Abstract
Ca2+/calmodulin-dependent protein kinase II (CaM-KII) regulates numerous physiological functions, including neuronal synaptic plasticity through the phosphorylation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptors. To identify proteins that may interact with and modulate CaM-KII function, a yeast two-hybrid screen was performed by using a rat brain cDNA library. This screen identified a unique clone of 1.4 kb, which encoded a 79-aa brain-specific protein that bound the catalytic domain of CaM-KII α and β and potently inhibited kinase activity with an IC50 of 50 nM. The inhibitory protein (CaM-KIIN), and a 28-residue peptide derived from it (CaM-KIINtide), was highly selective for inhibition of CaM-KII with little effect on CaM-KI, CaM-KIV, CaM-KK, protein kinase A, or protein kinase C. CaM-KIIN interacted only with activated CaM-KII (i.e., in the presence of Ca2+/CaM or after autophosphorylation) by using glutathione S-transferase/CaM-KIIN precipitations as well as coimmunoprecipitations from rat brain extracts or from HEK293 cells cotransfected with both constructs. Colocalization of CaM-KIIN with activated CaM-KII was demonstrated in COS-7 cells transfected with green fluorescent protein fused to CaM-KIIN. In COS-7 cells phosphorylation of transfected α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptors by CaM-KII, but not by protein kinase C, was blocked upon cotransfection with CaM-KIIN. These results characterize a potent and specific cellular inhibitor of CaM-KII that may have an important role in the physiological regulation of this key protein kinase.
Ca2+/calmodulin-dependent protein kinase II (CaM-KII) is a widely distributed protein kinase that is particularly abundant in neuronal tissues where it can constitute up to 1–2% of total protein (1). In vitro it can phosphorylate up to 40 proteins, including enzymes, ion channels, transcription factors, etc., and a number of these proteins appear to be physiological substrates. For example, CaM-KII is highly concentrated in the postsynaptic density of glutamatergic synapses where it phosphorylates and potentiates current through the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptor ion channel (AMPA-Rs) (2–4). This phosphorylation of AMPA-Rs occurs upon induction of long-term potentiation, a model of cellular learning and memory, in region CA1 of hippocampus and is thought to contribute to the postsynaptic current potentiation (2).
Oligomeric CaM-KII is comprised of multiple subunits, each composed of an NH2-terminal catalytic domain, a central regulatory motif that includes autoinhibitory (AID) and overlapping CaM-binding elements, and a COOH-terminal region involved in subunit assembly (1, 5). The kinase is maintained in an inactive form because of interaction of the AID with the catalytic domain. Binding of Ca2+/CaM conformationally disrupts the AID, allowing access of the catalytic domain to substrates. CaM-KII has the unusual property that upon activation by Ca2+/CaM it exhibits rapid intersubunit autophoshorylation on Thr286, generating constitutive kinase activity (6, 7). This constitutive activity prolongs the kinase function beyond the transient elevations of intracellular Ca2+, an essential characteristic for forms of synaptic plasticity such as long-term potentiation. A recent study shows that CaM-KII also can act as a sensor to decode the frequency of Ca2+ spikes (8).
It is likely that CaM-KII may interact with proteins, other than Ca2+/CaM, which can either anchor it to subcellular organelles or directly regulate its activity. For example, a 190-kDa protein may localize activated CaM-KII to the postsynaptic density (9, 10). In an attempt to identify proteins that interact with CaM-KII we used the yeast two-hybrid screen with the COOH terminus or the catalytic domain of CaM-KII as bait. This protocol identified a specific and potent cellular inhibitor protein for CaM-KII in brain. Inhibitory proteins that exert critical physiological roles are known for several protein kinases such as protein kinase A (PKA) (11), the cyclin-dependent kinases (12, 13), and the mitogen-activated protein kinase JNK (14).
EXPERIMENTAL PROCEDURES
Yeast Two-Hybrid Screen.
The screen was performed as described (15). The DNA sequence (1–830 bp) corresponding to the catalytic domain (amino acids 1–269) of the inactive β CaM-KII mutant (K43A) was inserted into the pBTM116 vector in-frame with the LexA-DNA binding domain. The COOH-terminal bait was constructed by using the sequence (975–1,650 bp) encoding the regulatory and association domains (amino acids 282–478) of the α subunit into the pBTM116 vector. The rat brain matchmaker cDNA library (CLONTECH) was constructed in a pGAD10, GAL4 activation domain vector. Interaction with the bait protein was monitored by prototrophy for histidine and/or β-galatosidase activity.
