Abstract
Purpose
A subset of patients with chronic myelogenous leukemia (CML) do not respond to the tyrosine kinase inhibitor (TKI) imatinib mesylate. Such primary imatinib resistance is distinguished from secondary resistance which reemerges after attainment of cytogenetic remission.
Patients and Methods
We studied gene expression patterns in total WBCs using a panel of 21 genes previously implicated in TKI handling, resistance, or progression comparing patients who had newly diagnosed TKI-naive CML that had optimal (n = 41), or suboptimal (n = 7) responses to imatinib, or primary resistance (n = 20). Expression patterns were compared to those in secondary TKI-resistant chronic phase CML without ABL1 kinase domain mutations (n = 29), and to lymphoid (n = 15) or myeloid blast phase disease (n = 12).
Results
Fifteen genes in the panel distinguished blast phase from chronic phase disease, and 12 genes distinguished newly diagnosed CML from TKI-resistant CML without ABL1 kinase domain mutations, but only a single gene, prostaglandin-endoperoxide synthase 1/cyclooxgenase 1 (PTGS1/COX1; P = .005), differentiated imatinib-responsive from primary imatinib-resistant CML. The association of primary imatinib resistance with higher transcript levels of the drug metabolism gene PTGS1 was confirmed in a separate data set of 68 newly diagnosed, imatinib-treated CML (P = .008). In contrast, up to 11 different genes were identified in a multivariate model that optimally discriminated secondary imatinib resistance lacking ABL1 kinase domain mutation from imatinib-responsive cases, likely related to the more complex pathogenesis of secondary resistance.
Conclusion
Gene expression profiling of CML at diagnosis for PTGS1 may be useful in predicting imatinib response and in selecting alternate therapy.
INTRODUCTION
The tyrosine kinase inhibitor (TKI) imatinib mesylate (Gleevec; Novartis Pharma, Basel, Switzerland) is an effective treatment for chronic myelogenous leukemia (CML) which acts by inhibiting the BCR-ABL kinase arising from the t(9;22) chromosomal translocation which is the hallmark of this leukemia. Therapeutic resistance to imatinib is seen in approximately 10% to 15% of patients and can be classified as primary or secondary depending on whether an initial decline in disease levels are observed or not.1 A major factor mediating secondary resistance is the emergence of acquired point mutations in the ABL kinase domain (KD) and BCR-ABL1 gene amplification, although other molecular mechanisms of resistance are also important.2
In contrast, the factors contributing to primary resistance are less well characterized.3 The initial response rates to imatinib are much lower in those patients presenting with CML already in accelerated phase (AP) or blast phase (BP) suggesting that factors mediating blast transformation compromise response to imatinib. Other postulated mechanisms of primary resistance among CML patients presenting in chronic phase (CP) include low activity of imatinib uptake transporter cation transporter 1 (OCT1)4–6 and increased activity of imatinib efflux transporters.7–9
The goal of this study was to evaluate the utility of a clinical-grade limited gene expression panel for predicting response to imatinib in newly diagnosed CML, and assessing the mechanisms of secondary imatinib resistance when ABL KD mutations were absent. We used a targeted approach selecting genes involved in the pharmacogenomics of imatinib and other TKIs, and in CML progression. We show that such an approach can identify genes differentially associated with primary and secondary imatinib resistance.
PATIENTS AND METHODS
Patient Characteristics and Therapy Response Criteria
The first CML study set was composed of diagnostic samples before the initiation of imatinib therapy from 68 patients presenting to University of Texas M. D. Anderson Cancer Center (Houston, TX) between June 2003 and February 2007, including all patients with excess samples who had suboptimal imatinib response or imatinib failure, and a randomly chosen group of patients with optimal imatinib response, secondary resistance, and blast transformation. A second set of 68 newly diagnosed CML patients was composed of patients with initial imatinib treatment before June 2003 or after February 2007, with all patients having at least 12 months of follow-up. Diagnostic work-up on all patients included CBC, bone marrow biopsy and aspiration, G-banded karyotypic study from short-term cultures of aspirate material, fluorescent in situ hybridization using a dual-fusion BCR-ABL1 probe on short-term culture of peripheral blood, and quantitative reverse transcription polymerase chain reaction (QRT-PCR) for the BCR-ABL1 fusion transcript on leukocytes from blood, as described.10,11 The study was performed according to an approved laboratory protocol and in accordance with the Declaration of Helsinki.
