: control vs UVC-exposed in each strain (three total); all pairwise strain comparisons under control conditions (three total); and all pairwise strain comparisons after UVC exposure (three total). The 2007 and 2008 data were not combined at the transcript level, as there was enough variation associated with the years that averaging them would have led to less rather than more statistical power. Rather, we carried out GO enrichment analysis separately for each year, and then compared the results at the level of altered GO biological processes. This approach has been shown to be an effective way of combining otherwise noisy microarray datasets [39]. Detailed descriptions of GOMiner and Cytoscape analyses are accessible as Supplemental methods. Hierarchical clustering, PCA analysis, and visualization were also carried out using GeneSpring versions 7.3 and GX10 (Agilent). Microarray data have been deposited in the National Center for Biotechnology Information's GEO and are accessible through GEO series accession number GSE15159.
3. Results
3.1. Lifespan, reproduction, and heat shock response do not detectably differ in xpa-1 and wild-type nematodes under control conditions
As described above, most other reports have indicated that xpa-1 nematodes appear to have fairly normal phenotypes under control conditions, and our own experience with this strain supports those observations. We carried out several experiments designed to examine the xpa-1 control phenotype in more detail. We measured reproduction, lifespan, heat shock response, and finally global gene expression (via microarray) in xpa-1 nematodes. Median lifespans of control wild-type (N2) and xpa-1 nematodes were not detectably different (Fig. 1; Suppl. Table 1). We also compared the number of offspring from N2 and xpa-1 nematodes, and observed no difference (Suppl. Fig. 1). Sublethal (40°C for 30 minutes) heat shock did not cause detectable DNA damage either immediately or 6 hours after exposure in either N2 or xpa-1, and caused a similar level of developmental delay in both strains (Suppl. Fig. 2).
Figure 1. NER is required for normal lifespan of UVC-exposed adult C. elegans.
Wild-type (red) and xpa-1 (blue) adult lifespans after chronic (daily) exposures to 0, 6, 12, 25, 50, or 100 J/m2 UVC irradiation. The average of 2 replicate experiments is presented.
3.2. NER is required for normal lifespan of UVC-exposed adult C. elegans
Lifespan was determined for adult C. elegans exposed daily to 6, 12, 25, 50, or 100 J/m2. Daily exposures were carried out to mimic the chronic exposures to genotoxins that organisms often experience outside of the laboratory environment. Although UVC does not penetrate the earth’s atmosphere, it is a convenient experimental tool that generates nuclear DNA damage that is repaired by the same pathway (NER) as important environmental genotoxins such as polycyclic aromatic hydrocarbons and mycotoxins. All UVC treatments greatly reduced xpa-1 lifespan while only 50 and 100 J/m2 reduced wild-type survival (Fig. 1). Exposure to 6 J/m2 UVC resulted in a greater decrease in lifespan in xpa-1 nematodes than did exposure to 100 J/m2 in wild-type nematodes (Suppl. Table 1).
While conducting lifespan experiments, a number of phenotypes were noted in xpa-1 nematodes (Fig. 2). Unexposed xpa-1 nematodes appeared to be slightly darker and longer than wild-type controls. In contrast, UVC-exposed xpa-1 nematodes exhibited a starved phenotype; they were shorter, thinner, and lighter than unexposed xpa-1 or wild-type nematodes at the same UVC doses. After 3 days of 25 J/m2 UVC, xpa-1 nematodes were observed to also have damaged intestinal and pharyngeal structure along with fluid-filled vacuoles and few, if any, embryos present in the gonad (Fig. 2, xpa-1 C25).
Figure 2. Images of phenotypes observed during lifespan studies.
Wild-type (upper panels) and xpa-1 (lower panels) nematodes after exposures to 0 or 25 J/m2 UVC irradiation either once on day 0 (single - S) or repeatedly for 3 days (chronic - C). Although untreated xpa-1 nematodes appeared slightly larger than wild-type, after 3 days of daily UVC exposure (C25), xpa-1 nematodes had a starved appearance and lack of embryos in the gonad. All images were taken at 100x magnification.
3.3. NER is required for normal growth and feeding of UVC-exposed adult C. elegans
The fact that UVC-exposed xpa-1 nematodes appeared to be smaller than wild-type might suggest that a reduction in feeding would be related to the reduction in lifespan. To confirm the growth differential, we quantified nematode size using a COPAS Biosort to measure optical extinction (EXT) as proxy for adult size (see Methods). Adult wild-type and xpa-1 nematodes were exposed to 0, 6, 12, 25, 50, and 100 J/m2 UVC either once (single UVC exposure) or daily (chronic UVC exposures). Wild-type nematodes were only slightly affected by 6 and 100 J/m2 doses of single UVC exposures, while all chronic exposures led to greater but similar decreases in EXT (Fig. 3, left panels). On the other hand, adult xpa-1 size was decreased in a dose-dependent fashion with all UVC doses leading to lower mean EXT values than controls after single or chronic UVC exposures (Fig. 3, right panels). Because UVC-exposed xpa-1 nematodes appeared starved and actually decreased in size throughout the growth experiment, feeding was also quantified using a Biosort. For feeding, nematodes were exposed to UVC on agar plates and red fluorescent microspheres were used to measure feeding rates (see Methods). The red fluorescence of individual nematodes was then measured with the Biosort and used to estimate feeding levels. Feeding levels in both strains varied between individuals and across experimental days, a trend that agreed with previous measurements of feeding [31]. General decreases in feeding levels with time, but no clear dose effects, were observed in wild-type nematodes (Fig. 4, left panels). Feeding levels were greatly affected in xpa-1 nematodes exposed to single doses greater than 12 J/m2 or any chronic doses of UVC (Fig. 4, right panels). Although some UVC-exposed xpa-1 nematodes ingested less red fluorescent microspheres than those starved over the course of the experiment, all fed nematodes, regardless of strain or UVC dose, were larger than unfed nematodes (Fig. 3). In fact, all unfed nematodes were dead by day 4 of the experiment. From these results, it seems unlikely that decreased growth after UV exposures could be entirely attributed to a lack of feeding.