Northern Analysis.
cDNA insert was isolated from the pGAD10 vector and labeled with [α-32P]dCTP by random priming. Total RNA from rat tissues was isolated with Trizol (GIBCO/BRL), and poly(A) RNA was further purified by using an Oligotex mRNA mini kit (Qiagen). Either 2 μg of poly(A) RNA or 20 μg of total RNA were subjected to electrophoresis on a formaldehyde-containing 1.2% agarose gel and then blotted onto Hybond-N (Amersham). The blot was hybridized in Rapid-H4B hybridization buffer (Amersham) with the radioprobe at 65°C for 2 hr and was washed once in 2× sodium chloride/sodium citrate (SSC), 0.1% SDS at 25°C for 15 min, and then twice in 1× SSC, 0.1% SDS at 65°C for 20 min.
Western Blot Analysis.
An adult male rat was euthanized, and tissues were harvested. The brain was quickly removed, and different regions were dissected. The samples were homogenized in lysis buffer containing 10 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.2 mM orthovanadate, 1% Triton X-100, 0.5% Nonidet P-40, 19 μg/ml of leupeptin, 10 μg/ml of pepstatin, 1 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride. Approximately 50 μg of lysate then was electrophoresed on a 15% SDS/PAGE. The gel was transferred onto Immobilon and blotted with the antibody. For preadsorption, 100 μl of a 100-μM solution of the peptide CaM-KIINtide was used to preincubate the antibody for 2 hr at 4°C before addition to the membrane.
Glutathione S-Transferase (GST) Precipitations.
Samples were incubated with purified recombinant GST/CaM-KIIN and glutathione-Sepharose in the appropriate buffer for 1 hr at 4°C, washed extensively, and eluted with 10 mM glutathione. The eluant was separated by SDS/PAGE and either stained with Coomassie blue or immunoblotted.
Immunoprecipitations.
For coprecipitation studies, samples were harvested in lysis buffer with the addition of 5 μg/ml of calpain inhibitor 1. They then were incubated with either a monoclonal CaM-KIIα antibody (Zymed) or a polyclonal CaM-KII antibody with or without 3 mM CaCl2 overnight at 4°C. Immunoprecipitates were washed extensively and eluted with SDS loading buffer, separated by SDS/PAGE, and immunoblotted. Immunoprecipitation of GluR1 was performed as described (2) by using antibodies from Robert Wenthold (National Institutes of Health).
Transient Transfections.
Either HEK 293 or COS-7 cells were transiently transfected by using Lipofectamine (GIBCO/BRL protocol). Briefly, either 10 μg (for HEK 293) of plasmid per 42 μl of Lipofectamine or 1 μg (for COS-7) of plasmid per 10 μl of Lipofectamine were transfected into 100- or 35-mm dishes, respectively. Cells were grown for 1–2 days and subsequently harvested.
Immunofluorescence.
COS-7 cells were transiently transfected, grown for 1 day, and fixed with 4% paraformaldehyde, 4% sucrose in PBS. Cells then were permeabilized with 0.025% Triton X-100 in PBS and stained with a phospho-T286 specific antibody to CaM-KII (Promega) and visualized with a Leitz DMRB scope.
Kinase Assays.
Standard CaM-KII assay conditions contained 0.8 mM CaCl2, 2 μM CaM, 1 mM DTT, 50 mM Hepes (pH 7.5), 10 mM Mg(OAc)2, and 0.2 mM [γ-32P]ATP (1,000–2,000 cpm/pmol) in 20- to 25-μl reactions. The assays were initiated by adding enzyme diluted in buffer (50 mM Hepes, pH 7.5/2 mg/ml BSA/10% ethylene glycol) to the reaction. 32P-incorporation was determined by spotting 10–15 μl of the reaction to Whatman P-81 phosphocellulose paper and subsequently washing in 75 mM phosphoric acid as described (16). For protein kinase C (PKC) phosphorylations in Fig. 3A, PKC (gift of John Scott, Vollum Institute) was activated by using 140 μM phosphatidylserine, 3.8 μM diacylglycerol, and 0.3 mM CaCl2. PKA catalytic subunit was generously provided by Richard Maurer (Oregon Health Sciences Univ.).