Sokal risk scores were calculated as described.12 Imatinib response criteria were as previously described.1 Complete hematologic response was defined as a WBC count of lower than 10 × 109/L, a platelet count lower than 450 × 109/L, no immature cells (blasts, promyelocytes, myelocytes) in the peripheral blood, and disappearance of all signs and symptoms related to leukemia (including palpable splenomegaly). Cytogenetic responses were defined as complete (0% t(9;22)/Philadelphia chromosome [Ph] positive), partial (pCyR, 1% to 35% Ph positive), minor (36% to 65% Ph positive), and minimal (66% to 95% Ph positive). A major cytogenetic remission included complete plus partial cytogenetic remissions (ie, < 35% Ph positive). Cytogenetic remission was judged by standard cytogenetic analysis in 20 metaphases. Major molecular response was defined as a BCR-ABL1/ABL1 transcript ratio of lower than 0.05% by QRT-PCR, representing more than 3-log reduction from the baseline for untreated patients in our laboratory. Complete molecular response was defined as undetectable levels of BCR-ABL1 transcript, representing at least 4.5-log reduction from baseline levels. Optimal treatment response at 12 months is defined by complete cytogenetic response, suboptimal treatment response at 12 months is defined by pCyR, and treatment resistant at 12 months is defined by less than pCyR (three patients assessed at slightly earlier time points 9, 10, and 11 months, respectively). ABL KD mutations were assessed in TKI-resistant samples using a nested PCR strategy covering codons 221 to 500 and a screening strategy as previously described.13,14
Selection of Genes for Transcript Profiling
The 24 genes in the panel included two normalizing genes (GUSB and 18S RNA) and 22 test genes with known influences on TKI entry, handling, and efflux as well as genes known to be related to disease progression in CML identified in previously published microarray studies. Genes influencing import, binding, and export of TKIs (and imatinib specifically) included the cation drug transporters OCT1 (SLC22A1), OCT2 (SLC22A2), and OCT3 (SLC22A3), drug metabolism genes including the P450 isoforms ABCB1 (multidrug resistance [MDR]-1), ABCC1, and ABCG2,15 and prostaglandin-endoperoxide synthase (PTGS)1 and PTGS2, and the blast marker CD34. Previously identified progression factors in CML included the granulocyte-macrophage colony-stimulating factor (CSF2),15 JAK/STAT signaling components JAK2, STAT5A, STAT5B,16 STAT3,17,18 the kinases ABL1, TEC, BTK,19 and LYN,20,21 and transcription factors CEBPA,22 RUNX1, and RUNX3.23 Also included were the three genes most highly associated with lack of response to imatinib in a previous microarray study of primary resistance, namely PTGS1, protein tyrosine phosphatase, nonreceptor type 22 (PTPN22), and frizzled homolog 7 (FZD7).24
Low-Density QRT-PCR Array
Expression profiling was done on RNA extracted from CML samples using a custom-designed TaqMan low-density QRT-PCR array containing one gene-specific forward and reverse primer pair and one TaqMan MGB probe (6-FAM dye-labeled) in each well (Applied Biosystems, Foster City, CA). Total RNA was extracted from WBCs following RBC lysis using the guanidium solubilization method (Trizol, Invitrogen, Carlsbad, CA) and complementary (c)DNA synthesized using Superscript III reverse transcriptase (Invitrogen) using random hexamers for priming. QRT-PCR was performed with 800 ng of cDNA from each sample, as described previously.25 Thermal cycling conditions were as follows: 2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. Measurements of the ABL1 genes on the arrays were correlated with the singe tube QRT-PCR BCR-ABL1 assays previously performed on these samples.
Statistical Analysis
The relative expression level of a particular gene of a given sample on the array was calculated by the delta (Δ) threshold cycle (Ct) method. Using the approach previously described for linear discriminant analysis (LDA),25 the ΔCt value was obtained by normalizing against the median Ct value of all 22 test genes for each sample except for OCT2 (SLC22A2) which was expressed at very low levels.
One-way analysis of variance (ANOVA) or t test were used to test against null hypothesis of no significant difference for any given gene expression among three treatment response groups, optimal, suboptimal, and resistant group, or between two groups when combining the optimal and the suboptimal into one group. Holm's method was applied to adjust P values of ANOVA and t-tests to correct multiple comparisons.26
LDA27 was used to model multiple gene effects regarding two response groups, resistant versus combination of optimal and suboptimal groups. A series of linear discriminant models were built to sequentially include the increasing number of genes, starting from one to 21 genes. The order of gene selection was decided by their t-statistics in training set. In order to perform a robust analysis, 3-fold cross validation with random assignment of training and test set was repeated 500 times for each model. The accuracy of training or of test set was measured by comparing predicted with observed group labels. Our statistical analyses, including unsupervised hierarchical clustering, were performed in R, version 2.7.0 (www.r-project.org), a freely available statistical software package.
RESULTS
Pathway-Based Gene Expression Panel Distinguishes CP CML From Lymphoid and Myeloid Blast Phases
The gene expression profiles of total WBCs from 68 patients with newly diagnosed CML (63 patients in CP, five patients in AP), 29 patients with secondary TKI-resistant CML without ABL1 KD mutations (23 patients receiving imatinib, six patients receiving other TKIs), 15 patients with CML in lymphoid BP (LBP, six newly diagnosed) and 12 patients with secondary myeloid BP (MBP) were compared. All of the genes in the transcript panel except OCT2 were significantly expressed in the samples tested. ABL1 levels detected in this panel were highly correlated with those detected in the single tube BCR-ABL1/ABL1 QRT-PCR assay (r = 0.81). Table 1 lists the median-normalized absolute expression levels for all genes for each sample group.
Table 1.
Shifts in Gene Expression With Blast Transformation
| Gene Expression | Median Normalized Expression Levels |
TKI-r CML From New CML |
TKI-r CML From CML-MBP |
TKI-r CML From CML-LBP |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| New CML (54 CP, 14 AP) | TKI-r CML | CML MBP | CML LBP | P | P adj* | P | P adj* | P | P adj* | |
| Most increased with blast transformation | ||||||||||
| ABCC1 | 0.479 | 0.910 | 1.617 | 2.697 | < .001 | < .001 | < .001 | .005 | < .001 | < .001 |
| CD34 | 0.166 | 0.008 | 3.194 | 4.67 | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 |
| FZD7 | 0.025 | 0.020 | 0.09 | 0.083 | .198 | .99 | .001 | .01 | .002 | .01 |
| Most decreased with blast transformation | ||||||||||
| CEBPA | 1.458 | 0.635 | 0.439 | 0.472 | < .001 | < .001 | < .001 | .005 | .001 | .01 |
| JAK2 | 2.762 | 1.604 | 0.720 | 0.884 | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 |
| LYN | 53.190 | 18.375 | 4.076 | 2.897 | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 |
| PTPN22 | 13.944 | 4.513 | 2.759 | 1.686 | < .001 | < .001 | < .001 | .005 | < .001 | < .001 |
| SLC22A1 | 0.180 | 0.328 | 0.02 | 0.040 | .01 | .084 | < .001 | < .001 | < .001 | < .001 |
| STAT3 | 6.364 | 6.916 | 4.286 | 4.877 | .446 | 1 | < .001 | .002 | .003 | .014 |
| STAT5B | 2.542 | 2.670 | 0.993 | 1.000 | .47 | 1 | < .001 | < .001 | < .001 | < .001 |
Abbreviations: TKI-r, tyrosine kinase inhibitor resistance; CML, chronic myelogenous leukemia; MBP, myeloid blast phase; LBP, lymphoid blast phase; CP, chronic phase; AP, accelerated phase.