3.4. DNA damage accumulation in C. elegans with chronic UVC exposures
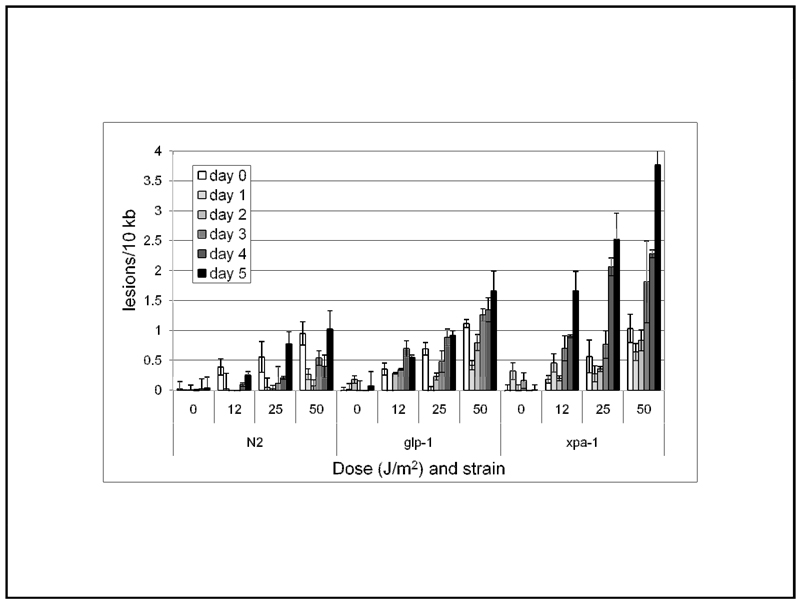
To compare the genotoxin-induced xpa-1 phenotypes that we observed with levels of DNA damage, we dosed adult C. elegans with UVC either once or daily, as described above. We sampled the exposed nematodes to determine the levels of DNA damage that resulted. 1-day old adult C. elegans were exposed to UVC and then frozen for QPCR immediately on day 0 or after 24 h of recovery. After freezing a portion of the nematodes, the remainder then received a daily UVC exposure. Thus, damage was measured immediately after the first exposure on day one, and immediately prior to subsequent exposures on subsequent days. A dose-dependent increase in lesion frequency was observed from 12–50 J/m2 for all strains: wild-type, glp-1 (a germline-deficient mutant strain used to eliminate the confounding effect of cell division; [23]), and xpa-1 (Fig. 5). A similar level of DNA damage was observed in all strains at the same UVC dose on day 0. However, the lesion frequency after daily exposure to 50 J/m2 increased between days 1 and 5 most rapidly for xpa-1, followed by glp-1 mutants. Wild-type nematodes showed the least time-dependent increase in DNA damage. The effects of strain, dose, and day were all statistically significant, as were the strain by treatment and strain by day interactions (p < 0.0001 in all cases, 3-factor ANOVA). The difference in damage accumulation between wild-type and glp-1 strains must be attributable to the lack of germ cell replication-dependent dilution of the damage in the wild-type strain, since we did not detect any difference in UV photoproduct repair capacity in wild-type and glp-1 mutants (in starved L1 larvae; [23]). The time-dependent accumulation of damage in the glp-1 mutants is also consistent with the lack of complete repair after 24 h that we previously observed [23]. The relatively high rate of increase in lesion frequency observed in xpa-1 mutants with chronic UVC exposure is probably attributable both to lack of repair and a decrease in dilution of damage by cell division as egg production was diminished in UVC-dosed xpa-1 mutants (data not shown). However, the small decrease in average damage level observed in the xpa-1 mutants on day 1, and the initially slow increase in damage (days 2–3), is likely explained by dilution of the damage, which is no longer possible by days 4 and 5.
Figure 5. DNA damage accumulates in mutant C. elegans after chronic UVC exposures.
Adult nematodes were exposed daily to 0, 12, 25, or 50 J/m2, and frozen 24 h later (immediately prior to dosing of remaining nematodes) for QPCR-based DNA damage analysis. The first dose was at 24 h post-L4 molt (“day 0”). DNA damage measured at day 0 was in nematodes sampled immediately after exposure; on subsequent days, nematodes were sampled 24h after the previous exposure and immediately prior to the following dose. The effects of strain, dose, and day were all statistically significant, as were the strain by treatment and strain by day interactions (p < 0.0001 in all cases, 3-factor ANOVA). n = 4–8 per column; error bars represent standard error of the mean.
3.5. Global gene expression is very similar in wild-type and xpa-1 strains under control conditions
Microarray analysis of global gene expression is a powerful way to detect subtle differences between strains because it allows the comparison of transcript levels of nearly all genes in the genome. We carried out microarray analysis of global gene expression in wild-type and xpa-1 young adults under control conditions and 3 h after exposure to 50 J/m2 UVC. The time point of 3 h was chosen to allow us to detect differential signaling in response to the initial damage, as opposed to the differential signaling that would occur at later times due to the persistence of damage in the xpa-1 but not the wild-type strain. A large portion of the nuclear DNA damage caused by UVC is still present at 3 h post-exposure even in NER-proficient nematodes [23]. However, we observed very few differences between the wild-type and xpa-1 strains under either control or UVC-exposed conditions. Specifically, the only gene that was expressed at a level convincingly higher (~10-fold) in xpa-1 than wild-type under both control and UV conditions is K07G5.3 (data not shown), which is located immediately adjacent to xpa-1 on chromosome I. Other than the xpa-1 gene itself, we detected only three genes expressed at a level convincingly lower (<4-fold) in xpa-1 than wild-type under both control and UV conditions: Y48G8AL.11 (haf-6), and Y19D10A.9 (clec-209) and/or F56A4.2 (data not shown). Y19D10A.9 and F56A4.2 are both represented on the array by two probes that cross-hybridize making it impossible to tell if one or both is lower. Thus, even at the level of global gene expression, there are very few differences between wild-type and NER-deficient nematodes.