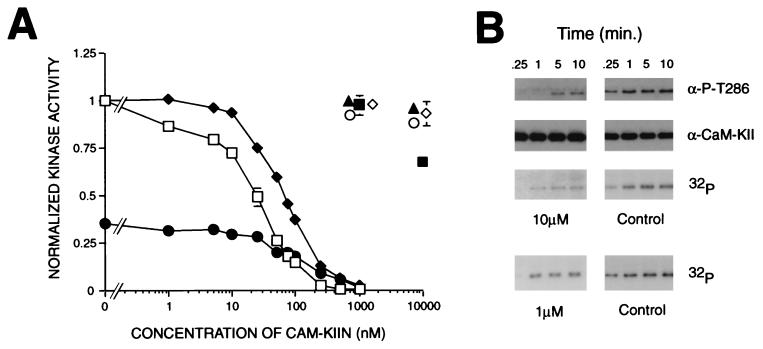
Figure 3.
Inhibition of CaM-KII by CaM-KIIN. (A) Concentration response curves of inhibition of kinase activity by CaM-KIIN and CaM-KIINtide. Autophosphorylated CaM-KII was assayed in the presence of Ca2+/CaM and varying concentrations of GST/CaM-KIIN (⧫) or CaM-KIINtide (□) for its ability to phosphorylate the peptide substrate syntide 2. Ca2+-independent activity (normalized to total activity) also was assayed in the presence of EGTA and CaM-KIINtide (•). Other kinases were assayed with their substrates by using either 0, 1, or 10 μM CaM-KIINtide and normalized to 0 μM CaM-KIINtide: PKC (1 nM, ▴), EGF receptor peptide (Sigma); PKA (100 nM, ○), RII peptide; CaM-KI (20 nM, ◊), and CaM-KIV (40 nM, ■) were activated by CaM-KK and then assayed for their abilities to phosphorylate syntide 2. (B) Effect of CaM-KIINtide on autophosphorylation of CaM-KII. Autophosphorylation reactions were performed at 5°C with 0 (Control), 1 or 10 μM CaM-KIINtide containing either cold ATP or [γ 32P]-ATP, stopped at the indicated times, and blotted with either antiphosphospecific Thr286 antibody (gift from L. Griffith, Brandeis University, Waltham, MA) or polyclonal CaM-KII antibody, or visualized by 32P incorporation, respectively.
Orthophosphate Labeling.
HEK 293 cells were transiently transfected with vectors expressing GluR1 and CaM-KII (His-282–Arg mutant) and 32P-labeled as described (2). PKC was activated by a 20-min treatment with 100 μM phorbol 12-tetradecanoate 13-acetate before harvesting.
RESULTS AND DISCUSSION
To identify proteins that interact with CaM-KII, a yeast two-hybrid screen was performed on a rat brain cDNA library. With the regulatory/association domains of α CaM-KII (residues 282–478) as bait, seven positive clones were identified. Five of these clones corresponded to the COOH termini of the various CaM-KII isoforms (data not shown), consistent with the known role of the COOH terminus as the subunit interaction domain (17). The remaining two clones corresponded to other portions of CaM-KII that might interact with the autoinhibitory domain. We next used as bait the catalytic domain of β CaM-KII (residues 1–269) with Lys-43 mutated to Ala to generate an inactive kinase (6). This was done because CaM-KII can stimulate transcription, and, by using an inactive mutant, we might stabilize interactions with unknown substrates. Sequence analyses showed that several positive clones corresponded to an insert size of 1.4 kb (Fig. 1A). This sequence has an initiation ATG that is consistent with a Kozak consensus sequence (18), a TAG stop codon, and a 3′ noncoding sequence including a poly(A) tail. The ORF of 237 nucleotides encoded a protein of 79 amino acids with a calculated molecular mass of about 8 kDa. This interacting protein appeared to be unique as no sequence homology was detected with known proteins by using the blast program. We have named this protein CaM-KIIN because, as shown below, it is a potent and specific CaM-KII inhibitor.
Figure 1.