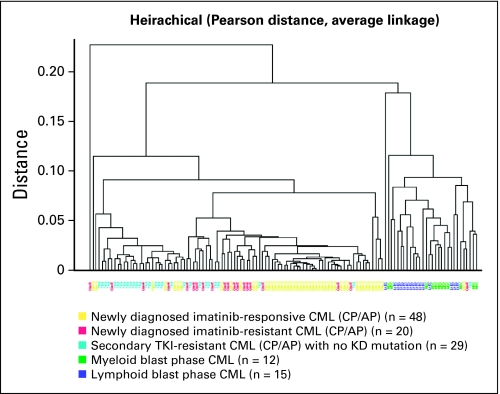
To examine the ability of this panel to distinguish CP CML at diagnosis from KD-unmutated, secondary TKI-resistant CML, LBP, and MBP, we performed unsupervised clustering analysis using 21 genes (normalizers 18S RNA and GUSB as well as OCT2/SLC22A2 were excluded). As shown in Figure 1, LBP and MBP could be easily distinguished in all but three cases from CML-CP/AP. In contrast, secondary TKI-resistant CML (light blue) clustered with newly diagnosed cases of CML that showed resistance to imatinib but generally away from those cases that responded to imatinib.
Fig 1.
Twenty-one gene targeted transcript panel can differentiate chronic from blast phases of chronic myelogenous leukemia (CML), and imatinib-responsive newly diagnosed CML from imatinib-resistant patients. Unsupervised hierarchical clustering of newly diagnosed imatinib-responsive (yellow) and imatinib-resistant (red) patients versus ABL KD-unmutated tyrosine kinase inhibitor (TKI)-resistant samples in chronic phase (CP) or accelerated phase (AP; light blue) and myeloid (green) and lymphoid blast phase (dark blue).
All but three (PTGS1, SLC22A3, and STAT5A) of the 21 genes in the panel were expressed at significantly different levels in TKI-resistant CP CML compared to LBP and MBP. Genes that were most upregulated with blast transformation were CD34 and ABCC1 (P < .0001), whereas genes whose expression was downregulated on blast transformation included transcription factors CEBPA, STAT3, STAT5A, kinases LYN, JAK2, phosphatase PTPN22, and SLC22A1 (OCT1). Most of these genes were also significantly different between newly diagnosed CML and KD-unmutated TKI-resistant CML (Table 1).
Correlations of Initial Imatinib Response With Patient and Tumor Characteristics
We separately analyzed gene expression profiles from the 68 newly diagnosed patients to determine factors that were associated with optimal and suboptimal responses and resistance to imatinib. Assessing response at 1-year, 41 patients (60.3%) had optimal response, seven patients (10.3%) had suboptimal response, and 20 patients (29.4%) were resistant to imatinib treatment (Table 2, first set). At start of imatinib treatment, 34 (90%) of 41 of the optimal response group, six (86%) of seven of the suboptimal group and 14 (70%) of 20 of the resistant group were in CP, with the rest in AP due to additional cytogenetic aberrations in 12 and low platelet count in two. No patients were in BP.
Table 2.
Patient Characteristics Among Newly Diagnosed Imatinib-Treated CML
| Characteristic | Optimal | Suboptimal | Resistant |
|---|---|---|---|
| Total patients | |||
| First set | 41 | 7 | 20 |
| Second set | 43 | 15 | 10 |
| Median age, years | |||
| First set | 46 | 51 | 36 |
| Range | 21-70 | 34-71 | 20-60 |
| Second set | 45 | 40 | 40 |
| Range | 18-77 | 24-71 | 32-84 |
| Stage at start of imatinib | |||
| First set | |||
| CP | 34 | 6 | 14 |
| AP | 7 | 1 | 6 |
| Second set | |||
| CP | 38 | 13 | 9 |
| AP | 5 | 2 | 1 |
| Median BCR-ABL1/ABL1 QRT-PCR, % | |||
| First set | 90.9 | 65.6 | 30.2 |
| Second set | 45.0 | 45.2 | 43.6 |
| Clonal evolution at presentation | |||
| First set | 6 | 1 | 4 |
| % | 15 | 14 | 20 |
| Second set | 2 | 0 | 1 |
| % | 5 | 10 | |
| Median imatinib dose over first year, mg/d | |||
| First set | 700* | 500† | 500‡ |
| Second set | 600 | 600 | 500 |
| Median follow-up (range, months) | |||
| First set | 45.0 | 43.3 | 35.0 |
| Range | 12-60.1§ | 15.4-54.3‖ | 7.3-55.4¶ |
| Second set | 60.9 | 60.9 | 43.6 |
| Range | 14.8-94.8 | 24.5-113.7 | 23.4-109.5 |
| Outcome first and second set combined | 3 DOD, 1 DOOD | 1 DOOD | 4 DOD, 2 DOOD |
| Hematologic parameters in first set | |||
| Median presenting WBC, ×109/L | 77 | 75.9 | 81 |
| Median presenting platelet count, ×109/L | 296 | 337 | 467 |
| Elevated PB blasts, > 5% | 2 | 1 | 5 |
| Elevated PB basophils, > 5% | 6 | 3 | 5 |
| Splenomegaly at presentation | 8 | 1 | 8 |
| Sokal risk score# | |||
| Low | 30 | 5 | 11 |
| Intermediate | 9 | 2 | 1 |
| High | 1 | 6 |
Abbreviations: CML, chronic myelogenous leukemia; CP, chronic phase; AP, accelerated phase; QRT-PCR, quantitative reverse transcription polymerase chain reaction; DOD, died of disease-related causes; DOOD, died of other disease; WBC, white blood cell; PB, peripheral blood; TKI, tyrosine kinase inhibitor; BP, blast phase.