3.6. Global gene expression in young adult C. elegans is dramatically different in glp-1than wild-type and xpa-1 strains
We also compared global gene expression in wild-type and xpa-1 to glp-1 young adult nematodes under control conditions and 3 h after exposure to 50 J/m2 UVC. The glp-1 strain was included to allow us to contrast the transcriptomic response in germ cell-deficient versus germ cell-bearing animals. Preliminary histological analysis, carried out to examine overall cell appearance and cell integrity in wild-type and xpa-1 nematodes 3 h post-exposure, did not detect any obvious signs of necrotic cell death (cellular destruction/lysis, cell swelling, excessive vacuolization, or fragmented organelles; data not shown). The microarray experiment was carried out twice in 2007, and twice in 2008, and different amplification and labeling protocols were used for the 2007 and 2008 samples. We refer to differences between the 2007 and 2008 experiments as “year” throughout this manuscript. The effect of “year” was noticeable in our gene expression results (e.g., Fig. 6). This is not surprising since such protocol differences are known to introduce some variability [39], and has the advantage of making it very likely that the gene expression differences that we observed are real.
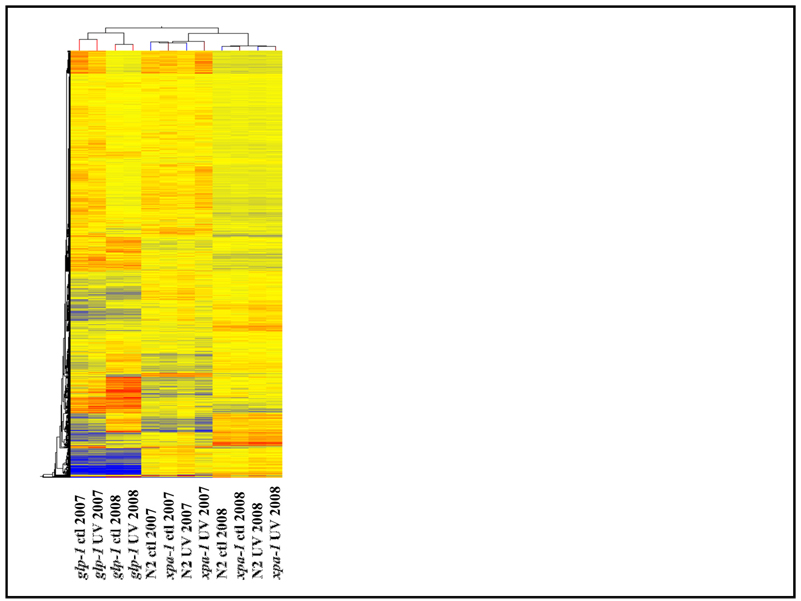
Figure 6. Global gene expression is comparable in the N2 and xpa-1 strains, but dramatically different in the glp-1 strain.
The glp-1 samples cluster completely separately from the N2 and xpa-1 samples. The effect of year (and amplification/labeling protocol) is greater than the effect of UVC exposure within the glp-1 samples, and greater than either UVC exposure or strain for the N2 and xpa-1 strains. UVC exposure affects global gene expression more than strain for the N2 and xpa-1 strains. This heat map was generated using GeneSpring; hierarchical clustering was performed on conditions and genes (including all genes that passed basic quality filtering).
As stated above, global gene expression was similar in the wild-type and xpa-1 strains, both with and without UVC exposure. Hierarchical clustering and principal components analysis both showed that the strain differences between wild-type and xpa-1 were less important than UV treatment in distinguishing global gene expression patterns (Fig. 6 and Suppl. Fig. 3). The similarity of these two strains is also evident in Supplemental Table 2, which highlights the small number of genes differentially expressed when comparing wild-type and xpa-1 samples in either control or UVC-exposed conditions. In contrast, global gene expression was highly divergent in the glp-1 strain from either wild-type or xpa-1, in both conditions (Fig. 6 and Suppl. Fig. 3).
3.7. Constitutive transcriptomic differences between glp-1 and wild-type or xpa-1
A full discussion of the extensive constitutive strain differences between glp-1 and other strains, which presumably largely reflect the differences between germ cells/early embryos and somatic cells, is beyond the scope of this manuscript. However, a subset of the differences is pertinent to the topic of DNA damage response. In keeping with our previous finding of much higher expression of many DNA repair genes in embryos vs. adults [23], the levels of expression of the great majority (~2/3) of known DNA repair-related genes in wild-type and xpa-1 gravid young adults were much higher than in glp-1 young adults, which under the conditions of this experiment lacked germ cells (Fig. 7 and Suppl. data file 1). These included NER genes as well as genes from all other major DNA repair pathways (base excision repair, double-strand break repair, mismatch repair) and other DNA damage responses (e.g., signaling and translesion synthesis). Nearly all of these genes were expressed at least 2-fold higher in wild-type and xpa-1 than glp-1, and many were at 5–30-fold higher in wild-type and xpa-1. The three exceptions to this trend were akt-2, pme-5, and gei-17, which were ~1.5→3-fold higher in glp-1 young adults than in wild-type or xpa-1. The actual fold differences between somatic and germ/embryonic cells must be larger than these, since our “germ cell” samples (wild-type and xpa-1 vs glp-1 young adults) are diluted by the presence of somatic cells.