Cloning and expression of CaM-KIIN. (A) DNA sequence of CaM-KIIN and deduced amino acid sequence. The underlined region denotes the inhibitory domain. Only part of the 3′ noncoding sequence is shown. (B) Tissue expression of CaM-KIIN message. A Northern blot was performed on poly(A) RNA (Left) and on total RNA (Right). The estimated size of the band is 1.4 kb (arrow). (C) Protein expression of CaM-KIIN. Western blot was performed by using a polyclonal antibody produced to a peptide within the underlined region of CaM-KIIN. (Upper Left) Rat tissues were harvested, and 50 μg of total protein was run per lane. (Upper Right) COS-7 cells were transiently transfected with CaM-KIIN or vector alone and harvested, and 20 μg of soluble protein was run per lane. (Lower) Same as Upper except the antibody was preadsorbed by preincubation with 100 μM of peptide antigen.
Tissue distribution of CaM-KIIN was assessed by performing a Northern analysis by using a radiolabeled probe. Screening both poly(A) RNA and total RNA showed that the message was expressed in rat forebrain, hippocampus, midbrain, cerebellum, and testis, but not in several other tissues examined (Fig. 1B). To compare protein expression with message levels, a polyclonal antibody was generated in rabbits to a COOH-terminal portion of CaM-KIIN (Fig. 1A, underlined sequence). Western blots of different brain regions showed a specific immunoreactive band at approximately 7 kDa and a less reactive protein at 18–19 kDa, both of which were absent using antibody preadsorbed with antigen, as well as a nonspecific band at 50 kDa (Fig. 1C). When the CaM-KIIN antibody was used for immunoprecipitation from a brain extract, a Western blot of the immunoprecipitate showed strong bands at 7 and 19 kDa (not shown), suggesting that the 19-kDa band may be another isoform of the inhibitor. Expression of the CaM-KIIN cDNA in COS-7 cells also identified a 7-kDa immunoreactive protein that comigrated with the 7-kDa immunoreactive protein in brain extracts (Fig. 1C, Right). Although the message for the clone is found within testis, CaM-KIIN protein was not detectable in this tissue. These results indicate an expression of CaM-KIIN that is much more restricted than CaM-KII. Although CaM-KII is 20- to 50-fold higher in brain, it can be detected in most other tissues (1). The restricted expression of CaM-KIIN suggests it has a unique function in brain.
We next tested whether CaM-KIIN could interact with CaM-KII both in vitro and in cells. CaM-KIIN was expressed as a bacterial GST-fusion construct and purified by using glutathione-Sepharose. When the GST/CaM-KIIN was incubated with purified recombinant α CaM-KII (19) and then isolated by glutathione-Sepharose, CaM-KII was copurified only if it had been preactivated in the presence of Ca2+/CaM (Fig. 2A, Left). After formation of the complex, subsequent removal of Ca2+/CaM by the addition of excess EGTA reversed the interaction between CaM-KII and CaM-KIIN (Fig. 2A). This finding suggests that the interaction required a portion of the kinase catalytic domain that was masked in the absence of Ca2+/CaM. There may be little isoform specificity for the interaction because the catalytic domain of the β subunit was used as bait, but CaM-KIIN also can interact with the α subunit. However, there is specificity for the CaM-KII family because the GST/CaM-KIIN did not bind to CaM-KIV (60 kDa) in the absence or presence of Ca2+/CaM (Fig. 2A). This finding is significant because the catalytic domains of CaM-KII and CaM-KIV exhibit 45% identity in amino acid sequence (20) and have considerable overlap in substrate specificities. To determine whether CaM-KIIN could interact with endogenous CaM-KII, GST/CaM-KIIN was incubated with a soluble rat brain extract in the absence or presence of Ca2+/CaM. Interacting proteins were isolated by glutathione-Sepharose, washed extensively, and separated by SDS/PAGE. Coomassie staining identified a major protein at 50 kDa and some fainter bands at about 60 kDa that interacted with CaM-KIIN only in the presence of Ca2+/CaM (Fig. 2A). These proteins were reactive to polyclonal CaM-KII antibody, confirming that they represent the α and β isoforms (Fig. 2A). The absence of other major bands indicates the specificity of this interaction for CaM-KII.
Figure 2.