Twelve patients were initially on 400 mg/day of imatinib, one patients on 300, six patients on 600, and 23 on 800 mg/day (including five patients also taking pegylated interferon and recombinant granulocyte-macrophage colony-stimulating factor, usually for short duration); nine of these patients had subsequent dose reductions due to toxicities, five patients had dose increases due to persistent BCR-ABL1 transcript levels.
Six of seven patients were initially on 400 mg/day; one patient on 800, with dose limiting toxicities precluding dose escalation in five patients.
Twelve patients were initially on 300 to 500 mg/day of imatinib (most on 400 mg), seven patients were on 800 mg per day (including three taking pegylated-intron/sargramostim). In eight patients, dose escalation was attempted, in the remaining there were dose-limiting toxicities. Two patients with resistant disease at the 1 year had transient responses in the first 3 to 6 months of treatment.
Four patients developed secondary imatinib resistance (with 1 BP, 1 AP, 2 CP) at a median of 24 months after initial therapy, with two switched to a new TKI.
Three patients developed secondary imatinib resistance (2 AP, 1 CP) and were switched to a new TKI (2 dasatinib, 1 bosutinib) at 22, 24, and 33 months post-imatinib start, three patients were continued on imatinib, and one patient was lost to follow-up.
Eleven patients were switched to a new TKI (dasatinib in three, bosutinib in seven, and nilotinib in one) after a mean duration of imatinib of 16.9 months (range, 6 to 42.7 months), three had stem cell transplant, and five had dose escalation with imatinib. Two patients with resistant disease died before 1 year; two were lost to follow-up.
One optimal response and two resistant patients could not be scored.
The median WBC counts and the median platelet counts at presentation and numbers of patients with elevated blood blasts or basophils were not significantly different between the three groups. However, a higher percentage of patients in the resistant group had high risk Sokal scores (30% v 3% for optimal and 0% for suboptimal) due to the higher incidence of splenomegaly in this group. The median BCR-ABL1/ABL1 percentages in the analyzed samples, 90.9%, 65.6%, and 30.2% respectively, were significantly higher in the optimal and suboptimal response groups compared to the resistant group (Table 2).
The pattern of imatinib dosing over the first year, and outcome for each group are summarized in the legend to Table 2, with a higher median imatinib dose in the optimal response group largely due to dose-limiting toxicities precluding escalation in the other groups. Eleven of the primary resistance cases (suboptimal or failure) were assessed at 12- to 18-month time points for BCR-ABL1 KD mutations before TKI change. Three of these patients showed mutations (E255K, F359V, and E459K); retrospective analysis did not show these mutations in the baseline samples.
PTGS1 Expression Differentiates Imatinib-Responsive and -Resistant Groups
The relative expression levels of all 21 test genes from newly diagnosed CML samples were compared based on optimal response, suboptimal response, and resistance to imatinib assessed at the 1-year time point (Appendix Table A1, online only). The P values of overall difference among the three response groups for specific genes (by ANOVA) were similar to those obtained by t-tests when combining optimal and suboptimal imatinib response groups so further comparisons were done grouping optimal/suboptimal versus resistant patients.
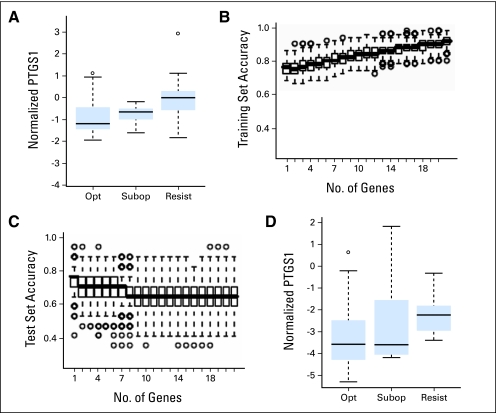
When values were corrected for multiple gene testing effects, only PTGS1 was significantly differentially expressed in the imatinib-failure/resistant group (Fig 2A). The significant association of increased PTGS1 expression with resistance to imatinib was confirmed by a multivariate LDA model that was repeatedly run in 500 three-fold cross-validations using randomly assigned training and test sets. As shown in Figures 2B and 2C, a single gene model was identified as the optimal for accuracy of prediction, with PTGS1 identified as that gene in 96% of runs.
Fig 2.