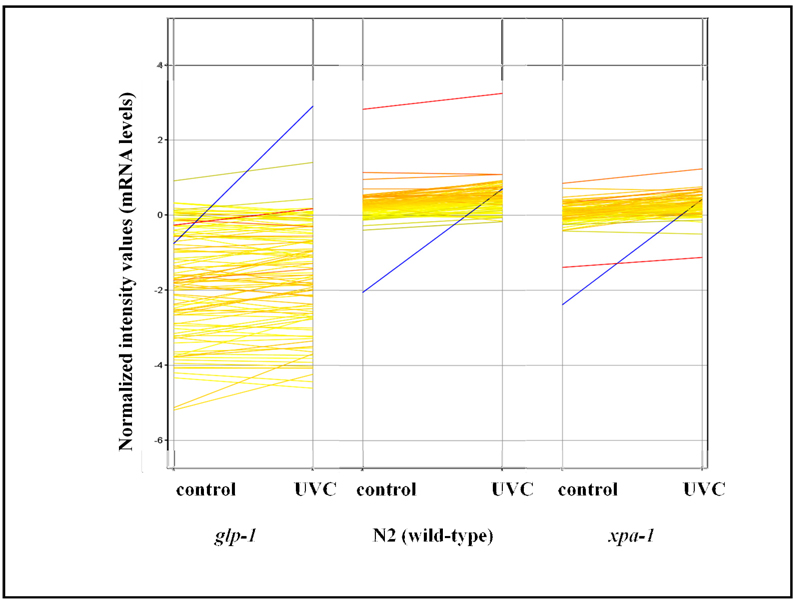
Figure 7. Expression of most DNA repair-related genes was not induced three hours after UVC exposure, and is in general low in germ cell-deficient nematodes.
Many DNA repair genes are expressed at a lower level in glp-1 than N2 and xpa-1 young adults (all at 25°C, so that glp-1 nematodes lack germ cells). The only significantly UVC-induced DNA repair-related gene was pme-4 (shown in blue). See Supplemental data files 1 and 10 for exact strain-to-strain and UVC-induced fold-change values.
Detailed GOMiner and Cytoscape (jActiveModules and BiNGO) analyses of strain-to-strain differences under both UV and control conditions can be found in Supplemental data files 2–5 (GOMiner) and 6–7 (BiNGO).
3.8. UVC induces specific changes in the transcriptomes of wild-type, xpa-1, and glp-1 young adults 3 h post-exposure
Our microarray analysis also allowed us to examine the transcriptomic response to UVC in wild-type, xpa-1, and glp-1 young adults. These responses were not identical, but did overlap significantly. Differentially expressed transcripts (defined as absolute fold change value > 1.3, a log ratio p-value < 0.05 by Rosetta Resolver, and a log(10) intensity measurement > −0.4) along with fold-change, p-values and GO annotation are listed for each strain and year in Supplemental data file 8. Hierarchical clustering and principal components analysis carried out using only differentially expressed transcripts again showed that the strain differences between wild-type and xpa-1 were minimal, and the glp-1 samples continued to cluster separately (Suppl. Fig. 4 and Suppl. Fig.5).
To identify with more confidence transcripts that are UVC-regulated in both somatic and germ cells, we identified transcripts that were UVC-regulated (as defined above) in both years in each strain (Suppl. data file 9), and then identified transcripts that were present in at least 2 of those 3 lists, and showed the same direction of change after UVC in all 3 strains. These UVC-regulated genes are presented in Table 1. Many of these genes are potentially associated with a stress response of some sort, including metabolism genes (a cytochrome P450 and several UDG glucuronosyl transferases), metal-responsive genes (cdr-4, numr-1), a p-glycoprotein, a fungus-responsive gene (cnc-4), a number of “downstream of daf” [40]genes (dod-22, dod-24, dod-17), two proteolysis-related genes (rpm-1, cul-6), and two DNA damage-responsive genes (pme-4 and crn-2).
Table 1. Transcripts regulated by UVC in all strains tested.
All transcripts listed were up- or down-regulated in the same direction in all 3 strains (N2, glp-1, xpa-1), and met our criteria for differential expression (absolute fold change value > 1.3, a log ratio p-value < 0.05, and a log(10) intensity measurement > −0.4) in at least 2 of the 3 strains.