Binding of CaM-KII to CaM-KIIN. (A) GST/CaM-KIIN precipitation of CaM-KII. Either recombinant CaM-KIV, CaM-KII, or soluble brain extract was incubated with GST/CaM-KIIN in the presence of Ca2+/CaM or EGTA, precipitated with glutathione-Sepharose, washed, and separated by SDS/PAGE. Samples then were visualized by Coomassie staining or by Western blot with anti-CaM-KII antibody. The positions of CaM-KII (upper arrow) and GST/CaM-KIIN (lower arrow) are indicated. The Western blot of the soluble brain extract precipitation (Right) visualized both the α and β subunits of CaM-KII. (B) Coimmunoprecipitation of CaM-KII and CaM-KIIN. Rat forebrain supernatant was split into 200-μl aliquots and incubated with either protein A Sepharose alone (first two lanes) or with a mAb CaM-KIIα in the presence of 3 mM CaCl2 or 1 mM EGTA. After centrifugation the immune complex was separated by SDS/PAGE and immunoblotted with the CaM-KIIN antibody (Upper Left) or with preadsorption with the antigenic peptide (Upper Right). The membrane then was immunoblotted with the polyclonal CaM-KII Ab showing both the α and β isoforms (Lower). (C) Coimmunoprecipitation of CaM-KIIN with CaM-KII from transfected cells. HEK 293 cells were transiently transfected with CaM-KIIN, CaM-KII (His-282–Arg), or together and subsequently lysed. CaM-KII was immunoprecipitated from the soluble cell extract with 3 mM Ca2+ and separated by SDS/PAGE. The samples then were immunoblotted with anti-CaM-KIIN. (D) Colocalization of active CaM-KII and CaM-KIIN with transfected cells. COS-7 cells were transiently transfected with the indicated combinations of GFP, GFP/CaM-KIIN fusion protein (GFP-KIIN), wild-type CaM-KII, or activated CaM-KII (H282R). Cells were visualized for GFP (Upper) or for activated CaM-KII by using a Thr286 phosphospecific-antibody (Lower).
Interaction between endogenous CaM-KII and endogenous CaM-KIIN was investigated by using coimmunoprecipitation experiments. Soluble rat brain extract was incubated with a CaM-KIIα mAb in the presence or absence of 3 mM Ca2+, and the immune complex was separated by SDS/PAGE and immunoblotted with the CaM-KIIN antibody. Only in the presence of Ca2+ was the 7-kDa CaM-KIIN coprecipitated along with the 19-kDa protein (Fig. 2B) that also was seen in the Western blot of forebrain in Fig. 1C. Both of these bands were eliminated with preadsorption of the CaM-KIIN antibody using the antigenic peptide (Fig. 2B, Right). This 19-kDa protein suggests the existence of another isoform of CaM-KIIN, similar to the multiple isoforms of PKA inhibitor (21). Interaction between CaM-KIIN and CaM-KII was further demonstrated in HEK 293 cells transfected with cDNAs for CaM-KIIN and/or for the constitutively active CaM-KII (His-282–Arg) mutant (22). Using a polyclonal CaM-KII antibody, there was only coprecipitation of CaM-KIIN from the cell extracts when CaM-KII was coexpressed (Fig. 2C).
Although the previous experiments demonstrate that CaM-KII and CaM-KIIN interact in vitro, they do not document their interaction within the cell. This question was investigated by using transient transfection experiments in COS-7 cells. CaM-KIIN was fused to green fluorescent protein (GFP) (CLONTECH) (GFP-KIIN) and transiently transfected either with wild-type CaM-KII or the constitutively active His-282–Arg mutant. Thr286 autophosphorylation is indicative of activated CaM-KII (1), and this species was detected by using a phospho-T286-specific antibody (Fig. 2D). GFP alone (not shown) or coexpressed with active (H282R mutant) CaM-KII (Fig. 2D, Right) was localized in both the nucleus and cytosol. GFP-KIIN when expressed alone or with wild-type CaM-KII also was distributed in both the nucleus and cytosol (Fig. 2D, compare left panels). However, when coexpressed with activated CaM-KII (i.e., the His-282–Arg mutant), GFP-KIIN was excluded from the nucleus (Fig. 2D, compare center panels). Because CaM-KIIα is found predominantly in the cytosol (23), this result indicates that the GFP-KIIN bound to the activated CaM-KII in the cytosol and was sequestered from freely diffusing into the nucleus. However, we cannot exclude the possibility that activated CaM-KII phosphorylated some protein that altered the localization of CaM-KIIN.