Higher prostaglandin-endoperoxide synthase 1 (PTGS1) transcript levels are associated with primary imatinib resistance in two different data sets. (A) Comparison of median-normalized PTGS1 transcript levels in total WBCs from the first set of newly diagnosed imatinib-naïve chronic myelogenous leukemia (CML) samples that showed optimal imatinib response (opt), suboptimal imatinib response (subopt) or imatinib failure/resistance (resist), as assessed at 1 year. (B, C) The contribution of each of 21 test genes to prediction accuracy of imatinib response at 1 year among the first set of 68 newly diagnosed patients was determined by linear discriminant analysis (LDA). Plotted are the results of 500 three-fold cross validations with each box demonstrating the distribution of the prediction accuracies including increasing numbers of genes. Accuracies of prediction by LDA in randomly chosen (B) training sets and (C) test sets. Accuracy for the test sets decline with inclusion of more than one gene, with PTGS1 identified as the predictor gene in 481 (96%) of 500 simulations. (D) Comparison of GUSB-normalized PTGS1 transcript levels in total WBCs from a different set of 68 newly diagnosed imatinib-naïve CML samples. PTGS1 transcript levels are again higher in the imatinib resistant/failure group (resist), as assessed at 1 year (P = .008, t-test). (A-D) Box and whisker plots to demonstrate data distributions. For all figures, the boxes show the interquartile range (IQR) with bar in box indicating median values. Values with whiskers indicate a range of 1.5 times of IQR, with circles representing outliers.
We next assessed the significance of PTGS1 transcript levels in RNA extracted from total blood WBCs before imatinib therapy in a different set of 68 patients with CML, including 43 optimal, 15 suboptimal responders, and 10 with imatinib resistance/failure (Table 2, second set). Demographic and hematologic features and BCR-ABL1/ABL1 percentages in the analyzed samples were similar between the three groups (Table 2 and not shown). When normalized to GUSB levels, elevated PTGS1 transcript levels were once again associated with imatinib resistance as compared to optimal responders (P = .0083, t-test; Fig 2D).
Differences Between the Gene Expression Profiles of Primary and Secondary Imatinib Resistance
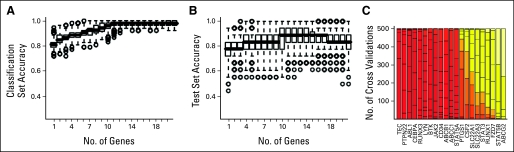
We examined whether gene expression patterns associated with secondary imatinib resistance in the absence of ABL KD mutation were similar to those seen in primary imatinib resistance. Genes whose expression was most significantly higher in secondary imatinib resistance compared to newly diagnosed optimal imatinib responders included ABCB1, ABCC1, STAT5A, and RUNX3. Genes whose expression was lower in secondary imatinib-resistant samples included PTPN22, CEBPA, TEC, JAK2, and LYN (Table 3). As shown in Figure 3A, 11 genes were required to optimally distinguish secondary TKI-resistant samples from newly diagnosed optimal responders using a LDA model, with higher level of RUNX3 transcript and lower levels of TEC, PTPN22, ABL1, and CEBPA being the most frequent discriminators in repeated cross-validations (Fig 3C). Similarly, a multivariate comparison of primary and secondary resistant samples identified multiple genes that distinguished these samples (not shown). We interpret these findings to indicate that secondary TKI-resistance is more complex and multifactorial than primary resistance, with some factors (eg, decreased expression of LYN, JAK2, PTPN22, and CEBPA) that are associated with secondary TKI resistance being common to blast phase transformation.
Table 3.
Genes Differentiating Secondary Imatinib Resistance From Newly Diagnosed CML With Imatinib Response
| Gene | Median Normalized Expression Levels |
t-test P | t-test P (adjusted) | |
|---|---|---|---|---|
| CML With Secondary Imatinib Resistance (n = 23) | CML With Imatinib Response (n = 48) | |||
| TEC | 0.207 | 0.588 | < .001 | < .001 |
| ABL1 | 0.864 | 1.437 | < .001 | < .001 |
| PTPN22 | 4.733 | 14.450 | < .001 | < .001 |
| CEBPA | 0.658 | 1.592 | < .001 | < .001 |
| RUNX3 | 1.662 | 0.683 | < .001 | < .001 |
| LYN | 18.375 | 53.190 | < .001 | < .001 |
| BTK | 1.325 | 2.104 | < .001 | < .001 |
| JAK2 | 1.648 | 2.734 | < .001 | < .001 |
| CD34 | 0.012 | 0.177 | < .001 | < .001 |
| STAT5A | 1.643 | 1.000 | < .001 | < .001 |
| ABCB1 | 0.206 | 0.045 | < .001 | < .001 |
| ABCC1 | 0.884 | 0.473 | < .001 | < .001 |
| CSF2 | 0.002 | 0.001 | .0252 | .2015 |
| PTGS1 | 0.599 | 0.378 | .0097 | .087 |
| SLC22A1 | 0.328 | 0.175 | .0365 | .2554 |
| FZD7 | 0.022 | 0.029 | .1548 | .5621 |
| SLC22A3 | 0.000 | 0.000 | .0685 | .4111 |
| STAT3 | 6.229 | 6.592 | .093 | .465 |
| RUNX1 | 3.567 | 3.546 | .1405 | .5621 |
| STAT5B | 2.828 | 2.532 | .7363 | 1 |
| ABCG2 | 0.007 | 0.007 | .9292 | 1 |
Abbreviation: CML, chronic myelogenous leukemia.
Fig 3.