| Gene | UVC regulation |
function/description |
|---|---|---|
| C01B7.6 (rpm-1) | down | E3 ubiquitin ligase |
| C05E11.5 (amt-4) | down | ammonium transporter protein |
| D1046.5 | down | Predicted membrane protein |
| F25E5.8 | down | uncharacterized |
| T28D6.3 | down | uncharacterized |
| Y51H4A.7 | down | Urocanate hydratase |
| C01B4.8 or Y19D10A.5 | up | Permease of th major facilitator superfamily |
| C08E3.6 (fbxa-163) | up | uncharacterized F-box-containing protein |
| C09H5.2 (catp-3) | up | Na+/K+ ATPase, alpha subunit |
| C09H5.5(str-144) | up | 7-transmembrane olfactory receptor |
| C10H11.6 (ugt-26) | up | UDP-glucuronosyl transferase |
| C12D8.11 (rop-1) | up | Uncharacterized |
| C18C4.3 (ugt-48) | up | UDP-glucuronosyl transferase |
| C24B9.9 (dod-3) | up | downstream of daf-16 |
| C47E12.1 (srs-2) | up | Seryl-tRNA synthetase |
| C49G7.10 | up | uncharacterized |
| CD4.2 (crn-2) | up | cell death-related nuclease |
| F08F3.7 (cyp-14A5) | up | cytochrome P450 |
| F08F8.5 (numr-1) | up | nuclear-localized metal-responsive gene |
| F55G11.5 (dod-22) | up | downstream of daf-16 |
| H23L24.5 (pme-4) | up | poly(ADP-ribose) glycohydrolase (PARG) |
| K01D12.11(cdr-4) | up | cadmium responsive gene |
| K02F3.4 (zip-2) | up | bZip transcription factor |
| K08E7.7 (cul-6) | up | cullin (ubiquitin ligase component) |
| K10D11.1 (dod-17) | up | downstream of daf-16 |
| R09B5.9 (cnc-4) | up | uncharacterized fungus-induced gene |
| T21E8.3 (pgp-8) | up | p-glycoprotein |
| Y38E10A.5 (clec-4) | up | c-type lectin |
| Y39G10AR.6 (ugt-31) | up | UDP-glucuronosyl transferase |
| Y45G5AM.3 | up | uncharacterized |
| Y49C4A.8 (ugt-29) | up | UDP-glucuronosyl transferase |
| Y58A7A.3 | up | uncharacterized |
| Y58A7A.4 | up | uncharacterized |
| Y58A7A.5 | up | uncharacterized |
We also searched for transcripts that were differentially regulated by UVC in different strains (all 3 pairwise comparisons for glp-1, wild-type, and xpa-1). Although some such genes could be identified based on our criteria for differential expression (Suppl. Table 2), closer inspection of these genes demonstrated that nearly all were in fact regulated in a similar fashion in all strains, and were only included in this table due to narrowly missing one of our criteria for differential expression in one of the strains. We detected only four genes that were qualitatively UVC-regulated in an apparently strain-specific fashion: T22E5.1 (uncharacterized) was down-regulated, and fipr-26 (F53B6.8, uncharacterized) and hoe-1 (E04A4.4, required for germline proliferation) were up-regulated, by UVC in glp-1 but not wild-type and xpa-1 young adults. Conversely, C31A11.5 (uncharacterized) was not upregulated in glp-1 but was between 3- and 10-fold upregulated in wild-type and xpa-1, respectively. We did not test for quantitatively different regulation (e.g., genes that met our statistical criteria for differential regulation in all strains, but for which the degree of up- or down-regulation was different).
3.9. Gene ontology (GO) biological processes altered 3 h post-UVC exposure
Since our list of UVC-regulated genes based on a minimal 1.3-fold change was relatively short (Table 1), we also carried out a jActiveModules network analysis. This algorithm can identify subnetworks highly enriched in regulated genes even if the fold-changes are not large, since it is based only on p-values [37]. We analyzed the resulting enriched subnetworks for GO enrichment, and identified the following biological processes as containing many genes that are coordinately UVC-regulated by a small amount (usually less than 2-fold), in all strains: ubiquitin-mediated proteolysis, glycolysis, transcription, translation, cell division, gamete generation, molting cycle, chromatin organization/DNA packaging and DNA-dependent DNA replication. As an example, a subnetwork enriched in UVC-regulated genes associated with cell cycle progression and gamete generation is shown in Supplemental Figure 6. In addition, genes involved in transcription, translation, cell division, gamete generation, chromatin organization/DNA packaging and DNA-dependent DNA replication were expressed at significantly (2→10-fold) lower levels in glp-1 than wild-type or xpa-1, while glycolysis genes were slightly higher in glp-1 nematodes. The genes in these processes that were consistently UVC-regulated were generally regulated similarly in all of the strains. DNA repair was identified as an altered biological process in the wild-type, but not xpa-1 or glp-1 dataset. However, close examination of DNA repair-related genes showed that only pme-4 was convincingly UVC-regulated in every strain (Fig. 7 and Suppl. data file 10). BiNGO GO enrichment analyses of jActiveModules results are available as Supplementary data files 11–13. GOMiner analysis of UVC-induced gene lists is not included because in no case did more than 4 differentially expressed genes (defined as fold change > 1.3, log ratio p-value < 0.05 by Rosetta Resolver, and log(10) intensity measurement > −0.4) map to a given GO biological process.
3.10. Inducible repair of total UVC damage was not detected in young adult C. elegans
Some DNA repair processes are inducible in some organisms (discussed below); it is not known whether NER or other DNA repair processes are inducible in C. elegans. As described above, our analysis of GO biological processes enriched in up-regulated transcripts after UVC did not detect a strong induction of DNA repair gene transcripts. To test more directly the hypothesis that NER would be inducible in C. elegans, we measured repair of UVC-induced DNA damage after pre-exposure to a low dose of UVC in young adult C. elegans. glp-1 nematodes were raised at 25°C to preclude germline cell proliferation and thus cell division-associated dilution of damaged DNA [23]. Young adults were exposed to 6 J/m2 UVC, and then allowed to recover for 18 h prior to a subsequent exposure to 0, 6, 12, 25, 50, or 100 J/m2. Subsets were then frozen at 0, 6, 12, 24, 48, and 72 h post-exposure in order to test the hypothesis that the pre-exposure to UVC would result in faster repair. We did not find strong evidence for inducibility of repair under these conditions (Fig. 8). The main effect of pre-treatment was not significant (p = 0.30). While there appeared to be a trend towards increased repair at 25 and 50 J/m2, we could not carry out specific post-hoc comparisons because neither the interaction of pretreatment with dose nor that of pretreatment with time point and dose were significant (p = 0.15 and 0.66, respectively) (3-factor ANOVA on dose, time point, and presence/absence of pretreatment).