What is the functional relevance of the interaction between CaM-KII and CaM-KIIN? In vitro kinase assays were performed to test whether CaM-KIIN was a substrate for CaM-KII. A phosphorylation time course using 20 nM CaM-KII showed no significant 32P-incorporation into 4 μM CaM-KIIN after 45 min at 30°C (data not shown). In these experiments 32P-autophosphorylation of CaM-KII was observed, but it was significantly suppressed by the presence of CaM-KIIN. This result suggested that CaM-KIIN might be an inhibitor of CaM-KII activity, so a concentration response curve was performed (Fig. 3A). GST/CaM-KIIN exhibited an apparent IC50 of 50 nM toward CaM-KII using its peptide substrate syntide 2 (Fig. 3A) whereas GST alone gave no inhibition up to concentrations of 10 μM (not shown). Both the Ca2+-independent as well as total activities of autophosphorylated CaM-KII were inhibited (Fig. 3A), and an identical inhibition was obtained for CaM-KII not previously autophosphorylated. COOH-terminal truncations of CaM-KIIN indicated that its inhibitory potency resided largely in 28 residues near its COOH terminus (data not shown), so a peptide was synthesized corresponding to this sequence (Fig. 1A, underlined sequence). This peptide, CaM-KIINtide (KRPPKLGQIGRSKRVVIEDDRIDDVLK), had a similar IC50 of 50 nM for both the total and the Ca2+-independent activities of CaM-KII (Fig. 3A). Inhibition of kinase autophosphorylation was further investigated by using 1 or 10 μM of CaM-KIINtide and detecting CaM-KII autophosphorylation by either 32P-incorporation or with an antibody specific for autophosphorylation of Thr286 (Fig. 3B). Reactions were performed at 5°C in an effort to limit autophosphorylation to Thr286. Although there was a dose-dependent inhibition of autophosphorylation, it was not as potent as inhibition of substrate phosphorylation. This higher inhibitory potency toward phosphorylation of an exogenous substrate versus CaM-KII autophosphorylation on Thr286 (Fig. 3B) is not surprising because the latter is an intramolecular, intersubunit reaction within the oligomeric protein (6, 7). Preliminary results suggest that inhibition of CaM-KII by CaM-KIINtide was not competitive toward peptide substrate (data not shown).
Is CaM-KIIN a specific inhibitor of CaM-KII as suggested by the fact that GST/CaM-KIIN did not interact with CaM-KIV (Fig. 2A)? Concentrations of 1 or 10 μM of CaM-KIINtide gave no significant inhibition of PKC, PKA, and CaM-KI and only 30% inhibition of CaM-KIV at 10 μM (Fig. 3A). At 10 μM, GST/CaM-KIIN did not inhibit CaM-KIV (not shown), suggesting that the peptide may have lost some specificity relative to the full-length protein. Furthermore, 10 μM of the peptide had no effect on the activation of either CaM-KI and CaM-KIV by CaM-KK (data not shown), indicating no inhibition of CaM-KK. This high specificity of CaM-KIIN and CaM-KIINtide will make it a very useful probe for identifying specific CaM-KII reactions. One of the major limitations of previous peptide inhibitors of CaM-KII, based on its autoinhibitory domain sequence, is their lack of specificity at higher concentrations (24, 25). Therefore, CaM-KIINtide was tested for inhibition of CaM-KII activity in extracts of rat forebrain, goldfish brain, and Drosophila. For all three extracts CaM-KIINtide showed similar IC50 values ranging from 100 to 400 nM toward phosphorylation of syntide 2 (data not shown).