Multiple genes distinguish imatinib responsive from secondary imatinib-resistant chronic myelogenous leukemia (CML). The contribution of the 21 test genes to accurately distinguishing ABL1 KD-unmutated secondary resistant CML-CP/AP from imatinib responsive baseline CML-CP/AP samples was determined by LDA. The prediction accuracies were estimated using randomly chosen test and training sets in 500 three-fold cross-validations are shown in (A) for training sets and in (B) for test set. The median accuracy of a model for discriminating imatinib response from secondary imatinib resistance improves by inclusion of up to 11 genes. The symbols in this Figure are as in Figures 2B and 2C. (C) The frequency of particular genes identified in these increasing gene model are shown on a bar plot, with overall frequency in all models represented by color (red to white). Prostaglandin-endoperoxide synthase (PTGS1) is not identified commonly in the models as a discriminator of secondary imatinib resistance.
DISCUSSION
Using a limited panel of genes selected based on prior studies of imatinib resistance and CML progression, we present a transcript profiling approach to simultaneously distinguish all phases of CML and predict primary TKI resistance and secondary TKI resistance in patients without detectable KD mutation. We included genes that were identified as the most differentially expressed genes in a prior microarray study of primary imatinib resistance (eg, FZD7, PTNP22, and PTGS1),24 or implicated in differential handling of imatinib (ABC transporter genes, and the OCT family of transporters), or repeatedly identified in studies of CML resistance. This approach provided a rapid route to clinical assay development that validates genes identified as discriminants in previous studies while assessing their interactions and ability to provide additional utility in subclassification and prognostication.
Using unsorted WBCs, this transcript panel could separate nearly all cases of CP CML from lymphoid and myeloid BP and most cases of imatinib-responsive from imatinib-resistant CML in an unsupervised clustering algorithm, validating the relevance of these previously implicated genes. In newly diagnosed CML, we identified a strong differentially increased expression of PTGS1 in imatinib-resistant patients; a finding that was confirmed in a second test set of 68 patients. PTGS1 has been previously shown to be upregulated in imatinib-resistant diagnostic CML samples,24 imatinib-resistant CML cell lines,28 and to be transcriptionally upregulated by BCR-ABL itself in vitro.29 Using cross-validation multivariate analysis, we show that this single gene provides nearly all of the predictive power for primary resistance in our test gene set.
While ATP binding–cassette type drug transporters did not significantly correlate with primary resistance, they were identified as discriminators of secondary resistance and as markers of blast phase. ABCB1 (MDR-1; P-glycoprotein) and ABCG2 are known to be highly expressed on primitive hematopoietic stem cells and have been shown to mediate drug resistance in many settings, including for TKI.30 Transcript levels of ABCG1 have also been associated with TKI resistance in vitro31 and in modeling studies.32 We show that increased ABCB1 levels were highly correlated with secondary resistance, with statistically significantly increased levels of ABCG2 also noted in blast phase. These associations are similar to what has been noted in sorted blasts populations,15 and suggest that transcript profiling of RNA from unsorted leukocytes can be used routinely in place of sorted material.
The solute carrier (SLC) family 22 cation drug transporters have been more specifically associated with imatinib handling. Using cell line models, activity of the OCT1/SLC22A1 transporter has been shown to mediate resistance in vitro to imatinib, and was correlated with clinical response.4,5 Differential binding and handling of other TKIs, such as dasatinib, by OCT1 has also been shown. Since dasatinib and nilotinib have been widely used in patients who are resistance or intolerant to imatinib, it has been suggested that profiling of the SLC22 family of transporters may be useful in selecting initial therapy in CML.5,33 However, we did not note strong correlations of OCT 1/SLC22A1 or OCT3/SLC22A3 transcript levels with imatinib resistance indicating that in vitro drug activity assays are not directly correlative with expression of the genes in primary samples. OCT1 transcript levels were noted to be decreased in blast phase disease.
We also examined whether primary or secondary imatinib resistance may be related to increased expression of genes which have been previously associated with blast transformation in CML. To attempt to isolate the factors associated with secondary resistance independent of ineffective blockade of BCR-ABL kinase activity, we included CML-CP/AP patients who lacked detectable ABL KD mutations at time of TKI shift. We identified increased expression of the transcription factors STAT5A and RUNX3 as significantly correlated with such secondary resistance as compared to newly diagnosed patients, although such increased expression was at odds with their role as tumor suppressors in blast transformation.23 Features of such secondary imatinib resistance that were shared with blast transformation included decreased expression of the kinases LYN and JAK2, the phosphatase PTPN22, and the transcription factor CEBPA. These findings support the ability of limited transcript profiling both to separate imatinib-responsive and -resistant subsets of CML in CP, and to highlight important molecular events associated with progression to BP of disease.
Acknowledgment
We thank Kevin Coombes, PhD, for assistance with data analysis.
Appendix
Table A1.