Figure 8. Inducible repair of UVC-induced DNA damage was not detected in young adult C. elegans.
DNA lesions detected in glp-1 young adults either untreated or pre-treated with a single 6 J/m2 UVC exposure, and then 18 h later exposed to a challenge dose of 0, 6, 12, 25, 50, or 100 J/m2 as indicated. Nematodes were frozen at 0, 6, 12, 24, 48, and 72 h post-dosing. Note that the y-axes all indicate lesions/10kb but have different scales; the data are graphed to highlight relative rates of repair. The effects of dose, timepoint, and interactions of dose by timepoint and dose by pretreatment were all significant (p < 0.004 in all cases, 3-factor ANOVA); all other factors and interactions were not significant (p > 0.15 in all cases, 3-factor ANOVA). n = 4–8 per column; error bars represent standard error of the mean.
4. Discussion
There have been mixed reports on the effects of mutations in the xpa-1 gene on lifespan in C. elegans strains. Hyun et al. [17] recently reported a markedly shorter lifespan both for the ok698 deletion allele of xpa-1 (used in the current report) and for RNAi-based knockdown of XPA-1. On the other hand, previous reports using both xpa-1(ok698) and rad-3(mn157), a strain carrying an independently isolated deletion mutation of xpa-1 [19,20], detected no difference [18,19]. In these experiments, we detected no difference in lifespan under control conditions.
However, our results are in agreement with those of Hyun et al. [17] with respect to the dramatic decrease in lifespan in xpa-1 nematodes following adult UVC exposure. We further demonstrate that adult growth and feeding are exquisitely sensitive to UVC damage in the context of NER deficiency, and that these phenotypes are associated with a dramatic increase in DNA damage accumulation. Our lowest dose of UVC (6 J/m2), led to as much or more reduction in lifespan, growth, and feeding in xpa-1 nematodes as did our highest dose (100 J/m2) in wild-type animals. The lack of growth in adults could be due to decreased feeding resulting from a loss in transcriptional competence [19], or DNA damage-mediated inhibition of endoreduplication upon which adult growth is dependent [41]. We found that feeding was strongly inhibited in UVC-exposed xpa-1 young adults, although it cannot have been completely abolished since starved young adults do not survive past 3 days.
Even using microarray detection of global gene expression, we detected very few differences between the wild-type and xpa-1 strains. K07G5.3, which is located immediately adjacent to xpa-1 on chromosome I, was the gene that was most convincingly differentially expressed, and seems most likely to reflect dysregulation due to the xpa-1 mutation. Similarly, while there was a clear transcriptional response to UVC in all strains, there were also few differences between wild-type and xpa-1 nematodes in that response, suggesting that immediate damage-induced signaling is not dramatically altered in xpa-1 nematodes. We deliberately chose an early time point to avoid detecting transcriptomic differences resulting from the greater persistence of DNA damage in xpa-1 nematodes; a significant amount of UVC-induced DNA damage persists at three hours even in NER-proficient nematodes [23]. These results suggest that the dramatic gene-environment interactions observed in xpa-1 nematodes can be attributed to the persistence of DNA damage in this strain, rather than to differential signaling mediated by xpa-1 under control conditions. It is also possible, however, that the UVC-induced xpa-1 phenotypes result from altered signaling at a different time point, or non-transcriptionally-mediated signaling.
Our results show that not just lifespan, but adult growth, feeding, heat shock resistance, egg production, and even global gene expression in xpa-1 nematodes are nearly identical to wild-type under control conditions. Thus, the level of background DNA damage that occurs in nematodes grown under our laboratory conditions is low enough that no phenotype could be detected using these measures. This suggests that the selective pressure that maintains NER in this short-lived organism must result from genotoxic stressors not encountered under controlled laboratory conditions, or require multiple generations to be evident.
The inducibility of DNA repair is important to characterize in C. elegans given the increasing use of this species for studies of DNA damage and repair. NER is inducible in bacteria and yeast [42–47], but inducibility of NER proteins or repair kinetics appears to be less robust and possibly species-specific in higher eukaryotes [48,49]. This raises the important question of whether C. elegans, a simple eukaryote, is more like the very simple eukaryote yeast, or more like the higher eukaryotes for which it is often used as a model, with respect to DNA damage inducibility. We detected the induction of very few DNA repair genes, and in fact very few DNA damage-responsive genes in general three hours post-UVC exposure, although DNA repair genes are induced by 1–3h in yeast [50,51]. This lack of induction of DNA repair genes was observed both in germ cell- and embryo-containing young adults (wild-type and xpa-1 strains) and in young adults composed entirely of terminally differentiated somatic cells (glp-1 strain). Consistent with these results, Greiss et al. [52] also detected induction of almost no DNA repair-related genes in C. elegans exposed to ionizing radiation. Furthermore, although induction of some DNA repair genes is p53-dependent in mammals, Derry et al. [53] detected no genes that were UVC-induced in a cep-1 (C. elegans p53)-dependent fashion.
Since it is possible that our microarray experiment missed an important time point for the detection of transcriptional regulation, or that NER might be inducible through non-transcriptional mechanisms, we also directly tested repair kinetics, but observed no induction. There are caveats associated with this observation. First, we measured repair in both strands of a transcribed gene, rather than specific repair of either transcribed or non-transcribed regions of the genome. Thus, if only transcription-coupled or global genomic repair were induced [48], it is possible that such a signal would be undetectable. Second, we did not measure repair in germ cells or early embryos, which have much higher levels of DNA repair gene expression (this manuscript and [23]). The question of whether DNA repair is faster and/or more inducible at earlier lifestages is an important one that should be addressed in future work. Finally, it is conceivable that a different pre-exposure dose might be more effective.