The in vitro data show that CaM-KIIN and CaM-KIINtide are inhibitors of CaM-KII autophosphorylation and activity toward an exogenous peptide substrate. We next investigated its inhibitory properties and specificity by using a protein substrate in vitro and in transfected cells. It previously has been shown that the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit GluR1 ion channel is potentiated through phosphorylation by CaM-KII and by PKC on Ser831 in its intracellular COOH terminus (2, 26, 27). To determine whether CaM-KIINtide would inhibit the phosphorylation of a protein substrate, the GST fusion of the COOH terminus of GluR1 (residues 816–889) was used in in vitro kinase assays with either PKC or CaM-KII as catalysts. CaM-KIINtide (1 μM) completely inhibited phosphorylation of GST/GluR1816–889 by CaM-KII and most of CaM-KII autophosphorylation (Fig. 4A). PKC catalyzed weaker phosphorylation of GST/GluR1816–889, and this was not blocked by CaM-KIINtide. Next, HEK 293 cells were transfected with GluR1 without or with cotransfection with CaM-KII (His-282–Arg mutant) and/or CaM-KIIN. Cells transfected without CaM-KII were stimulated with phorbol esters to activate PKC as this has been shown to result in the phosphorylation of GluR1 (26, 27). Cells were 32P-labeled, harvested, immunoprecipitated with GluR1 antibody, and analyzed by SDS/PAGE/autoradiography. As reported previously, the low basal phosphorylation of GluR1 was strongly stimulated by coexpression of the constitutively active CaM-KII (2, 26, 27), and this phosphorylation was blocked by expression of CaM-KIIN (Fig. 4B, Left). Furthermore, activation of PKC by 100 μM phorbol 12-tetradecanoate 13-acetate resulted in phosphorylation of GluR1 (26, 27), but this phosphorylation could not be inhibited by transfection of CaM-KIIN (Fig. 4B, Right). These results document that CaM-KIIN can act as a specific inhibitor of CaM-KII within the cell.
Figure 4.
Effect of CaM-KIIN on GluR1 phosphorylation by CaM-KII and PKC. (A) Kinase reactions, performed at 30°C for 20 min with the indicated mixtures of 0.2 mM [γ 32P]-ATP, 20 nM PKC catalytic fragment (PKM, Calbiochem) or 20 nM CaM-KII, 4 μM GST or 4 μM GST/GluR1816–889 with or without 1 μM CaM-KIINtide, were separated by SDS/PAGE and visualized by autoradiography. GST/GluR1816–889 migrated as a doublet. (B) HEK 293 cells, transfected with GluR1 without or with cotransfected CaM-KII (H282R) were 32P-labeled. Some cells without CaM-KII transfection were stimulated with phorbol 12-tetradecanoate 13-acetate to activate PKC for 20 min. Cells were harvested, and GluR1 was immunoprecipitated, analyzed by SDS/PAGE/autoradiography (Upper), and subjected to GluR1 Western blot (Lower).
A number of protein kinases have inhibitory proteins that exert important physiological regulation. For example, the specific inhibitor of PKA (11) has been very useful in identifying PKA-mediated reactions in vitro and in transfected cells, and this inhibitor appears to act physiologically to promote nuclear export of PKA (28, 29). Likewise, the inhibitor proteins of the cyclin-dependent kinases are critical in cell cycle control (12, 13), and the cytosolic inhibitor of JNK functions to retain this kinase in the cytosol (14). In this study we have identified a potent and specific inhibitor of CaM-KII and have demonstrated its ability to inhibit CaM-KII-, but not PKC-, mediated phosphorylations in vitro and in transfected cells. Further studies will be necessary to assess the physiological role of this inhibitor in CaM-KII actions in brain. Because one can detect high CaM-KII activity in brain extracts in the presence of Ca2+/CaM, conditions where CaM-KIIN interacts with and inhibits CaM-KII, it is obvious that the levels of expression of CaM-KIIN are much lower than those of CaM-KII. This finding implies that CaM-KIIN may physiologically regulate a specific pool of CaM-KII, and studies are in progress to examine this hypothesis. It is anticipated that CaM-KIIN and CaM-KIINtide also will be important molecular probes in identifying reactions mediated by CaM-KII. For example, we have shown that intradendritic injections of CaM-KIINtide prevented the induction of activity-dependent synaptic potentiations at mixed synapses on the goldfish Mauthner cell (A. E. Pereda, T. D. Bell, B.H.C., A. J. Czernik, A. C. Nairn, T.R.S., and D. S. Folder, unpublished work).
Acknowledgments
We thank Drs. Richard Goodman and John Scott for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM41292 (T.R.S.) and Training Grant NS07381 (B.H.C.).