Comparison of Expression of Progression/Resistance Factors in Imatinib-Responsive Versus Imatinib-Resistant Newly Diagnosed CML
| Gene | Median-Normalized Expression Levels |
ANOVA P | ANOVA P (adjusted) | t-test P | t-test P (adjusted) | |
|---|---|---|---|---|---|---|
| Optimal/Suboptimal | Resist | |||||
| PTGS1 | 0.378 | 0.931 | .001 | .024 | < .001 | .005 |
| LYN | 53.190 | 63.258 | .128 | 1.000 | .984 | 1.000 |
| PTPN22 | 14.450 | 9.120 | .135 | 1.000 | .047 | .932 |
| CEBPA | 1.592 | 0.993 | .161 | 1.000 | .063 | 1.000 |
| RUNX3 | 0.683 | 1.000 | .189 | 1.000 | .069 | 1.000 |
| FZD7 | 0.029 | 0.018 | .197 | 1.000 | .146 | 1.000 |
| CSF2 | 0.001 | 0.002 | .220 | 1.000 | .401 | 1.000 |
| STAT5A | 1.000 | 1.045 | .267 | 1.000 | .512 | 1.000 |
| BTK | 2.104 | 1.881 | .308 | 1.000 | .126 | 1.000 |
| RUNX1 | 3.546 | 3.955 | .339 | 1.000 | .176 | 1.000 |
| STAT5B | 2.532 | 2.995 | .352 | 1.000 | .720 | 1.000 |
| JAK2 | 2.734 | 2.843 | .439 | 1.000 | .205 | 1.000 |
| TEC | 0.588 | 0.561 | .486 | 1.000 | .243 | 1.000 |
| CD34 | 0.177 | 0.133 | .590 | 1.000 | .306 | 1.000 |
| ABL1 | 1.437 | 1.666 | .619 | 1.000 | .656 | 1.000 |
| ABCG2 | 0.007 | 0.014 | .711 | 1.000 | .449 | 1.000 |
| SLC22A1 | 0.175 | 0.197 | .851 | 1.000 | .579 | 1.000 |
| ABCC1 | 0.473 | 0.539 | .914 | 1.000 | .886 | 1.000 |
| STAT3 | 6.592 | 6.156 | .928 | 1.000 | .846 | 1.000 |
| SLC22A3 | 0.000 | 0.000 | .954 | 1.000 | .768 | 1.000 |
| ABCB1 | 0.045 | 0.072 | .955 | 1.000 | .776 | 1.000 |
Abbreviations: CML, chronic myelogenous leukemia; ANOVA, one-way analysis of variance; ANOVA P, P value of ANOVA test comparing three groups; ANOVA P adjusted, P value adjusted for multiple testing by Holm's method; t-test P, P value of t-test comparing resistant and response (suboptimal + optimal combined) groups; t-test P adjusted, P value of t-test adjusted for multiple testing by Holm's method.
Footnotes
Supported by a developmental grant from the Leukemia SPORE (1P50CA100632) awarded by the National Cancer Institute, Department of Health and Human Services.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: Elias Jabbour, Novartis Research Funding: Jorge E. Cortes, Novartis; Hagop M. Kantarjian, Novartis; Dan Jones, Novartis Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Wenyong W. Zhang, Jorge E. Cortes, Dan Jones
Financial support: Hagop M. Kantarjian, Dan Jones
Administrative support: Hagop M. Kantarjian
Provision of study materials or patients: Jorge E. Cortes, Elias Jabbour, Hagop M. Kantarjian, Dan Jones
Collection and assembly of data: Wenyong W. Zhang, Neelima G. Reddy, Elias Jabbour
Data analysis and interpretation: Wenyong W. Zhang, Jorge E. Cortes, Hui Yao, Li Zhang, Neelima G. Reddy, Dan Jones
Manuscript writing: Wenyong W. Zhang, Hui Yao, Dan Jones
Final approval of manuscript: Wenyong W. Zhang, Jorge E. Cortes, Hui Yao, Li Zhang, Neelima G. Reddy, Elias Jabbour, Hagop M. Kantarjian, Dan Jones
REFERENCES
- 1.Brugiatelli M, Bandini G, Barosi G, et al. Management of chronic lymphocytic leukemia: Practice guidelines from the Italian Society of Hematology, the Italian Society of Experimental Hematology and the Italian Group for Bone Marrow Transplantation. Haematologica. 2006;91:1662–1673. [PubMed] [Google Scholar]
- 2.Druker BJ. Circumventing resistance to kinase-inhibitor therapy. N Engl J Med. 2006;354:2594–2596. doi: 10.1056/NEJMe068073. [DOI] [PubMed] [Google Scholar]
- 3.Crossman LC, Mori M, Hsieh YC, et al. In chronic myelogenous leukemia white cells from cytogenetic responders and non-responders to imatinib have very similar gene expression signatures. Haematologica. 2005;90:459–464. [PubMed] [Google Scholar]
- 4.White DL, Saunders VA, Dang P, et al. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): Reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood. 2006;108:697–704. doi: 10.1182/blood-2005-11-4687. [DOI] [PubMed] [Google Scholar]
- 5.White DL, Saunders VA, Dang P, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: Higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110:4064–4072. doi: 10.1182/blood-2007-06-093617. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Giannoudis A, Lane S, et al. Expression of the uptake drug transporter hOCT1 is an important clinical determinant of the response to imatinib in chronic myelogenous leukemia. Clin Pharmacol Ther. 2008;83:258–264. doi: 10.1038/sj.clpt.6100268. [DOI] [PubMed] [Google Scholar]
- 7.Clark RE, Davies A, Pirmohamed M, et al. Pharmacologic markers and predictors of responses to imatinib therapy in patients with chronic myelogenous leukemia. Leuk Lymphoma. 2008;49:639–642. doi: 10.1080/10428190701858823. [DOI] [PubMed] [Google Scholar]
- 8.Illmer T, Schaich M, Platzbecker U, et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia. 2004;18:401–408. doi: 10.1038/sj.leu.2403257. [DOI] [PubMed] [Google Scholar]
- 9.Burger H, van Tol H, Brok M, et al. Chronic imatinib mesylate exposure leads to reduced intracellular drug accumulation by induction of the ABCG2 (BCRP) and ABCB1 (MDR1) drug transport pumps. Cancer Biol Ther. 2005;4:747–752. doi: 10.4161/cbt.4.7.1826. [DOI] [PubMed] [Google Scholar]