The lifestage-dependent difference in expression of DNA repair as well as other DNA damage-responsive genes is dramatic. Transcription of a high proportion of the components of the DNA damage response is much lower in glp-1 than in gravid wild-type or xpa-1 young adults, despite the dilution of the germ cells/early embryos by somatic cells. Interestingly, one exception to this pattern was genes involved in nonhomologous end-joining, of which only one was higher in germ cell-containing nematodes (cku-80, ~3.5-fold higher). This is consistent with a previous study that found that nonhomologous end-joining is important in non-cycling somatic cells [13]. While our experiments cannot distinguish between germs cells and early embryos, such experiments would be interesting given the fact that cell cycle checkpoint activation is silenced in C. elegans in early embryos, but not in the germline [12]. Importantly, we previously showed that there was no detectable difference in UVC damage repair kinetics between wild-type and glp-1 nematodes when measured in starved L1s, prior to germ cell proliferation [23].
It would appear that under normal conditions, DNA damage-responsive proteins are present at sufficient levels in adult somatic cells to carry out their functions, as evidenced by the fact that UVC damage is repaired in adults [23], and checkpoint proteins function in adult somatic cells [54,55]. However, it is possible that levels become insufficient with time, as suggested by our observation of decreased repair in older adults [23]. This insufficiency might be exacerbated by stressful environmental conditions (e.g., exposure to genotoxins), and is particularly interesting in the context of the observation by Weidhaas et al. [54] that checkpoint proteins, best known for conferring susceptibility to DNA damage-induced apoptotic cell death in germ cells, also confer resistance to DNA damage-induced necrotic cell death in somatic cells.
Conclusion
NER was not detectably inducible by UVC in C. elegans, but germ cells and/or early embryos have a much higher level of expression of NER and other DNA damage response-related genes than do adults consisting only of terminally differentiated somatic cells. NER-deficient C. elegans were nearly indistinguishable from the wild-type strain, even after examination of global gene expression, under control conditions. However, the need for NER in this organism is demonstrated by the dramatic reduction in lifespan, growth, and feeding after low levels of DNA damage. NER deficiency in C. elegans, as in humans, constitutes a dramatic example of a gene-environment interaction.
Supplementary Material
Acknowledgements
We thank Julie Rice and Daniel Snyder for technical assistance with COPAS Biosort experiments and C. elegans maintenance, Danica Ducharme of the NIEHS microarray facility for microarray work, and Marjolein Smith for statistical analysis of growth and feeding data. This work was supported by National Institutes of Health [P42 ES10356 to J.M.]; the National Toxicology Program [to W.B.]; and the Intramural Research Program of the National Institute of Environmental Health Sciences [to J.F.]. All nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources.
Abbreviations
- eEXT
optical extinction
- UVC
ultraviolet C radiation
- NER
nucleotide excision repair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Balajee AS, Bohr VA. Genomic heterogeneity of nucleotide excision repair. Gene. 2000;250:15–30. doi: 10.1016/s0378-1119(00)00172-4. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 3.Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 5.Costa RM, Chigancas V, Galhardo Rda S, Carvalho H, Menck CF. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 7.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 8.Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M. Combined functional genomic maps of the C elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 9.O'Neil N, Rose A. WormBook. 2005. DNA repair, in: T.C.e.R. Community. (Ed.) http://www.wormbook.org. [Google Scholar]
- 10.Leung MC-K, Williams PL, Benedetto A, Au C, Helmke KJ, Aschner M, Meyer JN. Caenorhabditis elegans: an Emerging Model in Biomedical and Environmental Toxicology. Toxicological Sciences. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Molecular Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 12.Holway AH, Kim SH, La Volpe A, Michael WM. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J Cell Biol. 2006;172:999–1008. doi: 10.1083/jcb.200512136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- 15.Park HK, Yook JS, Koo HS, Choi IS, Ahn B. The Caenorhabditis elegans XPA homolog of human XPA. Molecules and Cells. 2002;14:50–55. [PubMed] [Google Scholar]
- 16.Denver DR, Feinberg S, Steding C, Durbin M, Lynch M. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics. 2006;174:57–65. doi: 10.1534/genetics.106.059840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyun M, Lee J, Lee K, May A, Bohr VA, Ahn B. Longevity and resistance to stress correlate with DNA repair capacity in Caenorhabditis elegans. Nucleic Acids Res. 2008;36:1380–1389. doi: 10.1093/nar/gkm1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson TE, Hartman PS. Radiation effects on lifespan in Caenorhabditis elegans. Journals of Gerontology. 1988;43:B137–B141. doi: 10.1093/geronj/43.5.b137. [DOI] [PubMed] [Google Scholar]
- 19.Astin JW, O'Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues. RAD-3 and WWP-1, DNA Repair (Amst) 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Hartman PS, Herman RK. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics. 1982;102:159–178. doi: 10.1093/genetics/102.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman PS. Uv Irradiation of wild-type and radiation-sensitive mutants of the nematode Caenorhabditis elegans - fertilities, survival, and parental effects. Photochemistry and Photobiology. 1984;39:169–175. doi: 10.1111/j.1751-1097.1984.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 22.Coohill T, Marshall T, Schubert W, Nelson G. Ultraviolet mutagenesis of radiation-sensitive (rad) mutants of the nematode Caenorhabditis elegans. Mutation Research. 1988;209:99–106. doi: 10.1016/0165-7992(88)90025-5. [DOI] [PubMed] [Google Scholar]