ABBREVIATIONS
- CaM-KII
Ca2+/calmodulin-dependent protein kinase II
- PKA
protein kinase A
- GST
glutathione S-transferase
- PKC
protein kinase C
- GFP
green fluorescent protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FO41854).
References
- 1.Braun A P, Schulman H. Annu Rev Physiol. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]
- 2.Barria A, Muller D, Derkach V, Griffith L C, Soderling T R. Science. 1997;276:2042–2045. doi: 10.1126/science.276.5321.2042. [DOI] [PubMed] [Google Scholar]
- 3.Pettit D L, Perlman S, Malinow R. Science. 1994;266:1881–1885. doi: 10.1126/science.7997883. [DOI] [PubMed] [Google Scholar]
- 4.Lledo P M, Hjelmstad G O, Mukherji S, Soderling T R, Malenka R C, Nicoll R A. Proc Natl Acad Sci USA. 1995;92:11175–11179. doi: 10.1073/pnas.92.24.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soderling T R. Biochem Biophys Acta. 1996;1297:131–138. doi: 10.1016/s0167-4838(96)00105-7. [DOI] [PubMed] [Google Scholar]
- 6.Mukherji S, Soderling T R. J Biol Chem. 1994;269:13744–13747. [PubMed] [Google Scholar]
- 7.Hanson P I, Meyer T, Stryer L, Schulman H. Neuron. 1994;12:943–956. doi: 10.1016/0896-6273(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 8.DeKoninck P, Schulman H. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 9.McNeill R B, Colbran R J. J Biol Chem. 1995;270:10043–10049. doi: 10.1074/jbc.270.17.10043. [DOI] [PubMed] [Google Scholar]
- 10.Strack S, Choi S, Lovinger D M, Colbran R J. J Biol Chem. 1997;272:13467–13470. doi: 10.1074/jbc.272.21.13467. [DOI] [PubMed] [Google Scholar]
- 11.Walsh D A, Perkins J P, Krebs E G. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 12.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 13.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 14.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 15.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 16.Roskoski R J. Methods Enzymol. 1985;99:3–6. doi: 10.1016/0076-6879(83)99034-1. [DOI] [PubMed] [Google Scholar]
- 17.Shen K, Meyer T. J Neurochem. 1998;70:96–104. doi: 10.1046/j.1471-4159.1998.70010096.x. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brickey D A, Colbran R J, Fong Y L, Soderling T R. Biochem Biophys Res Commun. 1990;173:578–584. doi: 10.1016/s0006-291x(05)80074-9. [DOI] [PubMed] [Google Scholar]
- 20.Sikela J M, Law M L, Kao F T, Hartz J A, Wei Q, Hahn W E. Genomics. 1989;4:21–27. doi: 10.1016/0888-7543(89)90309-1. [DOI] [PubMed] [Google Scholar]
- 21.Van Patten S M, Howard P, Walsh D A, Maurer R A. Mol Endocrinol. 1992;6:2114–2122. doi: 10.1210/mend.6.12.1491692. [DOI] [PubMed] [Google Scholar]
- 22.Brickey D A, Bann J G, Fong Y L, Perrino L, Brennan R G, Soderling T R. J Biol Chem. 1994;269:29047–29054. [PubMed] [Google Scholar]
- 23.Srinivasan M, Edman C F, Schulman H. J Cell Biol. 1994;126:839–852. doi: 10.1083/jcb.126.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith M K, Colbran R J, Soderling T R. J Biol Chem. 1990;265:1837–1840. [PubMed] [Google Scholar]
- 25.Hvalby O, Hemmings H G, Paulsen O, Czernik A J, Nairn A C, Godfraind J M, Jensen V, Raastad M, Storm J F, Andersen P, Greengard P. Proc Natl Acad Sci USA. 1994;91:4761–4765. doi: 10.1073/pnas.91.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammen A L, Kameyama K, Roche K W, Huganir R L. J Biol Chem. 1997;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- 27.Barria A, Derkach V, Soderling T R. J Biol Chem. 1997;272:32727–32730. doi: 10.1074/jbc.272.52.32727. [DOI] [PubMed] [Google Scholar]
- 28.Wen W, Harootunian A T, Adams S R, Feramisco J, Tsien R Y, Meinkoth J L, Taylor S S. J Biol Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- 29.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]