- 10.Jabbour E, Kantarjian H, Jones D, et al. Characteristics and outcomes of patients with chronic myelogenous leukemia and T315I mutation following failure of imatinib mesylate therapy. Blood. 2008;112:53–55. doi: 10.1182/blood-2007-11-123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes J, Talpaz M, O'Brien S, et al. Molecular responses in patients with chronic myelogenous leukemia in chronic phase treated with imatinib mesylate. Clin Cancer Res. 2005;11:3425–3432. doi: 10.1158/1078-0432.CCR-04-2139. [DOI] [PubMed] [Google Scholar]
- 12.Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood. 1984;63:789–799. [PubMed] [Google Scholar]
- 13.Jones D, Thomas D, Yin CC, et al. Kinase domain point mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia emerge after therapy with BCR-ABL kinase inhibitors. Cancer. 2008;113:985–994. doi: 10.1002/cncr.23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortes J, Jabbour E, Kantarjian H, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myelogenous leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110:4005–4011. doi: 10.1182/blood-2007-03-080838. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Zhao Y, Smith C, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Cai D, Brendel C, et al. Adaptive secretion of granulocyte-macrophage colony-stimulating factor (GM-CSF) mediates imatinib and nilotinib resistance in BCR/ABL+ progenitors via JAK-2/STAT-5 pathway activation. Blood. 2007;109:2147–2155. doi: 10.1182/blood-2006-08-040022. [DOI] [PubMed] [Google Scholar]
- 17.Valdez BC, Murray D, Ramdas L, et al. Altered gene expression in busulfan-resistant human myeloid leukemia. Leuk Res. 2008;32:1684–1697. doi: 10.1016/j.leukres.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coppo P, Flamant S, De Mas V, et al. BCR-ABL activates STAT3 via JAK and MEK pathways in human cells. Br J Haematol. 2006;134:171–179. doi: 10.1111/j.1365-2141.2006.06161.x. [DOI] [PubMed] [Google Scholar]
- 19.Hantschel O, Rix U, Schmidt U, et al. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci U S A. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donato NJ, Wu JY, Stapley J, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101:690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Meng F, Lu H, et al. Lyn regulates BCR-ABL and Gab2 tyrosine phosphorylation and c-Cbl protein stability in imatinib-resistant chronic myelogenous leukemia cells. Blood. 2008;111:3821–3829. doi: 10.1182/blood-2007-08-109330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari-Amorotti G, Keeshan K, Zattoni M, et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPalpha. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miething C, Grundler R, Mugler C, et al. Retroviral insertional mutagenesis identifies RUNX genes involved in chronic myelogenous leukemia disease persistence under imatinib treatment. Proc Natl Acad Sci U S A. 2007;104:4594–4599. doi: 10.1073/pnas.0604716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villuendas R, Steegmann JL, Pollan M, et al. Identification of genes involved in imatinib resistance in CML: A gene-expression profiling approach. Leukemia. 2006;20:1047–1054. doi: 10.1038/sj.leu.2404197. [DOI] [PubMed] [Google Scholar]
- 25.Abruzzo LV, Lee KY, Fuller A, et al. Validation of oligonucleotide microarray data using microfluidic low-density arrays: A new statistical method to normalize real-time RT-PCR data. Biotechniques. 2005;38:785–792. doi: 10.2144/05385MT01. [DOI] [PubMed] [Google Scholar]
- 26.Holm S. A simple sequentially refective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 27.Venables WN, Ripley BD. Modern Applied Statistics with S. ed 4. New York City, NY: Springer; 2002. [Google Scholar]
- 28.Tipping AJ, Deininger MW, Goldman JM, Melo JV. Comparative gene expression profile of chronic myelogenous leukemia cells innately resistant to imatinib mesylate. Exp Hematol. 2003;31:1073–1080. [PubMed] [Google Scholar]
- 29.Flamant S, Kortulewski T, Dugray A, et al. Osteopontin is upregulated by BCR-ABL. Biochem Biophys Res Commun. 2005;333:1378–1384. doi: 10.1016/j.bbrc.2005.05.203. [DOI] [PubMed] [Google Scholar]
- 30.Kamath AV, Wang J, Lee FY, et al. Preclinical pharmacokinetics and in vitro metabolism of dasatinib (BMS-354825): A potent oral multi-targeted kinase inhibitor against SRC and BCR-ABL. Cancer Chemother Pharmacol. 2008;61:365–376. doi: 10.1007/s00280-007-0478-8. [DOI] [PubMed] [Google Scholar]
- 31.Widmer N, Rumpold H, Untergasser G, et al. Resistance reversal by RNAi silencing of MDR1 in CML cells associated with increase in imatinib intracellular levels. Leukemia. 2007;21:1561–1562. doi: 10.1038/sj.leu.2404671. author reply 1562-1564, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Brendel C, Scharenberg C, Dohse M, et al. Imatinib mesylate and nilotinib (AMN107) exhibit high-affinity interaction with ABCG2 on primitive hematopoietic stem cells. Leukemia. 2007;21:1267–1275. doi: 10.1038/sj.leu.2404638. [DOI] [PubMed] [Google Scholar]
- 33.Brave M, Goodman V, Kaminskas E, et al. Sprycel for chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia resistant to or intolerant of imatinib mesylate. Clin Cancer Res. 2008;14:352–359. doi: 10.1158/1078-0432.CCR-07-4175. [DOI] [PubMed] [Google Scholar]