- 23.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartman PS, Hevelone J, Dwarakanath V, Mitchell DL. Excision repair of UV radiation-induced DNA damage in Caenorhabditis elegans. Genetics. 1989;122:379–385. doi: 10.1093/genetics/122.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodoyianni V, Maine EM, Kimble J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabitis elegans. Molecular Biology of the Cell. 1992;3:1199–1213. doi: 10.1091/mbc.3.11.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulston JE, Hodgkin J. Methods. In: Wood W, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- 27.Williams PL, Dusenbery DB. Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts. Toxicology and Industrial Health. 1988;4:469–478. doi: 10.1177/074823378800400406. [DOI] [PubMed] [Google Scholar]
- 28.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 29.Boyd WA, Smith MV, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. Application of a mathematical model to describe the effects of chlorpyrifos on Caenorhabditis elegans development. PLoS One. 2009;4:e7024. doi: 10.1371/journal.pone.0007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith MV, Boyd WA, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. A discrete time model for the analysis of medium-throughput C. elegans growth data. PLoS One. 2009;4:e7018. doi: 10.1371/journal.pone.0007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd WA, McBride SJ, Freedman JH. Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high-throughput screening assay. PLoS ONE. 2007;2:e1259. doi: 10.1371/journal.pone.0001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Houten B, Cheng S, Chen Y. Measuring gene-specific nucleotide excision repair in human cells using quantitative amplification of long targets from nanogram quantities of DNA. Mutation Research. 2000;460:91–94. doi: 10.1016/s0921-8777(00)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 34.Neher DA, Sturzenbaum SR. Extra-long PCR, an identifier of DNA adducts in single nematodes (Caenorhabditis elegans) Comparative Biochemistry and Physiology C-Toxicology & Pharmacology. 2006;144:279–285. doi: 10.1016/j.cbpc.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Liao VH, Freedman JH. Cadmium-regulated genes from the nematode Caenorhabditis elegans. Identification and cloning of new cadmium-responsive genes by differential display. Journal of Biological Chemistry. 1998;273:31962–31970. doi: 10.1074/jbc.273.48.31962. [DOI] [PubMed] [Google Scholar]
- 36.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18 Suppl 1:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 38.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, Cunningham ML, Deng S, Dressman HK, Fannin RD, Farin FM, Freedman JH, Fry RC, Harper A, Humble MC, Hurban P, Kavanagh TJ, Kaufmann WK, Kerr KF, Jing L, Lapidus JA, Lasarev MR, Li J, Li YJ, Lobenhofer EK, Lu X, Malek RL, Milton S, Nagalla SR, O'Malley J P, Palmer VS, Pattee P, Paules RS, Perou CM, Phillips K, Qin LX, Qiu Y, Quigley SD, Rodland M, Rusyn I, Samson LD, Schwartz DA, Shi Y, Shin JL, Sieber SO, Slifer S, Speer MC, Spencer PS, Sproles DI, Swenberg JA, Suk WA, Sullivan RC, Tian R, Tennant RW, Todd SA, Tucker CJ, Van Houten B, Weis BK, Xuan S, Zarbl H. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 40.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 41.Lozano E, Saez AG, Flemming AJ, Cunha A, Leroi AM. Regulation of growth by ploidy in Caenorhabditis elegans. Curr Biol. 2006;16:493–498. doi: 10.1016/j.cub.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 42.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crowley DJ, Hanawalt PC. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asad LM, Medeiros DC, Felzenszwalb I, Leitao AC, Asad NR. Participation of stress-inducible systems and enzymes involved in BER and NER in the protection of Escherichia coli against cumene hydroperoxide. Mutat Res. 2000;461:31–40. doi: 10.1016/s0921-8777(00)00020-3. [DOI] [PubMed] [Google Scholar]
- 45.Scott AD, Waters R. Inducible nucleotide excision repair (NER) of UV-induced cyclobutane pyrimidine dimers in the cell cycle of the budding yeast Saccharomyces cerevisiae: evidence that inducible NER is confined to the G1 phase of the mitotic cell cycle. Mol Gen Genet. 1997;254:43–53. doi: 10.1007/s004380050389. [DOI] [PubMed] [Google Scholar]
- 46.Al-Moghrabi NM, Al-Sharif IS, Aboussekhra A. UV-induced de novo protein synthesis enhances nucleotide excision repair efficiency in a transcription-dependent manner in S. cerevisiae. DNA Repair (Amst) 2003;2:1185–1197. doi: 10.1016/j.dnarep.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Singh RK, Krishna M. DNA damage induced nucleotide excision repair in Saccharomyces cerevisiae. Mol Cell Biochem. 2006;290:103–112. doi: 10.1007/s11010-006-9173-z. [DOI] [PubMed] [Google Scholar]
- 48.Hanawalt PC, Ford JM, Lloyd DR. Functional characterization of global genomic DNA repair and its implications for cancer. Mutat Res. 2003;544:107–114. doi: 10.1016/j.mrrev.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Nouspikel T. DNA Repair in Mammalian Cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson GW, Nicolet CM, Kalainov D, Friedberg EC. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986;83:1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci U S A. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics. 2008;9:334. doi: 10.1186/1471-2164-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derry WB, Bierings R, van Iersel M, Satkunendran T, Reinke V, Rothman JH. Regulation of developmental rate and germ cell proliferation in Caenorhabditis elegans by the p53 gene network. Cell Death Differ. 2007;14:662–670. doi: 10.1038/sj.cdd.4402075. [DOI] [PubMed] [Google Scholar]
- 54.Weidhaas JB, Eisenmann DM, Holub JM, Nallur SV. A Caenorhabditis elegans tissue model of radiation-induced reproductive cell death. Proc Natl Acad Sci U S A. 2006;103:9946–9951. doi: 10.1073/pnas.0603791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.